Feasibility of Raman spectroscopic identification of gall bladder cancer using extracellular vesicles extracted from bile†
Thu Thuy
Bui
a,
Eunjin
Jang
a,
Ji Hyun
Shin
b,
Tae Hun
Kim
b,
Hayoon
Kim
b,
Dongho
Choi
b,
Tung Duy
Vu
 c and
Hoeil
Chung
c and
Hoeil
Chung
 *a
*a
aDepartment of Chemistry and Research Institute for Convergence of Basic Science, Hanyang University, Seoul 04763, Republic of Korea. E-mail: hoeil@hanyang.ac.kr
bDepartment of Surgery, College of Medicine, Hanyang University, Seoul 04763, Republic of Korea
cFaculty of Chemistry, University of Science, Vietnam National University, Hanoi, Vietnam
First published on 11th July 2023
Abstract
Extracellular vesicles (EVs), which are heterogeneous membrane-based vesicles with bilayer cell membrane structures, could be versatile biomarkers for the identification of diverse diseases including cancers. With this potential, this study has attempted the Raman spectroscopic identification of gall bladder (GB) cancer by directly measuring the EV solution extracted from human bile without further sample drying. For this purpose, bile samples were obtained from four normal individuals and 21 GB polyp, eight hepatocellular carcinoma (HCC), and five GB cancer patients, and EVs were extracted from each of the bile samples. The Raman peak shapes of the EVs extracted from the GB cancer samples, especially the relative intensities of peaks in the 1560–1340 cm−1 range, were dissimilar to those of the samples from the normal, GB polyp, and HCC groups. The intensity ratios of peaks at 1537 and 1453 cm−1 and at 1395 and 1359 cm−1 of the GB cancer samples were lower and higher, respectively, than those of the samples of the remaining three groups. The differences of peak intensity ratios were statistically significant based on the Mann–Whitney U test. DNA/RNA bases, amino acids, and bile salts contributed to the spectra of EVs, and their relative abundances seemed to vary according to the occurrence of GB cancer. The varied metabolite compositions and/or structures of EVs were successfully demonstrated by the dissimilar peak intensity ratios in the Raman spectra, thereby enabling the discrimination of GB cancer.
Introduction
Extracellular vesicles (EVs) are heterogeneous membrane-based vesicles with cell bilayer membrane structures, and they are released into the extracellular domain. Vesicles with sizes ranging from 100 to 1000 nm are referred to as micro-vesicles (MVs); exosomes are vesicles with sizes of 30–100 nm. EVs carry diverse biomolecules such as proteins, RNAs, nucleic acids, lipids, and other metabolites.1,2 If the contents of individual biomolecules in EVs vary with the occurrence of a target disease or pathological condition, EVs could be versatile biomarkers for the diagnosis of diverse diseases including cancers. Owing to the potential utility of EVs in biochemical and medical studies, various analytical methods based on mass spectrometry,3 chromatography,4 and vibrational spectroscopy5 have been adopted for subsequent analysis.For a simple, rapid, and non-destructive analysis, vibrational spectroscopic methods are excellent choices. Analysis of aqueous EV samples by infrared (IR) spectroscopy is difficult due to the strong IR absorption of water which limits the observation of peaks related to EVs. Raman spectroscopy is a superior candidate since the interference of water is negligible, and the subsequent spectral acquisition is uncomplicated. With these merits, Raman spectroscopy has been readily adopted to measure EVs extracted from diverse bio-specimens, and the obtained spectral features were analyzed for biomedical study and disease diagnosis. For example, Raman spectroscopy has been successfully used in the investigation of biochemical differences in EVs from the plasma of amyotrophic lateral sclerosis patients,6 the stratification of patients with chronic kidney disease in type 2 diabetes using urinary EVs,7 the characterization of EVs from bovine placenta and peripheral blood mononuclear cells,8 the detection of prostate cancer through characterization of relevant EVs derived from prostate cancer cells and red blood cells,9 and the identification of the amyloid beta peptide in small EVs of the Alzheimer's disease cell culture model.10 That the extracted EVs in these examples were dried for the acquisition of Raman spectra is noteworthy. Drying the samples would make the acquired spectral features less representative of EVs in vivo, so direct measurement of EVs in aqueous solution is recommended.
Here, we have first attempted the Raman spectroscopic analysis of EVs extracted from human bile for disease diagnosis and explored the feasibility of identification of gall bladder (GB) cancer. This study is worthwhile as a continuing effort for EV-based Raman spectroscopic biomedical diagnosis. EVs in bile are potentially disease markers since their bio-molecular compositions and/or structures may vary according to the pathological conditions of patients. For this study, bile samples were obtained from four normal individuals and 21 GB polyp, eight hepatocellular carcinoma (HCC), and five GB cancer patients; EVs were extracted from each of the bile samples. HCC is the most common type of primary liver cancer,11 and GB cancer, with an incidence of approximately 1.2% of all cancers,12 has a dismal prognosis with a 5-year survival rate of 5–10%.13,14
Size distributions of the extracted EVs were analyzed, and the Raman spectra of the EVs in the extraction solutions were directly acquired without drying using a wide area coverage (WAC, laser illumination diameter: 1 mm) scheme. The use of the WAC scheme was to ensure representative sampling of all extracted EVs in the solution and to prevent photo-thermal damage of EVs by decreasing the laser power per area (LP/A). The peak shapes and peak intensity ratios in the EV spectra among the four groups were comparatively examined. Also, distributions of principal component (PC) scores of the spectra were analyzed to assess the mutual discriminability, especially of GB cancer.
Experimental
Collection of bile samples and extraction of EVs from bile
Thirty-eight bile samples were collected from four normal individuals and 21 GB polyp, eight HCC, and five GB cancer patients after cholecystectomy at Hanyang University Hospital (Seoul, Korea). The purpose and content of the research were thoroughly explained to each participant, and all experiments were performed in accordance with the guidelines of the Institutional Review Board and approved by the ethics committee at Hanyang University (HYI-18-227-15). Informed consent was obtained from the participants in this study. The bile samples were promptly frozen after acquisition and stored below −20 °C to prevent further denaturation. Each sample was thawed at room temperature before EV extraction. Participant data including age, gender, and body mass index (BMI) are presented in the ESI.† Isoleucine, lysine, alanine, proline, valine, cytosine, and thymine were purchased from Tokyo Chemical Industry (Japan), and leucine, tryptophan, glutamate, guanine, and adenine were obtained from Junsei Chemical (Japan). Glycodeoxycholate (GDCA), glycochenodeoxycholate (GCDCA), glycocholate (GCA), taurodeoxycholate (TDCA), and taurocholate (TCA) were supplied by Sigma-Aldrich (USA).EVs were isolated from each of the bile samples using an ExoLutE kit (Rosetta Exosome Inc., Seoul, Korea) according to the manufacturer's instructions. Briefly, 0.1 mL of each bile sample was diluted with 1.0 mL of RPMI (Roswell Park Memorial Institute)-1640 solution, and the diluted sample was filtered using a 0.45 μm syringe filter. Then, 0.4 mL of solution D and 2.5 mL of RPMI-1640 solution were added to the filtered solution. This solution mixture was incubated at room temperature for 1 hour with shaking. For enrichment of the sample, 0.125 mL of solution A, 0.5 mL of solution B, and 2.0 mL of solution C were added to the mixture; this new mixture was shaken at room temperature for 15 minutes. The resulting solution was centrifuged at 3000 rcf (6659 rpm) for 15 minutes at room temperature to deposit crude EVs at the bottom, and 0.15 mL of solution R was added to the extracted EVs. This final EV solution was used for subsequent Raman measurement. The procedure for EV extraction is graphically described in the ESI.†
Sizes of the extracted EVs were determined using a nanoparticle tracking analysis (NTA) system (Nanosight NS300, Malvern Instrument Ltd, Malvern, UK) equipped with a 405 nm laser (55 mW) and an sCMOS camera. The extracted EVs were re-suspended in Dulbecco's phosphate buffered saline (DPBS) for NTA. Videos of the extracted EV samples were analyzed using NTA software with a camera setting of 16 and a detection threshold level of 5. The monitoring trajectory of EV movement, particle number and size distribution of EVs were determined by averaging the acquired videos of triplicate measurements, which were each 90 seconds in duration.
Western blotting (WB) analysis
Each of the EV samples was re-suspended in 15 μL of Laemmli buffer and stored at 95 °C for 5 minutes. The solution was loaded for 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at 110 V, and the sample-loaded gel was transferred onto a nitrocellulose membrane. The membrane was immersed in a solution containing 0.1% Tween-20 and 5% BSA for 1 hour and then incubated overnight at 4 °C with the appropriate primary antibodies (anti-CD63, Novus Bio., CO, USA). Next, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 90 minutes at room temperature. WB bands were recognized using a Fusion-SL4 detection system (Vilber Lourmat, Germany) incorporating an enhanced chemiluminescence kit (Cytiva, Amersham, UK).Acquisition of Raman spectra of extracted EVs and pure components
A Raman spectrometer (RamanRxn1 unit, Kaiser Optical Inc.) equipped with a PhAT probe providing a laser illumination diameter of 1 mm was employed. The adoption of the WAC scheme was to fully cover the extracted EVs for representative sampling and to reduce the LP/A for the prevention of photo-thermal destruction of the EVs. To acquire the Raman spectra, a 10 μL suspension of extracted EVs was transferred to a reflective cylindro-conical metal groove and measured without drying. The diameter at the top of the groove was 3.4 mm and the groove depth at the center was 1.65 mm (refer to the picture and dimensions of the reflective groove in the ESI (Fig. S2†)). The use of the reflective groove was to collect more of the generated Raman photons, thereby improving sensitivity. The laser power measured at the sample location using a power meter (Thorlabs Inc., Newton, New Jersey, USA) was 92.4 mW; therefore, the LP/A was 1.18 × 10−4 mW μm−2. At this LP/A, photo-thermal destruction or other denaturation of EVs was not observed after Raman measurement. Each spectrum with a resolution of 4 cm−1 was collected by accumulating 30 scans with a laser exposure time of three seconds for each scan (a total acquisition time of 90 seconds).To quantitatively evaluate the potential sensitivity enhancement upon using the reflective groove, the Raman spectra of an EV sample extracted from one bile sample of the GB polyp group were acquired by dropping the sample into the reflective groove and on a flat Si wafer. The intensities of the sample peaks were approximately 4.6 times higher when using the reflective groove than those measured with the flat-substrate (refer to the corresponding Raman spectra in the ESI (Fig. S3†)). For each extracted EV solution from the employed bile samples, triplicate spectra were acquired and averaged; the average spectra of each sample were used for subsequent analysis. Also, the Raman spectra of the pure components were collected by direct laser irradiation onto each pure powder or liquid sample under the same conditions without further treatments such as dissolution in a solvent. Baseline correction using asymmetric least squares smoothing (AsLS), normalization of spectra, and principal component analysis (PCA) were performed using MATLAB R2017b (The MathWorks Inc., USA).
Results and discussion
Size distributions of extracted EVs from bile samples
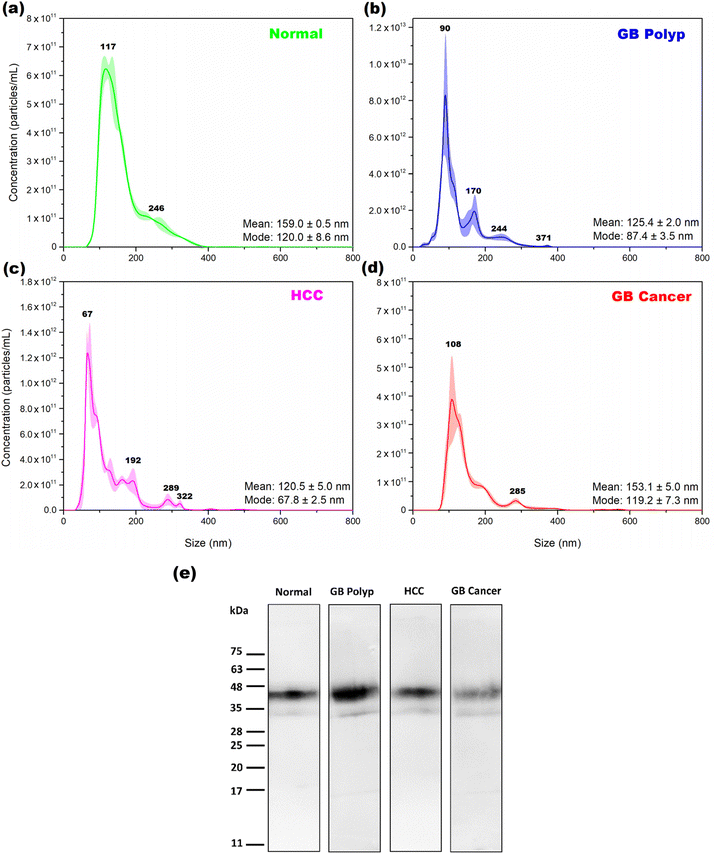
Fig. 1 shows the average size distributions of EVs extracted from one bile sample in each of the normal (a), GB polyp (b), HCC (c), and GB cancer (d) groups determined by NTA. The shading superimposed over the averages indicates the corresponding standard deviation in triplicate measurements. In each plot, the determined mean and mode sizes of the extracted EVs are denoted. The mode size is the size of the most populated EVs. The EVs with sizes from 40 to 400 nm are extracted, and sizes from 50 to 180 nm are the most dominant sizes in the measured EV samples. The size distributions of normal and GB cancer samples are relatively similar, showing a broad distribution over the 80–400 nm range, while dissimilar to those of the GB polyp and HCC samples from which the EVs with more distinct sizes were extracted. Based on the NTA results, both exosomes (30–100 nm) released by the fusion of multivesicular bodies containing intraluminal vesicles with the plasma membrane and microvesicles (100–400 nm) shed by outward blebbing of the plasma membrane are present together in the EV samples. The concentrations of EVs of the samples measured in (a), (b), (c), and (d) were 5.78 × 1013, 4.08 × 1014, 7.57 × 1013, and 2.59 × 1013 particles per mL, respectively. Each of the EV concentrations was estimated by calculating the area under the corresponding NTA peak.To confirm whether the isolated particles were EVs or not, WB was performed for the EV samples using CD63 as a biomarker. CD63, a kind of tetraspanin, is a constituent of exosomes, endosomes, and EVs.15 Therefore, CD63 detection in an extracted sample could be evidence for the presence of EVs. Fig. 1(e) shows the WB bands measured from each EV sample from the four groups. For all the measured samples, the bands clearly appeared in the 32–55 kDa range, indicating the successful extraction of EVs from the bile samples. The band intensity of the GB cancer sample is weaker than those of the other samples. As previously reported,16 GB cancer causes GB wall thickening, thereby degrading the water absorption ability of the GB wall. For this reason, the bile of the GB cancer patient is diluted due to the decreased water absorption by the thickened GB wall. Therefore, the concentration (particles per mL) of EVs extracted from the bile of a GB cancer patient is expected to be low. As demonstrated by the magnitude of the y-axis scale in Fig. 1(d), the number of EV particles obtained from the bile of GB cancer patients is lower than those in the other groups.
Raman spectral features of EV samples and examination of spectral differences
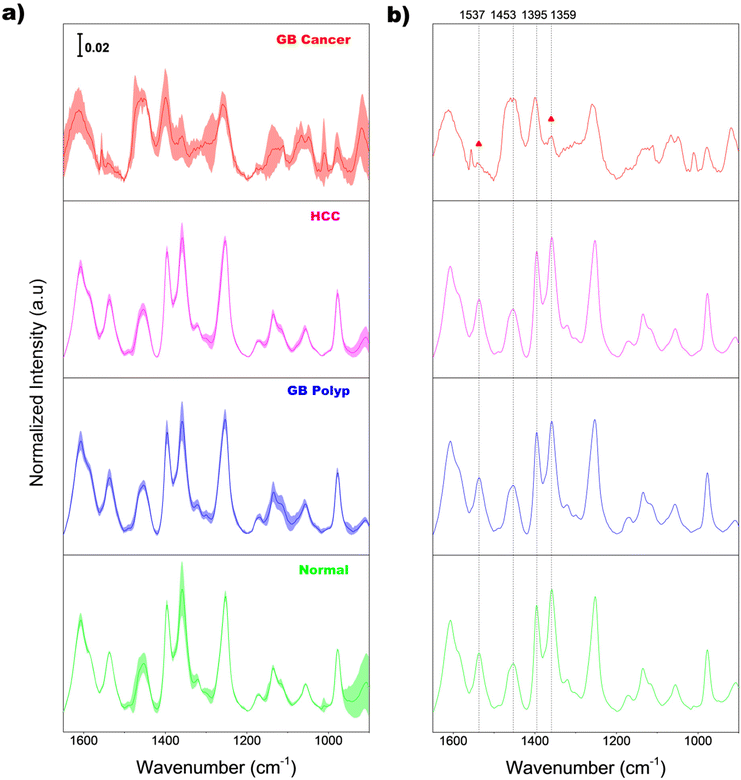
Fig. 2(a) shows the average normalized Raman spectra of EVs extracted from the bile samples of normal (light green), GB polyp (blue), HCC (magenta), and GB cancer (red) groups. Before averaging, the baseline of each raw spectrum was initially corrected by AsLS; then the intensities at each wavenumber in the AsLS-treated spectrum were individually divided by the peak area calculated under the 1650–900 cm−1 range for area normalization. The superimposed shading over each average spectrum indicates the standard deviations of the intensity at each wavenumber, equivalent to the sample-to-sample spectral variation in each group. The magnitude of the shading of the GB cancer group is greater than those of the other three groups, indicating the larger variation of EV composition among the GB cancer samples.For a clear comparison, the same average spectra without shading are also displayed in Fig. 2(b). Diverse peaks including 8–9 main peaks appear over the entire spectral range. The average EV spectrum of the GB cancer group has more noise since the concentrations of EVs in the corresponding samples are lower due to the dilution of bile. The vibrational assignments of 11 different peaks in the spectra are summarized in Table 1. As demonstrated, the observed peaks originate from various sources such as amino acids, proteins, lipids, and DNA/RNA bases and their backbones. Importantly, the overall spectral features of bile-extracted EVs in this study are considerably different from those of EVs extracted from different biological samples such as the plasma of amyotrophic lateral sclerosis patients,6 urine of patients with chronic kidney disease,7 peripheral blood mononuclear cells,8 and red blood cells of prostate cancer patients.9 Also, the band shapes of EVs in these studies were dissimilar to one another. This supports the assertion that the bio-molecular compositions and/or structures of EVs vary largely depending on the sources of raw biological samples.
| Position (cm−1) | Vibration assignment | Source | Ref. |
|---|---|---|---|
| Symbols and abbreviations: ν, stretching; δ, in-plane bending; def, deformation. | |||
| 1607 |
ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) C) |
Amino acids, DNA/RNA bases | 21 and 22 |
| 1537 | Amide II: –NH bending, ν(C–N) | DNA/RNA bases | 23–26 |
| 1453 | Def (CH2/CH3) | Lipids, amino acids, bile salts | 23 and 25 |
| 1395 | ν(COO−), def (CH2) | Amino acids, lipids | 22, 23, 25 and 27 |
| 1359 | ν(C–N) | Amino acids, bile salts | 22, 27 and 28 |
| 1253 | Amide III or def (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H) C–H) |
Amino acids | 22 and 24 |
| 1170 | ν(C–O) | Amino acids, DNA/RNA bases | 22 and 25 |
| 1135 | ν(C–N) | Proteins | 27 and 29 |
| 1056 | ν(C–O–C) | Lipids | 22 and 25 |
| 977 | ν(C–C), δ(C–H) | DNA/RNA backbone (ribose) | 22, 24 and 25 |
| 909 | ν(C–C), ν(C–O) | DNA/RNA backbone (ribose) | 24 |
As demonstrated in Fig. 2(b), the spectral features of the GB cancer group in the 1560–1340 cm−1 range are substantially different from those of the other three groups. Meanwhile, the peak shapes of the normal, GB polyp, and HCC groups are similar to one another. Notably, the intensities of peaks at 1537 and 1359 cm−1 (denoted with triangles) in the GB cancer spectrum are substantially lower than those in the spectra of the other three groups. For quantitative evaluation, the intensity ratios of peaks at 1537 and 1453 cm−1 (I1537/I1453), and at 1395 and 1359 cm−1 (I1395/I1359) calculated from all the sample spectra were analyzed and are displayed in Fig. 3(a) and (b), respectively. These four peaks are indicated with vertical dotted lines in Fig. 2(b) (also refer to their vibrational assignments in Table 1). The averages and standard deviations of the intensity ratio values obtained from each group are also shown in Fig. 3. Notably, the I1537/I1453 and I1395/I1359 values of the GB cancer group are much lower and higher, respectively, than those of the other three groups.
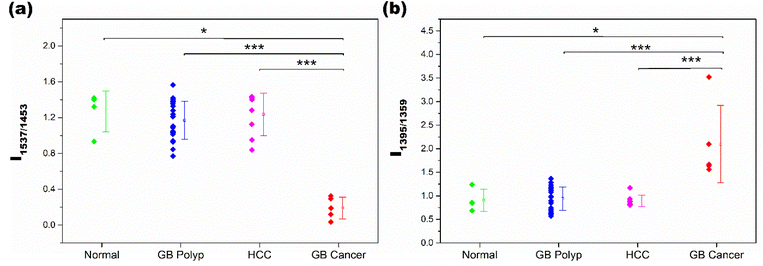 | ||
| Fig. 3 I 1537/I1453 (a) and I1395/I1359 values (b) calculated from the spectra of samples in the normal, GB polyp, HCC, and GB cancer groups. | ||
The difference of intensity ratio values between two groups was statistically evaluated using the Mann–Whitney U test, and the test was performed for all six two-group comparisons. The two groups showing significant differences are denoted with * or ***, corresponding to p-values smaller than 0.05 (95.0% confidence interval (CI)) or 0.001 (99.9% CI), respectively. The differences of I1537/I1453 values between the GB polyp and GB cancer samples, and the HCC and GB cancer samples are highly significant with CIs of 99.9%. The I1537/I1453 values of normal and GB cancer samples are also different with a CI of 95.0%. The I1537/I1453 values among the remaining three groups are not different; the p-values for these analyses are much greater than 0.05. Similar statistical results were obtained when the I1395/I1359 values of the groups were compared. The lower I1537/I1453 and higher I1395/I1359 values of GB cancer samples originated from the dissimilar composition of EVs in the measured samples.
To simply estimate the sensitivity of Raman measurement based on the WAC scheme, the EV sample extracted from one bile sample from the HCC group (the same sample measured in Fig. 1(c); concentration: 7.57 × 1013 particles per mL) was selected. Through continual dilution of the sample, a total of 12 samples with varying EV concentrations from 5.0 × 107 to 7.57 × 1013 particles per mL were prepared and the Raman spectra of each sample were acquired. Based on the observation of intensities of peaks at 1537, 1453, 1395 and 1359 cm−1 (refer to the corresponding spectra in the ESI (Fig. S4†)), it was sufficient to recognize the peaks when the concentration of EVs was 5.0 × 107 particles per mL. The detection of EVs at a level of around 107 particles per mL is thereby feasible by the proposed Raman measurement.
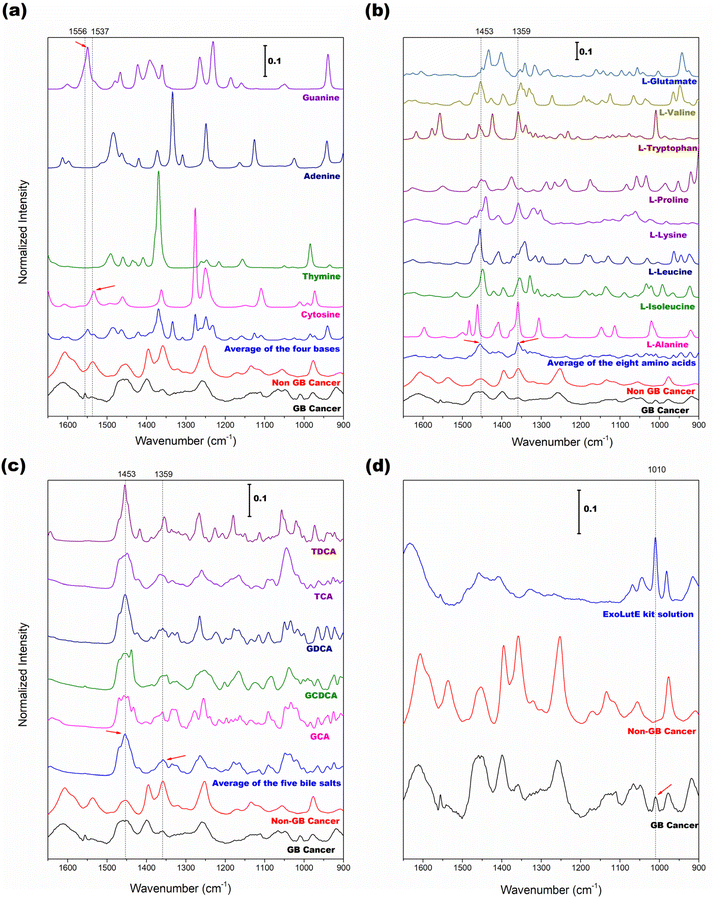
To further track the origins of the four selected peaks above, Raman spectra of the related pure components were collected, and their spectral features were compared with those of EV samples. Since DNAs and RNAs are the main components of EVs, examining the spectral features of the four DNA/RNA bases is beneficial. Fig. 4(a) shows the spectra of adenine, thymine, guanine, and cytosine along with the average spectra of EV samples of non-GB cancer (normal/GB polyp/HCC) and GB cancer groups. The average spectrum of the four bases is also displayed. The vertical bars indicate the wavenumbers at 1556 and 1537 cm−1. The small peak at 1556 cm−1 in the GB cancer spectrum is close to the guanine peak at 1549 cm−1 (marked with a right arrow). The peak at 1537 cm−1 in the non-GB cancer spectrum is close to the cytosine peak at 1534 cm−1 (indicated with a left arrow). Based on these observations, the occurrence of GB cancer would cause alteration of DNA sequences. The difference in sequences could be subsequently recognized in the spectra of EVs.
Protein is one component of EVs that would be abnormally expressed in cancers. For Raman spectroscopic fingerprinting of varied protein structures, protein peaks arising from the side chains and/or the amide-carboxyl groups of amino acids and nucleotides are typically examined. Therefore, the average spectra of eight amino acids, namely alanine, leucine, isoleucine, lysine, proline, tryptophan, valine, and glutamate, were analyzed. The results are shown in Fig. 4(b). The dotted vertical bars correspond to the wavenumbers at 1453 and 1359 cm−1 used for the evaluation of the peak intensity ratio. In the average spectrum of the eight amino acids, these two peaks are clearly observed (denoted with arrows) and also present in the average spectra of EV samples of the non-GB cancer and GB cancer groups. Fig. 4(c) presents the spectra of five bile salts (GDCA, GCDCA, GCA, TDCA, and TCA). The dotted vertical bars again correspond to the wavenumbers at 1453 and 1359 cm−1. Bile salts are the most abundant component of bile and remain in the extracted EV samples. In the average spectrum of bile salts, the 1453 and 1359 cm−1 peaks are also apparent (denoted with arrows). As demonstrated, amino acids (also protein) and bile salts contribute to the spectra of bile-extracted EVs.
Since the EVs in the extraction solution were directly measured without drying, the peaks of the extraction solution are expected to appear with the peaks of EVs. Therefore, knowledge of the spectral features of the extraction solvent is required to investigate its influence on the EV peaks. Fig. 4(d) shows the Raman spectrum of the extraction solution of the ExoLutE kit along with the average spectra of EV samples of non-GB cancer and GB cancer groups. The peaks of the extraction solution appear in the 1520–1280 and 1070–900 cm−1 ranges. The peaks in the 1520–1280 cm−1 range are broad and the corresponding intensities are relatively low, while the peaks in the 1070–900 cm−1 range are more intense and narrower. The dotted vertical line indicates the 1010 cm−1 peak of the extraction solution. Interestingly, the 1010 cm−1 peak is observed in the GB cancer spectrum (denoted with an arrow) but not in the non-GB cancer spectrum. Since the concentration of EVs is low in the case of GB cancer (refer to the NTA result in Fig. 1), the dominance of the solution peak is increased. This explains the appearance of the small 1010 cm−1 peak in the GB cancer spectrum. Based on these observations, the influence of solution peaks on the spectra of EVs, especially in the 1550–1200 cm−1 range, is tolerable.
The origins of peaks of the bile-extracted EVs and the different EV spectral features of the GB cancer samples are difficult to clearly explain. Nonetheless, DNA/RNA bases, amino acids, and bile salts contribute to the spectra of EVs, and their relative abundances vary according to the disease, especially GB cancer. As reported earlier by this group,17 the relative abundances of bile salts and lipids in the bile samples of GB cancers were different from those of other diseases. Increased concentrations of isoleucine and glutamate and decreased concentrations of leucine, valine, alanine, proline, and lipid in the blood samples of colorectal cancer (CRC) patients were reported.18 The concentrations of lysine, alanine, leucine, isoleucine, tryptophan, and valine were reduced, while the concentration of proline was elevated, in venous serum samples of CRC patients.19 Similarly, variations in relative abundances of amino acids in the blood samples of lung cancer patients have been illustrated.20 Although these reports were based on the investigations of cancers other than GB cancer, the assumption that there are GB cancer-induced variations in relative abundances of amino acids in the bile-extracted EVs is plausible.
Discriminability of GB cancer in the principal component (PC) score domain
To visualize the discriminability of GB cancer by an alternative method, PCA was performed using all the sample spectra. The distribution of subsequent PC scores was examined. Fig. 5(a) shows the first and second PC scores obtained from the spectra of bile-extracted EV samples. The percentages of spectral variation explained by the respective PCs are given in parentheses. These two PCs described 71.84% of the total spectral variation in the dataset. The scores of the GB cancer samples and those of the non-GB cancer samples are located on the left and right, respectively. Overlapping is absent, so the discrimination of the GB cancer group is clear. The first PC is highly descriptive in the differentiation of the GB cancer group from the other three groups. The first loading used for the PCA of the spectra is shown in Fig. 5(b). The weights on the peaks at 1537 and 1359 cm−1, employed for the comparison of peak intensity ratios, are relatively strong. This observation reinforces the usefulness of these peaks for the discrimination of GB cancer samples.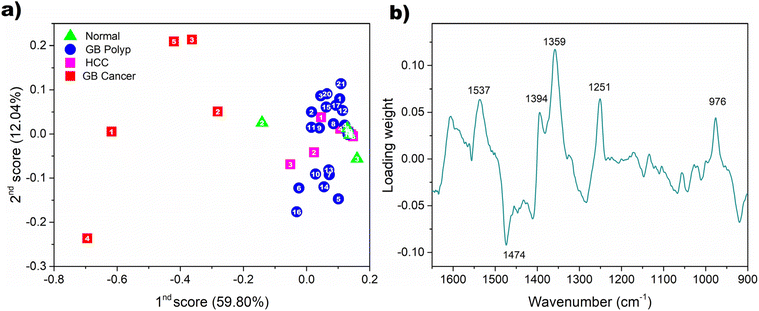 | ||
| Fig. 5 Distribution of the first and second PC scores obtained from all the spectra of bile-extracted EV samples (a) and the first loading used for PCA (b). | ||
Conclusions
We first explored the Raman spectroscopic fingerprinting of bile-extracted EVs for the identification of GB cancer. The EVs of human bile are versatile GB cancer markers. The Raman spectral features of EVs, especially the I1537/I1453 and I1395/I1359 values, of GB cancer samples were significantly different from those of the normal, GB polyp, and HCC samples. The varied metabolite compositions and structures of EVs due to the occurrence of GB cancer were directly reflected in the acquired Raman spectra, thereby enabling the discrimination of GB cancer. Also, the employed method directly measures the EVs in the extraction solution without drying, an additional advantage ensuring reliability of clinical assessment. Meanwhile, the discrimination between HCC and normal/GB polyp samples was not realized due to the similar EV composition of the corresponding samples. Unlike GB cancer, HCC does not appear to significantly alter the composition of EVs.Beyond this feasibility study, two further important issues need to be addressed to make the proposed Raman analysis of EVs clinically applicable. First, the compositions of bile-extracted EVs need to be determined to explain the observed spectral differences between non-GB cancer and GB cancer groups in detail. Second, a sample set with an increased number of bile samples is required for more reliable evaluation. In particular, inclusion of larger numbers of normal and GB cancer samples would be beneficial. However, collection of these samples is slow since normal bile samples are only obtainable from healthy organ donors for liver transplantation and GB cancer is not a disease with a high incidence rate. Research into resolving these two issues is underway in our group.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2021R1A2B5B02002412) and was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A06046728). Thu Thuy Bui was supported by a Hyundai Motor Chung Mong-Koo Global Scholarship.References
- M. Li, A. J. Rai, G. Joel DeCastro, E. Zeringer, T. Barta, S. Magdaleno, R. Setterquist and A. V. Vlassov, Methods, 2015, 87, 26–30 CrossRef CAS PubMed.
- D. Chalapathi, S. Padmanabhan, R. Manjithaya and C. Narayana, J. Phys. Chem. B, 2020, 124, 10952–10960 CrossRef CAS PubMed.
- G. Pocsfalvi, C. Stanly, A. Vilasi, I. Fiume, G. Capasso, L. Turiák, E. I. Buzas and K. Vékey, Mass Spectrom. Rev., 2016, 35, 3–21 CrossRef CAS PubMed.
- A. N. Böing, E. van der Pol, A. E. Grootemaat, F. A. W. Coumans, A. Sturk and R. Nieuwland, J. Extracell. Vesicles, 2014, 3, 23430 CrossRef PubMed.
- J. Mihály, R. Deák, I. C. Szigyártó, A. Bóta, T. Beke-Somfai and Z. Varga, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 459–466 CrossRef PubMed.
- C. F. Morasso, D. Sproviero, M. C. Mimmi, M. Giannini, S. Gagliardi, R. Vanna, L. Diamanti, S. Bernuzzi, F. Piccotti, M. Truffi, O. Pansarasa, F. Corsi and C. Cereda, Nanomedicine, 2020, 29, 102249 CrossRef CAS PubMed.
- A. Kamińska, M. Roman, A. Wróbel, A. Gala-Błądzińska, M. T. Małecki, C. Paluszkiewicz and E. Ł. Stępień, Nanomedicine, 2022, 39, 102468 CrossRef PubMed.
- H. Zhang, A. C. Silva, W. Zhang, H. Rutigliano and A. Zhou, PLoS One, 2020, 15, e0235214 CrossRef CAS PubMed.
- W. Lee, A. Nanou, L. Rikkert, F. A. W. Coumans, C. Otto, L. W. M. M. Terstappen and H. L. Offerhaus, Anal. Chem., 2018, 90, 11290–11296 CrossRef CAS PubMed.
- M. Imanbekova, S. Suarasan, T. Rojalin, R. R. Mizenko, S. Hilt, M. Mathur, P. Lepine, M. Nicouleau, N.-V. Mohamed, T. M. Durcan, R. P. Carney, J. C. Voss and S. Wachsmann-Hogiu, Nanoscale Adv., 2021, 3, 4119–4132 RSC.
- H. B. El-Serag and K. L. Rudolph, Gastroenterology, 2007, 132, 2557–2576 CrossRef CAS PubMed.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, CA-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- T. J. J. de Bitter, P. R. de Reuver, E. A. J. de Savornin Lohman, L. I. Kroeze, M. E. Vink-Börger, S. van Vliet, F. Simmer, D. von Rhein, E. A. M. Jansen, J. Verheij, C. M. L. van Herpen, I. D. Nagtegaal, M. J. L. Ligtenberg and R. S. van der Post, npj Precis. Oncol., 2022, 6, 83 CrossRef CAS PubMed.
- C. Are, H. Ahmad, A. Ravipati, D. Croo, D. Clarey, L. Smith, R. R. Price, J. M. Butte, S. Gupta, A. Chaturvedi and S. Chowdhury, J. Surg. Oncol., 2017, 115, 580–590 CrossRef PubMed.
- T. Akagi, K. Kato, M. Kobayashi, N. Kosaka, T. Ochiya and T. Ichiki, PLoS One, 2015, 10, e0123603 CrossRef PubMed.
- C. Eum, J. Park, S. Kumar, E. Jang, Y. Lee, S.-H. Nam, D. Choi and H. Chung, J. Anal. At. Spectrom., 2022, 37, 823–832 RSC.
- E. Jang, S. Jung, W. Sohng, D. Choi, G.-S. Hwang and H. Chung, Spectrochim. Acta, Part A, 2023, 286, 122030 CrossRef CAS PubMed.
- J. Gu, Y. Xiao, D. Shu, X. Liang, X. Hu, Y. Xie, D. Lin and H. Li, Dis. Markers, 2019, 2019, 3491852 Search PubMed.
- A. B. Leichtle, J.-M. Nuoffer, U. Ceglarek, J. Kase, T. Conrad, H. Witzigmann, J. Thiery and G. M. Fiedler, Metabolomics, 2012, 8, 643–653 CrossRef CAS PubMed.
- S. L. Lim, Z. Jia, Y. Lu, H. Zhang, C. T. Ng, B. H. Bay, H. M. Shen and C. N. Ong, Metabolomics, 2018, 14, 118 CrossRef PubMed.
- J. W. Chan, D. S. Taylor, T. Zwerdling, S. M. Lane, K. Ihara and T. Huser, Biophys. J., 2006, 90, 648–656 CrossRef CAS PubMed.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- S. Garip, A. C. Gozen and F. Severcan, Food Chem., 2009, 113, 1301–1307 CrossRef CAS.
- A. J. Hobro, D. M. Standley, S. Ahmad and N. I. Smith, Phys. Chem. Chem. Phys., 2013, 15, 13199–13208 RSC.
- F. Quilès, F. Humbert and A. Delille, Spectrochim. Acta, Part A, 2010, 75, 610–616 CrossRef PubMed.
- N. Simsek Ozek, I. B. Bal, Y. Sara, R. Onur and F. Severcan, Biochim. Biophys. Acta, Gen. Subj., 2014, 1840, 406–415 CrossRef CAS PubMed.
- J. Penders, A. Nagelkerke, E. M. Cunnane, S. V. Pedersen, I. J. Pence, R. C. Coombes and M. M. Stevens, ACS Nano, 2021, 15, 18192–18205 CrossRef CAS PubMed.
- H. Koch, S. Polepil, K. Eisen and S. Will, Phys. Chem. Chem. Phys., 2017, 19, 30533–30539 RSC.
- S. Shakeel, H. Nawaz, M. I. Majeed, N. Rashid, M. R. Javed, A. Tariq, B. Majeed, A. Sehar, S. Murtaza, N. Sadaf, G. Rimsha and I. Amin, Photodiagn. Photodyn. Ther., 2022, 39, 102949 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00806a |
| This journal is © The Royal Society of Chemistry 2023 |