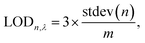Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Smartphone-based colorimetric detection system for portable health tracking†
Samira
Balbach
a,
Nan
Jiang
 *b,
Rosalia
Moreddu
*b,
Rosalia
Moreddu
 c,
Xingchen
Dong
ad,
Wolfgang
Kurz
a,
Congyan
Wang
a,
Jie
Dong
a,
Yixia
Yin
e,
Haider
Butt
f,
Martin
Brischwein
g,
Oliver
Hayden
g,
Martin
Jakobi
a,
Savas
Tasoglu
h,
Alexander W.
Koch
a and
Ali K.
Yetisen
c,
Xingchen
Dong
ad,
Wolfgang
Kurz
a,
Congyan
Wang
a,
Jie
Dong
a,
Yixia
Yin
e,
Haider
Butt
f,
Martin
Brischwein
g,
Oliver
Hayden
g,
Martin
Jakobi
a,
Savas
Tasoglu
h,
Alexander W.
Koch
a and
Ali K.
Yetisen
 *d
*d
aInstitute for Measurement Systems and Sensor Technology, Technical University of Munich, Munich 80333, Germany
bWest China School of Basic Medical Sciences & Forensic Medicine, Sichuan University, Chengdu 610041, China. E-mail: jiangnansophia@scu.edu.cn
cIstituto Italiano di Tecnologia, 16163 Genova, Italy
dDepartment of Chemical Engineering, Imperial College London, London SW7 2AZ, UK. E-mail: a.yetisen@imperial.ac.uk
eState Key Laboratory of Advanced Technology for Materials Synthesis and Processing, Wuhan University of Technology, Wuhan 430070, China
fDepartment of Mechanical Engineering, Khalifa University, Abu Dhabi 1277788, UAE
gHeinz Nixdorf Chair Biomedical Electronics, TranslaTUM, Technical University of Munich, Munich 81675, Germany
hDepartment of Mechanical Engineering, Koc University, Sariyer, Istanbul 34450, Turkey
First published on 8th September 2021
Abstract
Colorimetric tests for at-home health monitoring became popular 50 years ago with the advent of the urinalysis test strips, due to their reduced costs, practicality, and ease of operation. However, developing digital systems that can interface these sensors in an efficient manner remains a challenge. Efforts have been put towards the development of portable optical readout systems, such as smartphones. However, their use in daily settings is still limited by their error-prone nature associated to optical noise from the ambient lighting, and their low sensitivity. Here, a smartphone application (Colourine) to readout colorimetric signals was developed on Android OS and tested on commercial urinalysis test strips for pH, proteins, and glucose detection. The novelty of this approach includes two features: a pre-calibration step where the user is asked to take a photo of the commercial reference chart, and a CIE-RGB-to-HSV color space transformation of the acquired data. These two elements allow the background noise given by environmental lighting to be minimized. The sensors were characterized in the ambient light range 100–400 lx, yielding a reliable output. Readouts were taken from urine strips in buffer solutions of pH (5.0–9.0 units), proteins (0–500 mg dL−1) and glucose (0–1000 mg dL−1), yielding a limit of detection (LOD) of 0.13 units (pH), 7.5 mg dL−1 (proteins) and 22 mg dL−1 (glucose), resulting in an average LOD decrease by about 2.8 fold compared to the visual method.
Introduction
The modern approach to global health is largely focused on the development of portable diagnostic systems for infectious and chronic diseases.1–3 Rapid and low-cost testing platforms play an essential role in effectively controlling disease onset, progression and transmission.4,5 This concept has been particularly relevant in the past decades, with the worldwide diffusion of numerous diseases and infections.6–8 Besides infectious diseases, chronic diseases are a global health burden which would benefit from the development of effective point-of-care medical technologies and continuous monitoring methods.9Paper-based assays were regarded as promising in the last 50 years, due to their inexpensive nature and fast analysis time.10,11 However, only a few of them have hit the market and are employed on a daily basis, due to the lack of standardization, convenient integration, and readout methods. Paper microfluidic sensors were also developed to aid the diagnosis of metabolic disorders, diabetes, and cancer.12–15
The concept of at-home self-screening was explored from multiple perspectives throughout the years, leading to the development of different types of devices depending on the application.
The main criteria include the choice of the bodily fluid to test, the sensing technology, and the modality of interaction between the device and the fluid. Implantable devices or patch needles were demonstrated to aid the screening of blood and interstitial fluid.16 Dermal sensors based on colorimetric inks for acidosis and diabetes were demonstrated.17 Tears have recently emerged as a fluid that can provide information about the ocular health status of an individual, by expressing different patterns of biomarkers.18 Wearable contact lenses were investigated as the vehicle of integration, to monitor the ocular temperature and tear markers via colorimetric methods.19–23
Although the colorimetric analysis can be simply performed through visual comparison,24 this only provides qualitative information, and quantitative measurements heavily rely on bulky and costly equipment which inevitable increases the analysis time.25 Recent efforts in quantitative readouts of optical signals were put on the development of smartphone-based approaches, to simplify the real-time analysis of biomarkers whilst reducing the costs.26,27 However, the accuracy of smartphone readouts of colorimetric signals is affected by imaging angle and distance from the sample, ambient light condition, and parameters of the smartphone camera.24 For example, two smartphone applications were developed to readout urine test strips.26,27 The first approach was based on the transposition from the RGB to HSV space and allowed to minimize the effect of ambient lighting.26 However, it was solely calibrated using the reference points provided by the test strip manufacturer. This approach allows to automatize the readout method but keeps the same sensitivity of the visual readout. The second approach involved the use of the CIE 1931 chromaticity diagram for color visualization.27 However, it lacks in introducing a way of cross-correlating the smartphone app output with the proposed chromaticity diagram method.
Here, a customized smartphone application (Colourine) was developed for Android OS to quantify pH, glucose and proteins in commercial urine strips. The algorithm merges the two approaches discussed earlier to provide a mean of calculating urine analytes concentrations by introducing the nearest neighbor algorithm. The algorithm maps the newly acquired color in the CIE chromaticity space and compares it with the reference color. The flexibility of this approach allows the app to be programmed with a larger number of reference points compared to the ones provided in the commercial reference chart, hence it substantially increases the sensitivity. This method is coupled to the RGB-to-HSV transposition approach, which considers the intensity and saturation of the detected colors, and in turn allows to minimize the optical noise resulting from ambient light contributions. This is further strengthened by introducing a self-calibration step which is performed by asking the user to take a photo of the commercial reference chart prior to each measurement. The application outputs the concentration value and returns them in the form of trends. It enables tracking diagnostic records by storing patient information and previous test results. This technology may be extended to other sensing technologies, including fluorescence and diffraction, and find application as a readout method for continuous health tracking devices based on optical sensors.
Material and methods
Chemicals
pH buffer solutions were purchased from Merck (pH values 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0). D-(+)-Glucose (99.5%), bovine serum albumin (lyophilized powder, 96%), and phosphate-buffered saline (PBS) were purchased from Sigma-Aldrich and used without further purification. The Laboratory TranslaTUM provided deionized (DI) water. Cobas® Combur3Test® strips were purchased by Roche Diagnostics (GmbH, Mannheim, Germany).Preparation of buffer solutions
Protein and glucose buffer solutions were obtained by dissolving D-(+)-glucose or bovine serum albumin (BSA) in PBS at pH 7.40. BSA buffers were prepared to yield protein concentrations of 30 mg dL−1, 65 mg dL−1, 100 mg dL−1, 200 mg dL−1, 300 mg dL−1, 400 mg dL−1, and 500 mg dL−1. Glucose buffer solutions were prepared to yield concentrations of 2.8 mmol L−1, 4.12 mmol L−1, 5.5 mmol L−1, 11.26 mmol L−1, 17 mmol L−1, 27.5 mmol L−1, 36.02 mmol L−1, 44 mmol L−1, and 55 mmol L−1.Hardware
The smartphone app was implemented on a Samsung Galaxy A20e (Samsung Electronics, GmbH) running Android 10.0 OS. Color Name app was used to check the RGB colors captured with the smartphone camera to obtain the graphs.Algorithm development platform
The software was developed in C++, Java, and XML languages on Windows 10 OS. Image processing algorithms were developed in C++ using the integrated development environment Microsoft Visual Studio 2019 and the open-source library OpenCV 4.3.0. The layout for the user interface was created in XML and tested in Java, both running on API 28 in Android Studio 4.0. The C++ code algorithm was integrated in Android Studio by using Java Native Interface (JNI) framework. CMake was used as the complementary tool to JNI and provide a working interaction between Java and C++. The Android Native Development Kit (NDK) was used for development, debugging, and compile.Image acquisition and data pre-processing
The reference chart and test strip photographs taken by the user were filtered to isolate and define the color areas, i.e. the sensors. Morphological transformations were implemented in OpenCV. Rectangles were sorted into rows and columns to provide a multiplexed readout. The newly acquired colors were then compared with the reference chart.Concentration analysis using colorimetric signals
To quantify the digital color information based on the image, two analysis algorithms with color processing and comparison were implemented on the CIE 1931 chromaticity space, using the RGB (red, green, blue) and HSV (hue, saturation, value) color models. In the CIE space, a color is uniquely identified by its (x, y) coordinates and it is plot in the diagram. The color point was transferred from the RGB to the HSV space, and further plot in the CIE 2D chromaticity diagram. The newly acquired point was compared to the reference color points based on the nearest neighbor algorithm and linear proportion, where the distances are calculated. Hue states the color shade, hence it represents the primaries and their composition, indicated as a number from 0 to 360°. This also implies that the H value is independent of the color intensity, i.e. the luma is segregated from the chroma. The S value in the HSV space is expressed as a percentage 0 to 100 and it indicates the grey components. The higher the saturation, the lower the grey percentage. The V component in the HSV space expresses the color brightness with a percentage, where 0 is black and 100 is white. Since the luma can be affected by varying lighting conditions, the HSV color space was implemented to minimize the environmental background. In the RGB space, every component (red, green, blue) is related to a color contribution, making it harder to filter out external noise. For both evaluation algorithms, the estimated analyte concentration is returned as a result to the user via the user interface of the smartphone application. These results can be compared to the given reference values in the application to gather a diagnostic report. Details can be found in ESI and ESI Fig. S5 and S6.†Calculation of the limit of detection
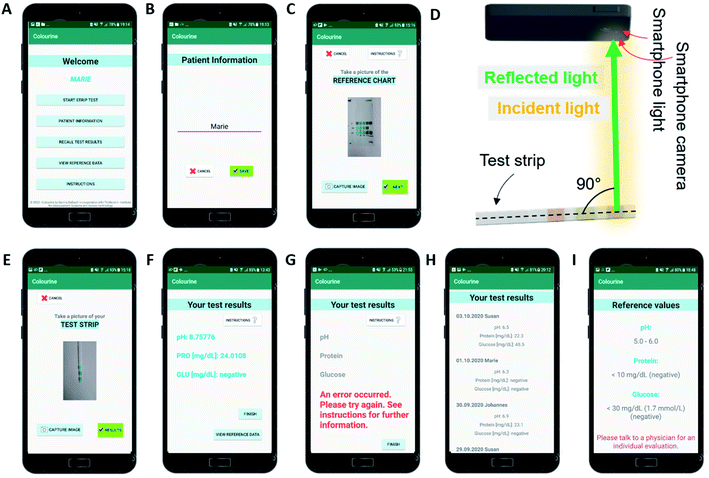
Individual LODs for each analyte (n) were calculated using the σ method:28where stdev is the standard deviation and m is the slope of the readout curve (linear trendline, y = mx + q) of the algorithm. The average of the CIE-based and HSV-based LODs was taken for each analyte.
Results
The Colourine smartphone application (app) comprises two key building blocks: the algorithm logic, based on the CIE and HSV color models, and the user interface (UI). The workflow requires the user to proceed by the following steps: capture the reference chart provided by the manufacturer and an unused test strip, perform a urinalysis test, and capture the tested strip. The results are obtained by comparing the tested strip color areas with the reference color areas and are displayed in the user interface.System development and calibration
The app was designed in Microsoft Studio and Android Studio to run on Android OS, using the open-source library OpenCV. Upon acquiring the images of the unused test strip and the color chart (Fig. 1A), the image processing is articulated into different steps. First, the algorithm filters out the background noise from the images, and extracts the regions of interest, i.e. the colored areas using the dilation function in OpenCV (Fig. 1B). Following this, a series of consequent morphological transformation is implemented. Each operation is performed on the latest image obtained by the algorithm. After extracting the colored areas, the expansion function is implemented to enlarge the rectangles (Fig. 1C), followed by a transposition from the RGB to the HSV space (Fig. 1D). The inRange function is then applied to the transposed image, which is converted to a binary image (Fig. 1E). The closing function is further implemented to conform the object by closing small white holes or black points (Fig. 1F), and the boundaries of the foreground objects are cut away with the erosion function (Fig. 1G). The further step involves the masking function, which applies a logic AND operation between each pixel from two images. This was implemented between the original image (Fig. 1A) and the latest image obtained by the algorithm (Fig. 1G). The result is a high-contrast image with clearly visible colored rectangles on a black background (Fig. 1H).The contours function is then implemented to ulteriorly define and isolate the colored areas, which are digitally stored as a chart made of rows and columns. These are obtained from Fig. 1G, implemented in Fig. 1H, and displayed in Fig. 1I. The border assignment is necessary to further associate each row/column to a specific analyte. The coherence between the original order of the detected colored rectangles and their corresponding analyte concentration is saved as indexes.
In the next step, the app is meant to detect the colors and assign them to the corresponding rectangles for subsequent evaluation. For this further processing, two algorithms are implemented independently. The first algorithm operates on the CIE-xy space, the second algorithm on the HSV-xy space. The smartphone application can be programmed either with the first or the second algorithm. In the CIE-xy algorithm the processed images (Fig. 1I) are converted from the RGB to the CIE-XYZ color space, from which the chromaticity coordinates x and y are calculated for each colored square. When the user performs a test with a urine strip and takes a photo, the test strip colors are plotted in the same CIE-xy diagram and compared to the reference colors. For each analyte, the distance between the newly imaged color and the reference colors in the CIE-xy plot is calculated by the algorithm by projecting the point to the line segment connecting the two nearest reference points. The concentration value corresponding to the reference color point having the shortest distance is given as output, yielding the theoretical “real-valued” index of the test point. This leads to the theoretical “real-valued” index of the test point. The indexes of the rectangles serve as x-values, the reference analyte concentrations serve as y-values. The evaluation curve for an analyte, based on the reference values, is either linear or a piecewise cubic hermite interpolating polynomial, with the reference values as supporting points.
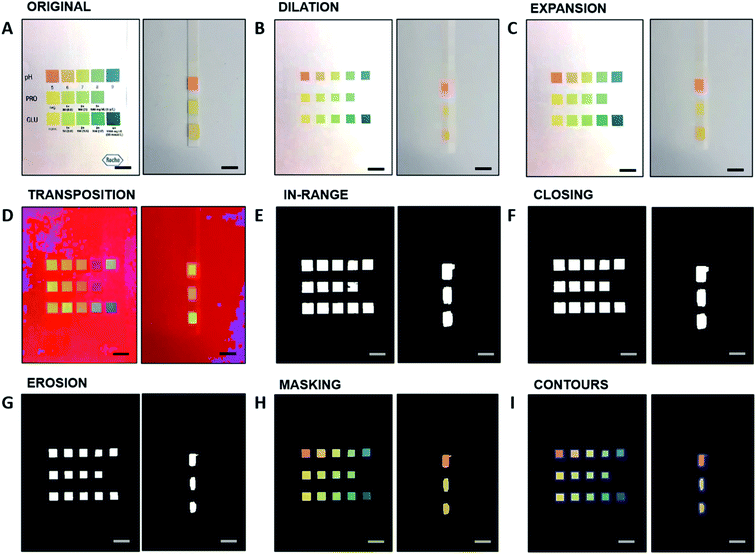
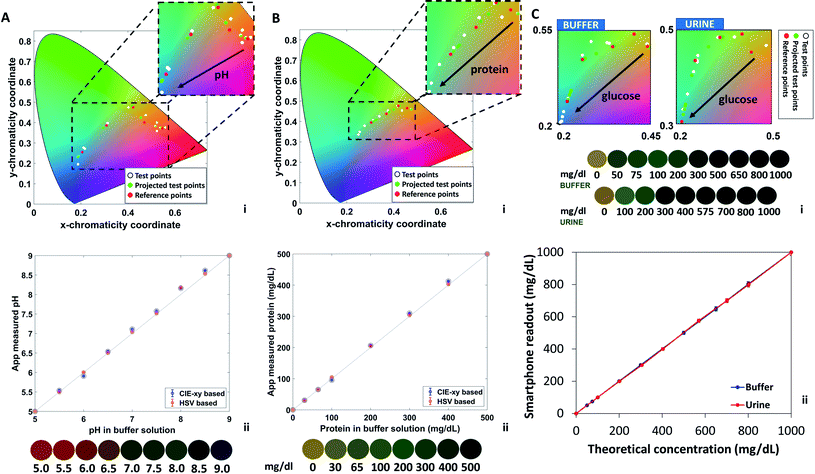
Fig. 2A–C depicts the calculated real-value index from the CIE-xy algorithm for pH (0–9 units), protein (0–500 mg dL−1), and glucose (0–1000 mg dL−1), respectively. Consequently, with the determined index of the test color point, an analyte concentration is assigned to the test color point by means of the corresponding evaluation function. Different analytes are discriminated by their position along the strip, and this operation is performed independently for each analyte, to provide a cumulative output for multiple biomarkers.
On the other hand, the HSV model acts primarily on the H-values, whereas the S and the V values serve as additional information to minimize the errors given by color saturation. The H-values of the reference rectangles for each analyte serve as x-value for the evaluation curve, while the reference analyte concentrations serve as y-values. The HSV model operates similarly to the CIE-xy model. The evaluation curves are displayed in Fig. 2D–F for pH, proteins, and glucose, respectively. These evaluation functions determine the resulting analyte concentration for a specific H-value of a colored area of the test strip.
User interface
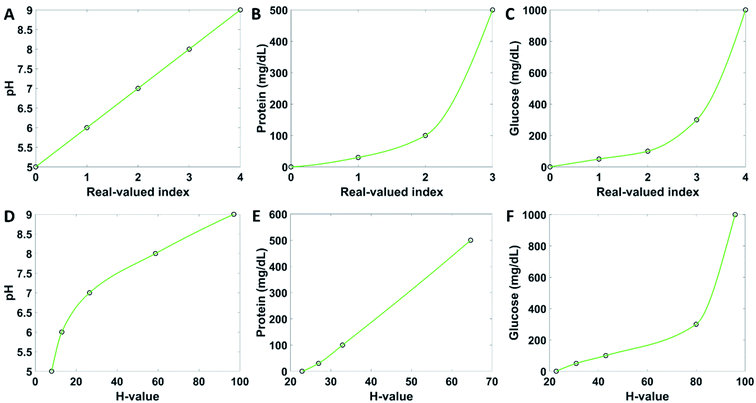
The user interface of the application guides the user to perform the colorimetric analysis of the test strips step by step. The home page of the application contains five buttons (Fig. 3A). The patient information button brings the user to a new window where they can upload their personal details, to which all future measurements can be assigned and stored in the form of trends (Fig. 3B). If the name has been used before, the user can choose an already existing profile. When a name is saved, it appears on the main page. The name saving step can be skipped and run the test as guest. The button start strip test bring the user to a page where to take a photo of the reference chart (Fig. 3C) by clicking the capture image button, which invokes the smartphone camera. When the picture is taken, its bitmap is shown on the same layout page. The photo must be taken at normal incidence, from 10 cm of distance (Fig. 3D). After confirming the photograph by choosing next, the same procedure starts for capturing an image of the test strip (Fig. 3E). The next button includes a feedback loop which prevents the user from going further before an image is taken and uploaded. When a picture of the test strip is taken successfully, the app brings out the results page, where the analyte concentrations are displayed (Fig. 3F). If the evaluation process failed, an error report is shown (Fig. 3G). By clicking the button finish, the results are saved, and the user gets back to the main page. The user may look at past results by clicking the recall test results button (Fig. 3H). The view reference data page allows to observe the healthy reference values (Fig. 3I). The page instructions contains information on how to carry on the measurements. These instructions comprise the testing procedure, particularly the angle at which the measurements should be taken.Quantitative analysis
Urine is a typical body fluid formed as metabolic waste in the human kidney.29 Detection of analytes in urine can directly report on health conditions at an early stage.30 The smartphone application was tested using commercial colorimetric strips for urinalysis, both buffer solutions (to test pH, proteins, glucose concentrations) and a urine-based solution (to test glucose concentration). Glucose detection was further implemented in a human urine sample. The buffer solution (200 μL) was deposited on the strips by drop casting using a pipette, the excess liquid was removed by shaking the strip, as per commercial protocol. Photographs were taken under constant ambient lighting of 300 lux, at normal incidence and at 10 cm of distance between the smartphone and the strip.Fig. 4 illustrates the algorithm results for pH, protein, and glucose detection from urine strips. Urinary pH varies from 4.5 to 8.0 units and it is correlated to the physiological health status. Prolonged alkaline urine is a potential indicator of infection with urea-splitting microbes; high acidic urine may be a sign of diabetic ketoacidosis, gout, and diarrhea.31Fig. 4A displays the pH-dependent chromaticity diagram (i) which includes the reference points, the test points, and the projected test points. The correlation between theoretical pH values and pH values measured by the algorithm is also shown (ii), with standard deviation of 0.05 units, yielding a LOD of 0.13 pH units. The inset shows the concentration-dependent color variations from red (pH 5) to blue (pH 9). Such colors were interpreted using the RGB and the HSV models. The trends of individual components upon pH changes are plot in ESI Fig. S1† for the (R, G, B) and the (H, S, V) spaces. The red component was found to decrease from 155/255 to 2/255 upon shifting the pH value from 5 to 9 units; the blue component was found to increase from 55/255 to 65/255 in the same range. The green component was found to fluctuate around an average value of 80/255, providing a nearly constant contribution. In the HSV model, the H and the V values were found to increase from 200 to 230 and from 0 to 100, respectively. The S value was found to decrease from 160 to 8 upon increasing the pH from 5 to 9 units.
The algorithm was tested in parallel for proteins levels in buffer solutions containing PBS and bovine serum albumin (BSA). The standard range of protein in human urine is 0–20 mg dL−1, and higher concentrations indicate proteinuria, which is a biomarker for kidney disease. Most healthy people have a daily protein secretion of less than 150 mg, while a pregnant woman level can exhibit protein levels in urine of about 300 mg.32Fig. 4B displays the concentration-dependent chromaticity diagram (i) which includes the reference points, the test points, and the projected test points. The correlation between theoretical protein values and protein values measured by the algorithm are also shown (ii), where the error bars are covered by the points and were found to yield a standard deviation of 3.08. Based on the plot, a LOD of 7.5 mg dL−1 was calculated. The standard protein sensor displays a color change from yellow to dark green when varying protein concentration from 0 to 500 mg dL−1. The color chart is displayed as an inset. Such colors were interpreted using the RGB and the HSV models. The trends of individual components upon protein changes are plot in ESI Fig. S2.† The red and green components were found to decrease from 150/255 to 40/255 and from 120/255 to 80/255 respectively, upon varying protein concentrations from 0 to 500 mg dL−1. The blue component was found to increase from 50/255 to 65/255 in the same range. In the HSV model, the V values were found to decrease from 145 to 80, the H component was found to increase from 20 to 80, and the S component exhibited a decrease from 160, reaching a minimum at 60 and further increased to 120.
As for protein and pH, the algorithm readout glucose concentration in parallel. Normally, only a small amount of glucose can be detected in healthy urine (0–0.8 mmol L−1). Increased glucose level is an indication of kidney diseases or injuries, and diabetes. Glycosuria is a common symptom of diabetes. Glucose was tested both in buffer solution and in human urine. Urine samples were obtained from a volunteer and used the same day. The reliability of the smartphone readout was double checked by testing the same urine samples using test strips. Different glucose concentrations in urine were obtained by adding glucose to the urine solution stepwise. Fig. 4C shows a zoom on the region of interest of the chromaticity plot in urine and buffer solutions (i). In both cases, the diagrams display the test points, the projected test points, and the reference colors. The inset shows the glucose-dependent color variations from yellow (0 mg dL−1) to green (1000 mg dL−1). The comparison between theoretical concentration of glucose in urine/buffer solution and the CIE-xy Colourine app readout is also shown (ii). In both cases, a high correlation was observed. From a standard deviation of 8.71, a LOD of 22 mg dL−1 was calculated. To better visualize the color contributions and their correlation in urine and buffer solutions, individual components are plot in ESI Fig. S3 and S4.† All components displayed the exact same trends, with little variations which were mainly visible at low glucose concentrations for R, G, H, and V, and at intermediate concentrations for B and S.
The algorithm allowed to decrease the LOD by 4 folds for pH, 2.5 folds for glucose, and 2 folds for proteins. This technique can be easily extended to multiple types of markers, providing a portable and facile tool for standardize the readout of test strips, as well as provide a digital database where the user can check their trends over time. Hence, another advantage of the readout method may play a role in turning a single snapshot test, such as the urine strip, in an actual continuous monitoring platform by storing the data.
The background noise given by the ambient light contribution remains the main challenge in point-of-care optical sensors. In this context, the Colourine app minimizes this contribution by implementing two features. On one hand, the background light is turned into a constant contribution by asking the user to take a photo of the reference chart and the bare test strip at the beginning of each test. The newly captured test is then compared with the reference, and this allow to subtract the background noise and operate at different light levels. Furthermore, the HSV space is introduced to compensate for the errors that remains in the RGB color space, considering color intensity and saturation.
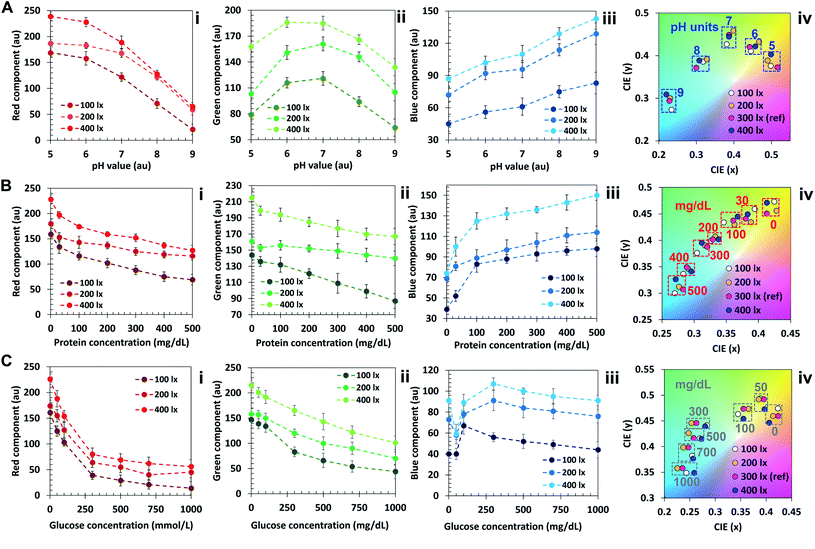
The algorithm was additionally tested at light levels of 100 lx, 200 lx and 400 lx, and the results were compared to the ones obtained at standard illumination (300 lx, Fig. 5). Fig. 5A–C(i–iii) display the comparison between concentration-dependent red, green and blue components at light levels of 100, 200 and 400 lx for pH (A), protein (B) and glucose (C). It is possible to observe that the curves show a similar trend with shifted intensity. This result is in accordance with previous works.20 At 400 lx, the R, G and B components are pronounced and closer to 255. At 200 lx their value decreases, reaching the minimum at 100 lx. It should be noted that, as shown in Fig. 5A–C(iv), the color diagram displays that the variations in saturation and hue are minimal, hence they can be associated to the correct nearest neighbor in the color plot despite ambient light variations in the range 100 to 400 lx.
In the protein color plot, the readout points display a higher proximity compared with pH and glucose, attributed to the narrower wavelength range of operation of the sensor. To obtain a reliable output outside this ambient lighting range, an additional step may be added in the algorithm, such that different calibration points are associated to different light levels. A lightmeter may be added to the app, and the reference points of interest could be selected based on the ambient light level.
To further improve this concept, a readout device may be designed and supplied together with the test strip package. A smartphone-compatible black box may be sufficient to optically isolate the test strip while taking a photo with flash on, making the smartphone torch the only light source. As an alternative, the light source may be incorporated into the readout device to compensate for differences between smartphones. In addition, by adjusting the wavelengths of light sources and optical filters in the readout device, the readout may be extended to other chromogenic dyes,33 fluorescent probes,34 diffraction gratings,35 and nanoparticles.36
Conclusion
Colourine smartphone app was developed to readout commercial urine test strips, based on the CIE-xy nearest neighbor algorithm, the RGB and the HSV color spaces. The app was tested to detect pH, proteins and glucose in buffer solutions as well as for glucose detection in human urine, resulting in a LOD decrease by about 4 folds for pH, 2.5 folds for glucose, and 2 folds for proteins, compared to the visual method. The transposition to the HSV color space and the initial step of operation which requires the user to take a photograph of the color chart, allowed to compensate for environmental background light. This was also demonstrated by testing the sensor at light levels of 100, 200, 300 and 400 lx. This technique may be easily extended to other optical readout applications, based on colorimetric and fluorescent sensors.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
A. K. Y. acknowledges the Engineering and Physical Sciences Research Council (EPSRC) for a New Investigator Award (EP/T013567/1). N. J. acknowledges the Fundamental Research Funds for the Central Universities(YJ202152). H. B. acknowledges KU-KAIST Joint Research Center (8474000220-KKJRC-2019-Health1) and Sandooq Al Watan LLC (SWARD, 8434000391-EX2020-044).References
- P. Yager, G. J. Domingo and J. Gerdes, Point-of-care diagnostics for global health, Annu. Rev. Biomed. Eng., 2008, 10, 107–144 CrossRef CAS PubMed.
- N. P. Pai, C. Vadnais, C. Denkinger, N. Engel and M. Pai, Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low-and middle-income countries, PLoS Med., 2012, 9(9), e9215 CrossRef PubMed.
- S. K. Vashist, P. B. Luppa, L. Y. Yeo, A. Ozcan and J. H. Luong, Emerging technologies for next-generation point-of-care testing, Trends Biotechnol., 2015, 33(11), 692–705 CrossRef CAS PubMed.
- P. K. Drain, E. P. Hyle, F. Noubary, K. A. Freedberg, D. Wilson, W. R. Bishai, W. Rodriguez and I. V. Bassett, Diagnostic point-of-care tests in resource-limited settings, Lancet Infect. Dis., 2014, 14(3), 239–249 CrossRef PubMed.
- G. Wu and M. H. Zaman, Low-cost tools for diagnosing and monitoring HIV infection in low-resource settings, Bull. World Health Organ., 2012, 90, 914–920 CrossRef PubMed.
- K. McIntosh, The SARS coronavirus: rapid diagnostics in the limelight, Clin. Chem., 2003, 49, 845–846 CrossRef PubMed.
- A. Nougairede, L. Ninove, C. Zandotti, X. de Lamballerie, C. Gazin, M. Drancourt, B. La Scola, D. Raoult and R. N. Charrel, Point of care strategy for rapid diagnosis of novel A/H1N1 influenza virus, PLoS One, 2010, 5(2), e9215 CrossRef PubMed.
- C. Wang, P. W. Horby, F. G. Hayden and G. F. Gao, A novel coronavirus outbreak of global health concern, Lancet Neurol., 2020, 395(10223), 470–473, DOI:10.1016/S0140-6736(20)30185-9.
- B. H. Weigl, T. Neogi and H. McGuire, Point-of-care diagnostics in low-resource settings and their impact on care in the age of the noncommunicable and chronic disease epidemic, J. Lab. Autom., 2014, 19(3), 248–257 CrossRef PubMed.
- M. M. Gong and D. Sinton, Turning the page: advancing paper-based microfluidics for broad diagnostic application, Chem. Rev., 2017, 117(12), 8447–8480 CrossRef CAS PubMed.
- J. Reboud, G. Xu, A. Garrett, M. Adriko, Z. Yang, E. M. Tukahebwa, C. Rowell and J. M. Cooper, based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities, Proc. Natl. Acad. Sci. U.S.A., 2019, 116(11), 4834–4842 CrossRef CAS PubMed.
- R. Hu, R. Gupta, Z. Wang, C. Wang, H. Sun, S. Singamaneni, E. D. Kharasch and J. J. Morrissey, Bioplasmonic paper–based assay for perilipin-2 non-invasively detects renal cancer, Kidney Int., 2019, 96(6), 1417–1421 CrossRef CAS PubMed.
- R. T. Robinson, Development of a Paper-Based Whole Blood Phenylalanine Assay for PKU Diagnosis and Monitoring in Low Resource Settings, Micromachines, 2016, 7(2), 28 CrossRef PubMed.
- D. Sechi, B. Greer, J. Johnson and N. Hashemi, Three-dimensional paper-based microfluidic device for assays of protein and glucose in urine, Anal. Chem., 2013, 85(22), 10733–10737 CrossRef CAS PubMed.
- L. C. Brazaca, J. R. Moreto, A. Martín, F. Tehrani, J. Wang and V. Zucolotto, Colorimetric Paper-Based Immunosensor for Simultaneous Determination of Fetuin B and Clusterin toward Early Alzheimer's Diagnosis, ACS Nano, 2019, 13(11), 13325–13332 CrossRef CAS PubMed.
- G. Dutta, A. Regoutz and D. Moschou, Enzyme-assisted glucose quantification for a painless Lab-on-PCB patch implementation, Biosens. Bioelectron., 2020, 167, 112484 CrossRef CAS PubMed.
- A. K. Yetisen, R. Moreddu, S. Seifi, N. Jiang, K. Vega, X. Dong, J. Dong, H. Butt, M. Jakobi and M. Elsner, Dermal Tattoo Biosensors for Colorimetric Metabolite Detection, Angew. Chem., 2019, 131(31), 10616–10623 CrossRef.
- R. Moreddu, D. Vigolo and A. K. Yetisen, Contact Lens Technology: From Fundamentals to Applications, Adv. Healthcare Mater., 2019, 8(15), e1900368 CrossRef PubMed.
- R. Moreddu, M. Elsherif, H. Adams, D. Moschou, M. F. Cordeiro, J. S. Wolffsohn, D. Vigolo, H. Butt, J. M. Cooper and A. K. Yetisen, Integration of paper microfluidic sensors into contact lenses for tear fluid analysis, Lab Chip, 2020, 20(21), 3970–3979 RSC.
- R. Moreddu, M. Elsherif, H. Butt, D. Vigolo and A. K. Yetisen, Contact lenses for continuous corneal temperature monitoring, RSC Adv., 2019, 9(20), 11433–11442 RSC.
- R. Moreddu, J. S. Wolffsohn, D. Vigolo and A. K. Yetisen, Laser-inscribed contact lens sensors for the detection of analytes in the tear fluid, Sens. Actuators, B, 2020, 317, 128183 CrossRef CAS.
- R. S. Riaz, M. Elsherif, R. Moreddu, I. Rashid, M. U. Hassan, A. K. Yetisen and H. Butt, Anthocyanin-Functionalized Contact Lens Sensors for Ocular pH Monitoring, ACS Omega, 2019, 4(26), 21792–21798 CrossRef CAS PubMed.
- R. Moreddu, V. Nasrollahi, P. Kassanos, S. Dimov, D. Vigolo and A. K. Yetisen, Lab-on-a-contact lens platforms fabricated by multi-axis femtosecond laser ablation, Small, 2021 DOI:10.1002/smll.202102008.
- S. Jain, R. Rajasingham, F. Noubary, E. Coonahan, R. Schoeplein, R. Baden, M. Curry, N. Afdhal, S. Kumar and N. R. Pollock, Performance of an optimized paper-based test for rapid visual measurement of alanine aminotransferase (ALT) in fingerstick and venipuncture samples, PloS One, 2015, 10(5), e0128118 CrossRef PubMed.
- S. Kim, E. Jung, M. J. Kim, A. Pyo, T. Palani, M. S. Eom, M. S. Han and S. Lee, A simple, fast, and easy assay for transition metal-catalyzed coupling reactions using a paper-based colorimetric iodide sensor, Chem. Commun., 2012, 48(70), 8751–8753 RSC.
- J. I. Hong and B. Y. Chang, Development of the smartphone-based colorimetry for multi-analyte sensing arrays, Lab Chip, 2014, 14(10), 1725–1732 RSC.
- L. Shen, J. A. Hagen and I. Papautsky, Point-of-care colorimetric detection with a smartphone, Lab Chip, 2012, 12(21), 4240–4243 RSC.
- A. Shrivastava and V. B. Gupta, Methods for the determination of limit of detection and limit of quantitation of the analytical methods, Chronicles Young Sci., 2011, 2, 21–25 CrossRef.
- N. A. Brunzel, Fundamentals of Urine and Body Fluid Analysis-E-Book. Elsevier Health Sciences, London, UK: 2016 Search PubMed.
- J. A. Simerville, W. C. Maxted and J. J. Pahira, Urinalysis: a comprehensive review, Am. Fam. Physician, 2005, 71(6), 1153–1162 Search PubMed.
- A. E. Kitabchi, R. Young, H. Sacks and L. Morris, Diabetic ketoacidosis: reappraisal of therapeutic approach, Annu. Rev. Med., 1979, 30(1), 339–357 CrossRef CAS PubMed.
- B. A. Julian, H. Suzuki, Y. Suzuki, Y. Tomino, G. Spasovski and J. Novak, Sources of urinary proteins and their analysis by urinary proteomics for the detection of biomarkers of disease, Proteonomics Clin. Appl., 2009, 3(9), 1029–1043 CrossRef CAS PubMed.
- E. A. Roberts, L. E. Morrison, L. J. Behman, R. Draganova-Tacheva, R. O'Neill and C. C. Solomides, Chromogenic immunohistochemical quadruplex provides accurate diagnostic differentiation of non-small cell lung cancer, Ann. Diagn. Pathol., 2020, 45, 151454 CrossRef PubMed.
- A. K. Yetisen, N. Jiang, A. Tamayol, G. U. Ruiz-Esparza, Y. S. Zhang, S. Medina-Pando, A. Gupta, J. S. Wolffsohn, H. Butt and A. Khademhosseini, based microfluidic system for tear electrolyte analysis, Lab Chip, 2017, 17(6), 1137–1148 RSC.
- N. Jiang, H. Butt, Y. Montelongo, F. Liu, S. Afewerki, G. L. Ying, Q. Dai, S. H. Yun and A. K. Yetisen, Laser Interference Lithography for the Nanofabrication of Stimuli-Responsive Bragg Stacks, Adv. Funct. Mater., 2018, 28(24), 1702715 CrossRef.
- L. Polavarapu, J. Pérez-Juste, Q.-H. Xu and L. M. Liz-Marzán, Optical sensing of biological, chemical and ionic species through aggregation of plasmonic nanoparticles, J. Mater. Chem. C, 2014, 2(36), 7460–7476 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ay01209f |
| This journal is © The Royal Society of Chemistry 2021 |