Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Amino acids as biomimetic crystallization agents for the synthesis of ZIF-8 particles†
Kang
Liang
*a,
Raffaele
Ricco
a,
Cara M.
Doherty
a,
Mark J.
Styles
a and
Paolo
Falcaro
*ab
aCSIRO Manufacturing Flagship, Private Bag 10, Clayton South, Victoria 3169, Australia. E-mail: Kang.Liang@csiro.au
bInstitute of Physical and Theoretical Chemistry, Graz University of Technology, Stremayrgasse 9, 8010 Graz, Austria. E-mail: paolo.falcaro@tugraz.at
First published on 16th February 2016
Abstract
Biomimetic mineralization of metal–organic frameworks (MOFs) exploits the use of biomolecules to control MOF crystallization processes. Here, we investigate 20 natural amino acids as biomimetic crystallization agents for the synthesis of ZIF-8 particles in aqueous solution. The morphology, size, and particle number of resultant MOF crystals were strongly dependent on the chemical nature of amino acid side chains that match closely the four classes of amino acids: nonpolar, polar neutral, polar negative and polar positive.
Metal–organic frameworks (MOFs) are an emerging class of porous materials comprised of inorganic and organic units linked together by strong bonds.1 Due to their flexible and modular synthetic approach achieved by simply combining different inorganic and organic building units, almost unlimited MOF structures can be prepared.2,3 This has made MOFs ideal candidates for many applications including gas storage and separation,4–6 catalysis,7,8 optics,9,10 microelectronics,11,12 sensing,13,14 decontamination,15,16 and biotechnology.17,18
Recently, a new research trend in MOFs has been focusing on the tuning of particle morphology and crystal size. Morphological control has proven to be one of the important factors for optimising the performance of MOFs.19 For instance, tuning both the size and polyhedral shape of MOF crystals affects the overall surface area and diffusion resistance,20 the self-assembly into highly ordered MOF-superstructures,21 as well as the efficacy and circulation time of MOF-based drug delivery carriers.22,23 To date, various strategies have been reported for controlling the MOF particle size and morphology: use of the coordination modulation strategy;24 addition of surfactants to various MOF systems has been shown to affect particle shape, size, and mesoporosity;25–30 controlled post synthetic chemical etching can lead to particles with diverse morphology;31,32 and inorganic nanoparticles have been shown to act as seeds to control the size of resulting MOFs.33 A strategy that remains largely unexplored is the use of biomolecules to control the morphology of MOF particles. Very recently, we introduced a biomimetic mineralization approach that utilises biomolecules as crystallization and directing agents for engineering a range of MOFs,34 showing interesting perspectives in biomolecule capturing, encapsulation, protection, controlled release, and even forensic science.34–36 The resultant MOF particles exhibited protein-dependent morphological features, with significant diversity when compared to standard MOF particles. However, only complex biomolecules such as proteins, enzymes and DNA as were used as biomimetic mineralization agents to control the morphology of the MOF crystals. In particular, particles with the shape of spheres, cubes, leaves, flowers and stars were obtained using Zeolitic imidazolate frameworks (ZIFs). ZIFs are scientific and technologically interesting MOFs due to their exceptional thermal and chemical stability.37,38 ZIFs are promising candidates for applications in gas storage,39 separation,40,41 catalysis,42,43 and sensing.44 Additionally, they could be used for biomedical applications because of the demonstrated negligible cytotoxicity.34,45
Here, for the first time we explore the use of the main protein building blocks, namely the 20 natural amino acids, as crystal and structural directing agents for ZIF-8. Distinctive crystal morphologies were obtained by adding different amino acids in an aqueous ZIF-8 precursor solution. Due to the simple and unique structure of the different amino acids, we were able to identify a correspondence between the ZIF-8 particle morphology and the standard classification amino acid used. This information will allow us to develop an understanding of the fundamental crystallization mechanisms of biomimetically mineralized MOFs.
Classically, amino acids are grouped into five categories based on the propensity of the side chain to be in contact with a polar solvent; hydrophobic (non-polar and non-polar aromatic), polar neutral, and polar charged (positive and negative). The five categories are proposed in Fig. 1a. Therefore we grouped the resulting ZIF-8 particles according to this amino acid classification method.
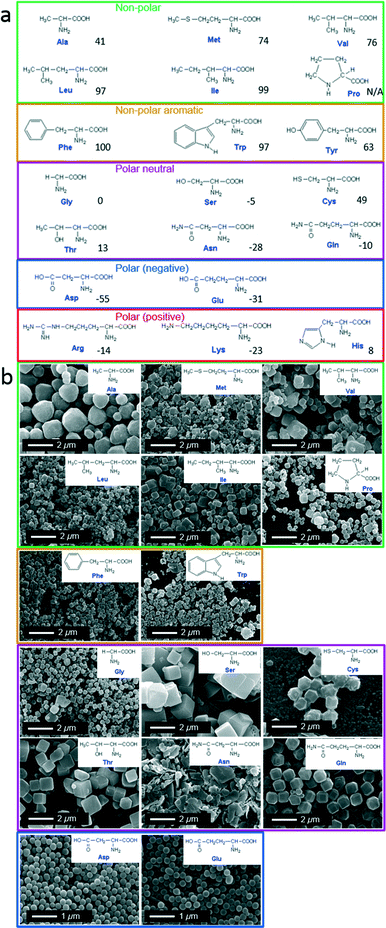 | ||
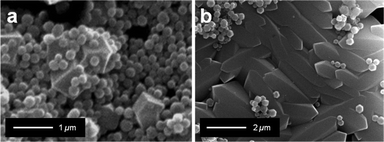
| Fig. 1 (a) Structure of 20 natural amino acids grouped according to nature of side chains: non-polar (green), non-polar aromatic (orange), polar neutral (purple), polar negative (blue), and polar positive (red). Hydrophobic value are given at pH 7.46 (b) SEM images of ZIF-8 particles prepared using different natural amino acids. | ||
In a typical experiment, each single amino acid was dissolved in an aqueous solution of 2-methylimidazole (HmIm) and then mixed with an aqueous solution of zinc acetate. After 10 min, the precipitates were washed and collected by centrifugation. In a control experiment, the mixing of aqueous solutions of HmIm and zinc acetate without the addition of amino acids, did not result in any observable particles (analysis performed using SEM). Powder X-ray diffraction (PXRD) measurements performed on washed samples obtained by adding amino acids showed diffraction patterns analogous to pure ZIF-8 (Fig. S1†). Scanning electron microscopy (SEM) revealed the morphological difference of ZIF-8 particles prepared with each different amino acid (Fig. 1b).
Out of the 20 amino acids, 4 amino acids (Lys, His, Arg, and Tyr) did not afford the formation of ZIF-8 particles. This was attributed to the highly positive charge on the Lys, His, and Arg, which could oppose Zn2+ ions thereby preventing ZIF-8 crystal growth. On the other hand, Tyr showed very low solubility in water, and likely could not participate in the crystallization process efficiently. The addition of all the other amino acids resulted in ZIF-8 particles with distinctive size and morphology. Increasing side chain hydrophobicity of non-polar amino acids resulted in a ZIF-8 morphological transition from spheroid to rhombic dodecahedron, then to truncated cubes. While, amino acids with aromatic side chains only induced the formation of truncated cubic ZIF-8 particles. In the case of polar neutral amino acids, side chains terminated with a hydroxyl group also induced ZIF-8 particles with a cubic morphology. Interestingly, negatively charged amino acids induced the formation of spherical ZIF-8 particles with a narrow size distribution.
The average particle size from each sample was calculated using ImageJ software on SEM images; the results are plotted in Fig. 2. In general, increasing the hydrophobicity of the non-polar amino acid side chain (Ala, Met, Val, Leu, Ile and Pro)46 decreased the ZIF-8 particle size from 1.25 μm to 0.5 μm. Amino acids with aromatic side chains produced relatively small ZIF-8 particles (ca. 300 nm). Although Gly produced the smallest ZIF-8 particles, amino acids with polar neutral side chains generally lead to a decreasing trend in the particle diameter with increasing side chain length. Moreover, the amino acids with negatively charged side chains (Asp and Glu) formed relatively small particles (ca. 200 nm) with narrower size distribution.
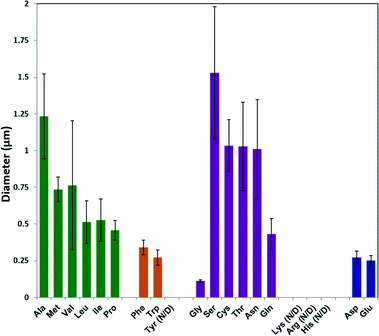 | ||
| Fig. 2 Diameters of ZIF-8 particles prepared using different natural amino acids, as determined by ImageJ from SEM images. | ||
To determine the total number of ZIF-8 particles produced using each different amino acid, NMR measurements were performed. In the ZIF-8 precursor solution, the decrease of the CH3 integral at 2.2–2.3 ppm of HmIm, as a result of ZIF-8 formation, was monitored against the unaffected CH3 signal at 1.7–1.8 ppm of the acetate ion (internal standard), and the amount of free HmIm in the precursor vs. time was calculated. As a result, the total weight of ZIF-8 particles was calculated by knowing these values (Table S1†). The number of particles was then determined by dividing the total weight by the average weight of the individual ZIF-8 particles calculated based on the diameters in Fig. 2 and under the approximation that the ZIF-8 particles are spherical (Methods section in ESI†). The estimated number of ZIF-8 particles produced per mL of precursor solution showed a trend opposite to the one observed for particle size: increasing hydrophobicity of the non-polar amino acid side chain resulted in an increase in the ZIF-8 particle number, as shown from Ala to Pro Fig. 3.46 Amino acids with aromatic side chains produced relatively high particle numbers. Gly produced the highest amount of particles, while the remaining polar neutral amino acids followed a similar trend. To confirm the number of ZIF-8 particles, we also performed elemental analysis on Zn2+ using the inductively coupled plasma (ICP) technique. The total amount of Zn2+ was determined by dissolving the ZIF-8 particles collected from each sample (Table S2†), and the calculated particle numbers were in good agreement with NMR results (Fig. S2†).
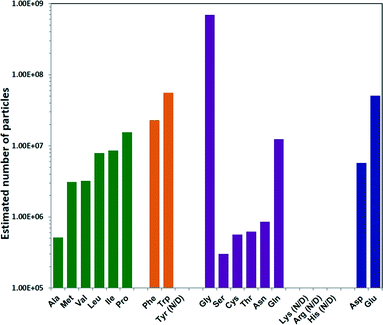 | ||
| Fig. 3 Estimated number of ZIF-8 particles per mL reaction mixture prepared using different natural amino. | ||
We further investigated the effects of mixed amino acids on the morphology of ZIF-8 particles. Equal molar amounts of Glu and Ser were mixed and used to induce the ZIF-8 growth. As pure Glu and Ser induced only spherical and cubic ZIF-8 particles respectively, the resulting ZIF-8 morphology of the mixed Glu and Ser showed a bimodal distribution of crystals with spherical and cubic morphology (Fig. 4a). Similarly, a mixture of Gly and Asn with the ZIF precursors induced the formation of crystalline particles with a bimodal distribution of ZIF-8 morphologies mimicking a mixture of ZIF-8 crystals prepared using Gly and Asn separately (Fig. 4b).
Conclusions
In summary, 20 natural amino acids were used as biomimetic crystallization agents for the controlled growth of ZIF-8 particles in aqueous solution. The resulting ZIF-8 particle size, morphology, and number were found to be strongly dependant on the nature of the amino acid side chains. Increasing the hydrophobicity of the non-polar amino acids led to ZIF-8 morphological transition from spheroid to rhombic dodecahedron to truncated cube, while reducing the particle size and increasing the particle number. Similar trends were observed for the polar neutral amino acids. Negatively charged amino acids produced small nearly monodisperse ZIF-8 spheres. Interestingly, the mixture of two different amino acids as a biomimetic mineralizing agent led to the formation of particles with a bimodal distribution, corresponding to the mixture of MOF crystals prepared using the two amino acids separately. The proposed results enable the precise tuning the morphological features of MOF particles. We believe that by combining these observations with simulation tools, the complicated biomimetic MOF crystallization mechanism could be revealed.Acknowledgements
K. L. acknowledges CSIRO OCE Science scheme and FST SPG grant. C. M. D. is supported by the Australian Research Council (ARC, DECRA Grant DE140101359). P. F. acknowledges the Australian Research Council (ARC, DECRA Grant DE120102451) and the AMTCP CSIRO scheme.Notes and references
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- H.-C. Zhou, J. R. Long and O. M. Yaghi, Chem. Rev., 2012, 112, 673–674 CrossRef CAS PubMed.
- H.-C. J. Zhou and S. Kitagawa, Chem. Soc. Rev., 2014, 43, 5415–5418 RSC.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- D. Banerjee, A. J. Cairns, J. Liu, R. K. Motkuri, S. K. Nune, C. A. Fernandez, R. Krishna, D. M. Strachan and P. K. Thallapally, Acc. Chem. Res., 2015, 48, 211–219 CrossRef CAS PubMed.
- M. R. Gonzalez, J. H. González-Estefan, H. A. Lara-García, P. Sánchez-Camacho, E. I. Basaldella, H. Pfeiffer and I. A. Ibarra, New J. Chem., 2015, 39, 2400–2403 RSC.
- L. Ma, J. M. Falkowski, C. Abney and W. Lin, Nat. Chem., 2010, 2, 838–846 CrossRef CAS PubMed.
- D. Farrusseng, S. Aguado and C. Pinel, Angew. Chem., Int. Ed., 2009, 48, 7502–7513 CrossRef CAS PubMed.
- Y. Cui, R. Song, J. Yu, M. Liu, Z. Wang, C. Wu, Y. Yang, Z. Wang, B. Chen and G. Qian, Adv. Mater., 2015, 27, 1420–1425 CrossRef CAS PubMed.
- H. S. Quah, W. Chen, M. K. Schreyer, H. Yang, M. W. Wong, W. Ji and J. J. Vittal, Nat. Commun., 2015, 6, 7954 CrossRef CAS PubMed.
- A. A. Talin, A. Centrone, A. C. Ford, M. E. Foster, V. Stavila, P. Haney, R. A. Kinney, V. Szalai, F. El Gabaly, H. P. Yoon, F. Léonard and M. D. Allendorf, Science, 2014, 343, 66–69 CrossRef CAS PubMed.
- I. Stassen, M. Styles, G. Grenci, H. Van Gorp, W. Vanderlinden, S. De Feyter, P. Falcaro, D. De Vos, P. Vereecken and R. Ameloot, Nat. Mater., 2015 DOI:10.1038/nmat4509.
- J. E. Mondloch, M. J. Katz, W. C. Isley, P. Ghosh, P. Liao, W. Bury, G. W. Wagner, M. G. Hall, J. B. DeCoste, G. W. Peterson, R. Q. Snurr, C. J. Cramer, J. T. Hupp and O. K. Farha, Nat. Mater., 2015, 14, 512–516 CrossRef CAS PubMed.
- P. Falcaro, F. Normandin, M. Takahashi, P. Scopece, H. Amenitsch, S. Costacurta, C. M. Doherty, J. S. Laird, M. D. H. Lay, F. Lisi, A. J. Hill and D. Buso, Adv. Mater., 2011, 23, 3901–3906 CrossRef CAS PubMed.
- E. Barea, C. Montoro and J. A. R. Navarro, Chem. Soc. Rev., 2014, 43, 5419–5430 RSC.
- C. M. Doherty, E. Knystautas, D. Buso, L. Villanova, K. Konstas, A. J. Hill, M. Takahashi and P. Falcaro, J. Mater. Chem., 2012, 22, 11470 RSC.
- S. Diring, D. O. Wang, C. Kim, M. Kondo, Y. Chen, S. Kitagawa, K. Kamei and S. Furukawa, Nat. Commun., 2013, 4, 2684 Search PubMed.
- C. M. Doherty, G. Grenci, R. Riccò, J. I. Mardel, J. Reboul, S. Furukawa, S. Kitagawa, A. J. Hill and P. Falcaro, Adv. Mater., 2013, 25, 4701–4705 CrossRef CAS PubMed.
- N. Stock and S. Biswas, Chem. Rev., 2012, 112, 933–969 CrossRef CAS PubMed.
- K. Li, D. H. Olson, J. Seidel, T. J. Emge, H. Gong, H. Zeng and J. Li, J. Am. Chem. Soc., 2009, 131, 10368–10369 CrossRef CAS PubMed.
- M. Sindoro, N. Yanai, A.-Y. Jee and S. Granick, Acc. Chem. Res., 2014, 47, 459–469 CrossRef CAS PubMed.
- R. C. Huxford, J. Della Rocca and W. Lin, Curr. Opin. Chem. Biol., 2010, 14, 262–268 CrossRef CAS PubMed.
- C.-Y. Sun, C. Qin, X.-L. Wang and Z.-M. Su, Expert Opin. Drug Delivery, 2013, 10, 89–101 CrossRef CAS PubMed.
- A. Umemura, S. Diring, S. Furukawa, H. Uehara, T. Tsuruoka and S. Kitagawa, J. Am. Chem. Soc., 2011, 133, 15506–15513 CrossRef CAS PubMed.
- S. K. Nune, P. K. Thallapally, A. Dohnalkova, C. Wang, J. Liu and G. J. Exarhos, Chem. Commun., 2010, 46, 4878–4880 RSC.
- X. Fan, W. Wang, W. Li, J. Zhou, B. Wang, J. Zheng and X. Li, ACS Appl. Mater. Interfaces, 2014, 6, 14994–14999 CAS.
- T. Xing, Y. Lou, Q. Bao and J. Chen, CrystEngComm, 2014, 16, 8994–9000 RSC.
- Y. Wu, M. Zhou, B. Zhang, B. Wu, J. Li, J. Qiao, X. Guan and F. Li, Nanoscale, 2014, 6, 1105–1112 RSC.
- Z. Zhang and M. J. Zaworotko, Chem. Soc. Rev., 2014, 43, 5444–5455 RSC.
- D. Bradshaw, S. El-Hankari and L. Lupica-Spagnolo, Chem. Soc. Rev., 2014, 43, 5431–5443 RSC.
- C. Avci, J. Ariñez-Soriano, A. Carné-Sánchez, V. Guillerm, C. Carbonell, I. Imaz and D. Maspoch, Angew. Chem., Int. Ed., 2015, 54, 14417–14421 CrossRef CAS PubMed.
- M. Hu, S. Furukawa, R. Ohtani, H. Sukegawa, Y. Nemoto, J. Reboul, S. Kitagawa and Y. Yamauchi, Angew. Chem., Int. Ed., 2012, 51, 984–988 CrossRef CAS PubMed.
- D. Buso, K. M. Nairn, M. Gimona, A. J. Hill and P. Falcaro, Chem. Mater., 2011, 23, 929–934 CrossRef CAS.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- K. Liang, C. J. Coghlan, S. G. Bell, C. Doonan and P. Falcaro, Chem. Commun., 2016, 52, 473–476 RSC.
- K. Liang, C. Carbonell, M. J. Styles, R. Ricco, J. Cui, J. J. Richardson, D. Maspoch, F. Caruso and P. Falcaro, Adv. Mater., 2015, 27, 7293–7298 CrossRef CAS PubMed.
- X.-C. Huang, Y.-Y. Lin, J.-P. Zhang and X.-M. Chen, Angew. Chem., Int. Ed., 2006, 45, 1557–1559 CrossRef CAS PubMed.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O'Keeffe and O. M. Yaghi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186–10191 CrossRef CAS PubMed.
- R. Banerjee, H. Furukawa, D. Britt, C. Knobler, M. O'Keeffe and O. M. Yaghi, J. Am. Chem. Soc., 2009, 131, 3875–3877 CrossRef CAS PubMed.
- S. R. Venna and M. A. Carreon, J. Am. Chem. Soc., 2010, 132, 76–78 CrossRef CAS PubMed.
- H. T. Kwon and H.-K. Jeong, J. Am. Chem. Soc., 2013, 135, 10763–10768 CrossRef CAS PubMed.
- U. P. N. Tran, K. K. A. Le and N. T. S. Phan, ACS Catal., 2011, 1, 120–127 CrossRef CAS.
- F. Zhang, Y. Wei, X. Wu, H. Jiang, W. Wang and H. Li, J. Am. Chem. Soc., 2014, 136, 13963–13966 CrossRef CAS PubMed.
- G. Lu and J. T. Hupp, J. Am. Chem. Soc., 2010, 132, 7832–7833 CrossRef CAS PubMed.
- J. Zhuang, C.-H. Kuo, L.-Y. Chou, D.-Y. Liu, E. Weerapana and C.-K. Tsung, ACS Nano, 2014, 8, 2812–2819 CrossRef CAS PubMed.
- O. D. Monera, T. J. Sereda, N. E. Zhou, C. M. Kay and R. S. Hodges, J. Pept. Sci., 1995, 1, 319–329 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detained synthesis and characterization of ZIF-8 crystals using amino acids. PXRD pattern. Calculation of particle number from NMR and ICP. See DOI: 10.1039/c5ce02549d |
| This journal is © The Royal Society of Chemistry 2016 |