E. coli surface display of streptavidin for directed evolution of an allylic deallylase†
Abstract
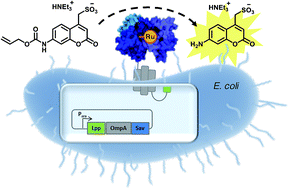
Artificial metalloenzymes (ArMs hereafter) combine attractive features of both homogeneous catalysts and enzymes and offer the potential to implement new-to-nature reactions in living organisms. Herein we present an E. coli surface display platform for streptavidin (Sav hereafter) relying on an Lpp-OmpA anchor. The system was used for the high throughput screening of a bioorthogonal CpRu-based artificial deallylase (ADAse) that uncages an allylcarbamate-protected aminocoumarin 1. Two rounds of directed evolution afforded the double mutant S112M–K121A that displayed a 36-fold increase in surface activity vs. cellular background and a 5.7-fold increased in vitro activity compared to the wild type enzyme. The crystal structure of the best ADAse reveals the importance of mutation S112M to stabilize the cofactor conformation inside the protein.


 Please wait while we load your content...
Please wait while we load your content...