Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Accelerating materials research with a comprehensive data management tool: a case study on an electrochemical laboratory†
Nico C.
Röttcher
 *ab,
Gun D.
Akkoc
*ab,
Gun D.
Akkoc
 ab,
Selina
Finger
ab,
Selina
Finger
 ab,
Birk
Fritsch
ab,
Birk
Fritsch
 a,
Jonas
Möller
a,
Jonas
Möller
 a,
Karl J. J.
Mayrhofer
a,
Karl J. J.
Mayrhofer
 ab and
Dominik
Dworschak
ab and
Dominik
Dworschak
 *a
*a
aForschungszentrum Jülich GmbH, Helmholtz Institute for Renewable Energy Erlangen-Nürnberg, Cauerstr. 1, 91058 Erlangen, Germany. E-mail: n.roettcher@fz-juelich.de; d.dworschak@fz-juelich.de
bDepartment of Chemical and Biological Engineering, Friedrich-Alexander-Universität Erlangen-Nürnberg, Cauerstr. 1, 91058 Erlangen, Germany
First published on 11th January 2024
Abstract
The pressing need for improved energy materials calls for an acceleration of research to expedite their commercial application for the energy transition. To explore the vast amount of material candidates, developing high-throughput setups as well as enhancing knowledge production by synthesis of published data is essential. Therefore, managing data in a clearly defined structure in compliance with the FAIR principles is imperative. However, current data workflows from laboratory to publication often imply poorly structured data, limiting acceleration in materials research. Here, we demonstrate a comprehensive data management tool structuring data throughout its life-cycle from acquisition, analysis, and visualization to publication by means of an SQL database. This guarantees a persistent relation between experimental data, corresponding metadata, and analysis parameters. Manual interaction required to handle data is reduced to a minimum by integrating data acquisition devices (LabVIEW), defining script-based data analysis and curation routines, as well as procedurally uploading data upon publication (Python). Keeping the link, published data can be traced back to underlying experimental raw and metadata. While we highlight our developments for operando coupled electrochemical experiments, the used approach is generally applicable for other fields given its inherent customizability. Design of such automated data workflows is essential to develop efficient high-throughput setups. Further, they pave the way for self-driving labs and facilitate the applicability of machine learning methods.
1 Introduction
The challenges of the energy transition require a significant speed-up in research on energy materials. To pave the way towards accelerated material discovery and optimization, currently novel systems are being developed ranging from high-throughput setups with human-in-the-loop1–3 to fully self-driving laboratories.4–6 Moreover, the rise of machine learning (ML) techniques supports this development by augmenting researchers and guiding experiments,7,8 optimizing processes,9 and even designing novel materials.10,11 As these methods heavily rely on the availability and quality of data, collaboration throughout the scientific community will further enhance these developments.12,13However, navigating in the haystack of previously published data suffers from the limitations of text-based search. Despite advances in scraping and tabulating data from the literature by using Natural Language Processing (NLP) techniques14–17 in tandem with data extraction techniques from figures, the quality of the obtained data is still limited due to incompleteness and lack of structure of published data.18 Moreover, due to societal bias, publication of unsuccessful attempts is often neglected, despite being essential not only for modeling but also to prevent other researchers from conducting redundant experiments.19–21 By structuring research data, the application of ML techniques can benefit twofold from simpler inclusion of unsuccessful experiments in a tabulated format as well as from more reliable extraction of input data.20 Thus, it is clear that publishing all data in a tabulated and structured format is one of the most pressing improvements in the publishing of scientific data.22
To overcome the lack of data quality, the FAIR principles were defined – a guideline for researchers on how to manage and share data to render it Findable, Accessible, Interoperable, as well as Reusable and in this way improve the quality of big data approaches.22 In specific, they highlight the importance of linking experimental data with enriched metadata, any data treatment and openly sharing original data parallel to publications. To fulfill these requirements, in many scientific fields structured databases have been developed such as ISA in the field of life science,23 Open Phacts in pharmaceutical research,24 and NOMAD for (computational) materials research.25 For experimental materials research in general, and electrochemistry in specific, the design of a common database is challenging given the vast diversity of experimental approaches. On this way, there are advances in defining a standardized ontology for battery research which sets the ground for sharing research data across different groups.26,27 On a more specialized scale, echemdb provides a database solution for cyclic voltammetry (CV) data.28 Beyond uploading data into databases, numerous data repository services have been established. For electrochemical research, however, there are only a few experimental publications taking steps toward publishing raw data and detailed descriptions of processing procedures.29–31 FAIR-compliant publishing of electrochemical data is far from being standard despite being essential to compare and reproduce experimental results.32,33 Insufficient time has been identified as a key barrier for researchers to share their data inter alia due to data preparation and structuring only on short notice before publication.34 In contrast, managing research data from the initial acquisition up to its publication as outlined in the FAIR principle in a structured way is key to enable open data sharing in a time-efficient manner. However, a comprehensive data workflow is still missing for materials science in general and electrochemistry in particular.35
Instead, conventional data management often relies on dynamically growing folder structures with customized file naming schemes for data and metadata files as well as handwritten notes. To keep files organized and links between metadata and data comprehensible, strategies for the proper naming of files and folders have been suggested.36,37 However, as ‘relevant’ parameters are different for researchers even in the same research group and are suspected to change in the course of a study of single researchers, different naming conventions will evolve. On top, the processing of data is often performed using click-based graphical user interfaces (GUIs), lacking scaleability and tracking of data lineage.
While this approach is flexible and easy to use at the beginning, the effort to keep information organized in multiple files without inherently linking experimental data, metadata, and processed data grows steadily in the course of an experimental study. Furthermore, considering the reuse of data by other (future) researchers within the same group and of any published data by the whole community, it is clear that understanding and tracking data lineage within a file-folder structure is a time-consuming task. However, if not undertaken, information that could be extracted from the data or by correlating to other data, is lost – an issue that has to be prevented, especially regarding the application of advanced data science methods.
Thus, a comprehensive data management strategy is key to ensure the quality of data and avoid time-consuming data (re)structuring at a later stage of an experimental study. To support researchers on the way toward comprehensively structured data, a wide range of electronic lab notebooks (ELNs) have been developed.25,38,39 ELNs assist in structuring data by creating templates for experiments with metadata being filled in pre-defined fields and data files being attached to the entry. A database in the backend manages the links between experimental data, metadata, and entries of physical items. Yet, these approaches do not a priori provide a data processing platform, require substantial user interaction, and are limited in customizability. Alternatively, the concept of laboratory-scale individualized databases is suggested.40–42
Based on this, here, we showcase a comprehensive workflow integrating data acquisition devices, analysis, visualization, and FAIR-compliant publishing of data already applied successfully in recent work.43 This tool is built on off-the-shelf software packages for data storage (SQL database) and processing data (Python44 and its diverse data analysis libraries45–49). Thus, this system offers high customizability and simplicity in development considering the maturity of the used software and support by active communities. While the results are widely applicable for research data management, we highlight examples from the field of electrochemistry related to energy materials.
2 Results and discussion
A common workflow of a researcher dealing with experimental data can be divided into three main steps, namely (i) the data acquisition, (ii) processing and analysis, (iii) its visualization and interpretation (see Fig. 1). Each of these steps involves accessing and generating data. To establish complete data lineage tracking, keeping reliable links from published data down to its raw (meta)data, all data is stored and organized in a central database. This database stores information from physical inventory items used during experiment acquisition, produced experimental data and its metadata, and processed data generated during process and analysis procedures.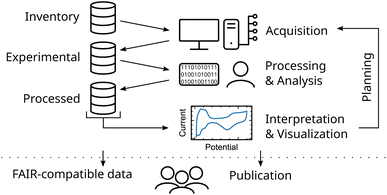 | ||
| Fig. 1 Illustration of a comprehensive research data workflow integrating acquisition, analysis, visualization and publication. | ||
Interpretation and visualization of this data produce new information and, therefore new data. Subsequently, new experiments can be planned by the researchers. Additionally, with structured data at hand, ML methods can be readily utilized to explore and exploit the experimental space. Furthermore, structured data is seamlessly added when publishing the experimental findings in compliance with the FAIR principles. This increases the trustworthiness of the interpretation and enables reusability of the data. Finally, structuring data already from the beginning of an experimental study is key to enhance scientific knowledge production.
2.1 Structure research data
In its essence, the structuring of data in a digital format is an abstraction of the physical level where the actual data is stored on the disk and the representation of the data for the user.50 To retrieve data it is not required to know where the data is physically stored but data is rather selected by any information known by the user. This is in contrast to an analog or similarly file-based storage where the hierarchy of folders follows a specific pattern of a few relevant parameters. For instance, considering the selection of an experiment, these relevant parameters can be the name of the experimenter, the date it was recorded, and the sample used. In a database, however, experiments can be selected by any metadata parameter stored with the experiment such as sample preparation parameters, its elemental composition, or the sample history.To achieve this abstraction, different database management systems are available. The most mature and applied solutions are relational databases with developments dating back to the 1970s.50,51 Relational databases organize data in a schema – a rigid network of tables with specified relations between their columns. Beyond that, there are more modern post-relational databases such as key-value, column, document-oriented, and graph databases which are more flexible in handling less structured data, making more versatile connections between data, and are easier to maintain upon changes of the data structures.52 However, in the scope of designing data workflows for experimental setups with specified parameters for metadata and result data, a relational database is an appropriate choice. Considering researchers being familiar with data in tabulated form, the design process of a relational database schema is intuitive and supports a clear definition of a complete metadata set for the experimental setups. Furthermore, in light of the widespread application of relational databases, there is a large community further supporting the design process by the vast availability of solutions for common problems. Nevertheless, as experimental setups and workflows might develop with time, revision of the database schema is required. This indicates the importance of the initial design process already considering possible developments.
Besides that, considering a wide spectrum of knowledge on programming languages in materials research-related degree programs, the interaction with a relational database – selecting, inserting, and updating entries – is intuitively integrated in a researcher's workflow given its simple but powerful structured query language (SQL). Furthermore, this interaction can be constrained, i.e., the change of raw data can be restricted, ensuring the integrity of the original measurement data. Finally, active learning techniques with common surrogate functions such as Gaussian Process Regression and Random Forest require tabulated data, thus, giving an intrinsic compatibility with relational databases.
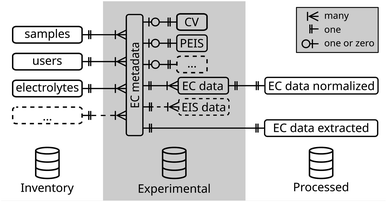
For the simple case of an electrochemical experiment, a database schema is exemplarily outlined in Fig. 2. This structure is divided into three categories (i) the inventory of physical objects used in the experiment, (ii) the experimental (meta)data itself, and (iii) processing data created during the analysis of the raw experimental data. A complete database schema which is used within our group is added as ESI file† and further described in the ESI in Section 1.†
For each experiment, a new entry in the ec metadata-table is created and linked via an identifier to the samples-table and, thus to its properties. The relation is defined as one-to-many relation, meaning an experiment can only include one sample, while many experiments can be conducted on one sample. Similarly, the user, the used electrolyte, and other metadata parameters are linked.
Often in electrochemistry, multiple techniques are run sequentially in a batch. Recording of metadata, thus, can be defined in a per-batch or per-technique manner. To enable tracking of changes of metadata parameters in between techniques of the same batch, such as the change of a gas purging through the electrolyte or the rotation rate in a rotating disk electrode (RDE) experiment, metadata is recorded on a single technique level. To identify techniques of the same batch, a batch identifier is added to the ec metadata-table.
Metadata parameters specific to the electrochemical technique, such as the scan rate of a CV experiment or the frequency range in an electrochemical impedance spectroscopy (EIS) experiment are stored in separate tables linked to the main metadata table. This schema is extended in the same way to cover a complete metadata set so that all parameters required to reproduce the experiment can be stored in one place. This includes also environmental parameters such as room temperature or relative humidity. Finally, the metadata is distributed over multiple connected tables. By definition of a database view, all metadata can be joined into a single table for each kind of experiment, which decreases complexity when selecting experiments, while keeping quality constraints by the definition of multiple tables.
For the acquired data further tables are defined. Data of the same kind is stored in a single table with an experiment identifier linked to its metadata, in contrast to a file-based system where each experiment produces a new file and a link is created by its name and folder path. In the case of electrochemical data, due to its different domains (time and frequency), a table for direct current (CV, CA, etc.) and another for alternating current experiments (EIS) is defined.
Besides the experimental data, also tables for processed data can be defined. In these, the results of an analysis, such as the extracted electrochemical active surface area (ECSA) from a CV experiment, are stored. Additionally, any parameters introduced during the analysis such as the integration limits are included. Linking processed data tables to the originating experiment ensures data lineage tracking.
Thus, in contrast to a file-based approach, a direct link among metadata, experimental data, and processed data is ensured by design. By that, structuring data in a relational database fulfills more easily the FAIR principles in comparison to a folder-file structure. In addition, organization of the data in a relational database enables simple finding and selecting of specific experiments by its metadata using SQL queries.
2.2 Experiment – database communication
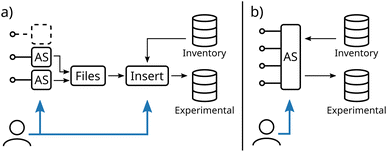
After the preparation of the database schema, the insertion of experimental data into the database can be established. Fig. 3 illustrates two different modes of communication between acquisition software (AS) and the database.In conventional workflows, proprietary acquisition software as delivered by the manufacturer is used to control the experiment. Experimental data and inherent metadata such as the electrochemical protocol are exported into files (see Fig. 3a). To establish the link between the experiment and additional metadata like the used electrodes or electrolyte, a second user interaction is required during the insertion of the file into the database. Similarly, in the case of ELNs, the measurement files are linked to a separately created entry. In addition, in the case of experiments with multiple devices usually multiple software has to be handled, as an integration of other devices is in most cases not possible. Consequently, multiple files with (meta)data have to be inserted increasing the complexity of this workflow.
In contrast, as depicted in Fig. 3b, customizing a single acquisition to communicate with multiple devices and directly with the database offers multiple advantages. (i) (Meta)data acquired by all devices are inherently linked, which simplifies the correlation of their results. (ii) A second step to link additional metadata to existing database inventory tables is not necessary as it can be set directly in the acquisition software before performing the experiment (see Fig. S1†). (iii) No user interaction is required to name, link, and transfer data, enabling remote operation of the experimental setup and, thus, achieving one of the main prerequisites of self-driven laboratories. As these customizations are not available within commercial potentiostat software, this approach relies on using a potentiostat with a programmable interface controlled by a custom-built acquisition software, based, for instance, on LabView or Python.53,54
2.3 Processing & analysis
The next step in the workflow is the processing and analysis of data. This process can be subdivided into (i) the selection of data and its corresponding metadata, (ii) performing the analysis, (iii) ensuring the quality of the analysis, and (iv) storing the results.By using a relational database, the selection of data can be linked to conditions an experiment has to fulfill to be suitable for an analysis routine. For instance, for the derivation of the ECSA, the selection of experiments can be restricted to the type CV and Pt-containing working electrodes. The clearly defined data structure simplifies sharing and standardization of analysis routines across users.
In performing the analysis, some research groups still rely on GUI-based data analysis software such as Origin or Excel. This is problematic in different regards:
• These software store a redundant copy of the raw data and thus the link to the original file can be lost.
• Analysis steps are not recorded, for instance, the ambiguous adjustment of analysis parameters such as potential boundaries for an integration of the current to derive the ECSA-specific charge or manual exclusion of data points to fit a Nyquist plot with an equivalent circuit.
• Required human interaction scales linearly with the amount of data to be analyzed because of unnecessary repetitions of the same steps over again.
In contrast, when performing script-based data analysis, the link between analysis parameters, analyzed data, and raw data can be consistently established by integrating the linking into the scripted routine. Thus, data lineage and the reproducibility of the analysis procedure are guaranteed. By sharing the processing scripts among researchers, the time to develop those scripts is reduced and discussion on the quality of the data analysis procedure can be rooted.
For such data analysis procedures, the open-source programming language Python is feasible, due to its active community46 and range of libraries based on efficient array programming47 that facilitates ready-to-use scientific data analysis procedures.46 Although most of the cutting-edge ML libraries such as XGBoost, TensorFlow, and Torch are coded in low level, hence fast programming languages (i.e., C++, Fortran), the Python wrappers are also actively maintained, usually by the official developers.55–58 While providing the extensive possibilities of a fully-featured programming language, Python is a good choice also for researchers with little programming experience thanks to its ease of use and community-backed comprehensive documentation and guides.
Nonetheless, a strict definition of an analysis procedure can fail for certain experiments depending on its complexity. For example, the unintended inclusion of outlying data points into a fitting procedure or the incorrect choice of an equivalent circuit model for impedance spectroscopy. User blindness can propagate such errors leading to wrong conclusions in subsequent interpretations. Therefore, a quality control step is essential for every analysis procedure. For a low amount of data, this can be a manual step, like visually verifying the suitability of a model to fit the raw data. For larger amounts of data and simple processing steps, specific quality parameters can be defined, and experiments discarded from further analysis if these don't match. For instance, the goodness of fit might be evaluated by the R2adj value. However, this value cannot differentiate between a statistical error (noisy signal) and a systematic error (wrong model), thus, care must be taken in defining these quality indicators. Alternatively, data quality can also be evaluated based on ML models comparing results with existing experiments in the local, relational database or from data repositories.59,60
Result data of the analysis as well as input parameters are stored in pre-defined tables in the database. To ensure data lineage tracking, these tables must (i) contain columns for any input parameter of the analysis, (ii) be linked to the original experiment (iii) as well as to the performed analysis script.61 Especially, if different versions of the analysis routine are developed, a column for the version of the applied script should be added.
To illustrate the advantages of script-based data analysis procedures, examples are outlined in Fig. 4. Therefore, we classify data analysis procedure into normalization, extraction, and correlation of data.
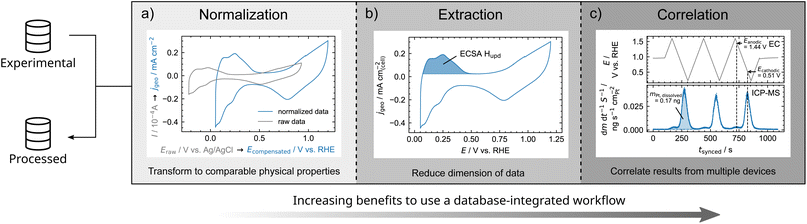 | ||
| Fig. 4 Classification of data analysis procedures with increasing benefits from normalization to extraction and correlation of data when implemented in a database-integrated workflow. This classification is illustrated by electrochemical experiments on polycrystalline Pt. (a) Normalization of current response of a CV by the geometric surface area of the working electrode as well as referencing electrode potential to the reversible hydrogen electrode (RHE) system. (b) Derivation of the ECSA from CV via the Hupd charge. (c) Correlation of time-resolved electrochemical potential and dissolution of Pt as determined by ICP-MS.62 Experimental details can be found in the ESI in Section 3.1.† | ||
In our labs, for instance, an electrochemical scanning flow cell (SFC) can be coupled to an ICP-MS. Electrochemical and mass spectrometric experiments are correlated by their timestamps and corrected for a delay time required to transport dissolved species to the downstream mass spectrometer. As dissolution is depending on the absolute surface area, the mass flow rate of dissolved species is normalized by the ECSA extracted from CV (see Fig. 4b). By this, dissolution peak onset, maximum, and shape can be correlated to the electrode potential. For instance, the electrochemical potential of peak dissolution during CV can be derived (see Fig. 4b and S3†). Thus, the electrochemical stability of energy-related materials can be examined in depth within minutes. Once the data workflow is established, also the analysis procedure of the data is performed within minutes and can be reused for any experimental study. This workflow was successfully applied to stability studies of bipolar plate materials for proton-exchange membrane water electrolyzers.43
2.4 Visualization of research data
To robustly interpret and clearly communicate experimental data, its visualization is of utmost importance.77 As for the developing of analysis procedures, also the visualization of data is benefiting from selecting experiments by specifying any combination of metadata parameters by means of an SQL query to the database. From the metadata table, corresponding experimental raw data can be retrieved. Next, any required analysis procedure can be performed. By outsourcing this into external functions, code is kept clean, and the usage of the same analysis procedure for several visualization routines is ensured, while tracking of data lineage is guaranteed. The actual visualization step is performed in a single command looping through all experiments selected in the first step. To plot different experiments, only the SQL query has to be adjusted regardless of the number of queried experiments. By this, the comparison of any experiments is fast and flexible. A detailed description of this procedure and an example of the underlying code can be found in the ESI in Section 4.†Having all metadata available, any style elements of the displayed graph such as legend labels, colors, line- or marker styles can be defined by metadata values. Thus, creating a color code for multiple graphs throughout a publication based on e.g. the material is simplified.
Furthermore, having the metadata of all experiments in a tabulated form, experiments can be thoroughly compared and in that way any (unintended) differences in the experimental procedure retraced. For electrochemical experiments, for instance, the electrochemical history of the sample can be compared. With such a comparison, the accordance of experimental parameters between experiments is quickly verified. Thus, the reliability of the data and their interpretation can be improved.
2.5 Publication of research data
Considering the publication of the visualized data, underlying data has to be prepared to be comprehensible for other researchers. As long as there are no suitable online databases available for the specific experiments performed to which data can be streamlined, researchers have to fall back on uploading data into data repositories in an accessible file format like comma-separated values. Still, with the clearly defined data structure at hand, this export functionality can routinely be performed adding no additional effort during the publication process. Considering the importance of the sample history, based on the experiments and thus samples selected, during export data of any additional experiments previously performed on the same sample can be simply included. Adding the analysis and visualization procedures to the data repository opens the possibility of retracing data lineage.To overcome the low comprehensibility of customized data structures uploaded to data repositories, there are services such as Binder which opens up the possibility to interactively explore figures, underlying data, and analysis routines of a published work.78 This service enables the execution of Python scripts by hosting Jupyter Notebooks uploaded to a Zenodo data repository. Therefore, it is seamlessly combined with the data workflow presented here. To showcase its applicability, data visualized in Fig. 4 is made available online, including raw data, experimental metadata, analyzed data and any analysis tools, routines to visualize the data as well as the underlying Python scripts.79,80 By this, the data lineage from acquisition up to publication can be interactively explored without the need of additional software installation. While this approach is convenient to showcase the development of the data management tool presented here, it is advantageous to enable FAIR access to research data, in general.
2.6 Case-study of implementing the system in our research group
The specific setup in our group is built around a centrally hosted MySQL database. Insertion of electrochemical data is performed directly via a home-built LabView Software (see Fig. 3b),53 while ICP-MS data is outputted by the manufacturer's software and inserted via a Python routine post-measurement (see Fig. 3a). The standard analysis routine includes normalization of current and potential data based on experimental metadata, similarly, normalization as well as calibration of dissolution data, smoothing of internal standard in the ICP-MS data, adjustment of the delay time between electrochemical and ICP-MS experiments, and integration of dissolution peaks. The routines are fully automated (normalization), require a visual control (calibration, noise reduction), or include GUI-aided manually adjustable parameters (delay time, integration). All routines are based on home-built Python scripts and are distributed and executed within Jupyter Notebooks centrally hosted on-site.44–46,48,49This enables cross-platform access via browser, outsourcing of data-intensive calculation to the server, and automatic backup of data and scripts. Technical limitations such as storage ease, capacity and processing speed are considered. Additionally, maintenance of Python libraries is handled centrally hence avoiding compatibility issues. This enables sharing and standardization of data analysis routines and visualization templates which in our experience drastically reduces barriers for researchers with little or no expertise in programming. Building our tool based on MySQL and Python/Jupyter relies on the software being open-source, free to use, and having a large community with huge support opportunities also by using modern Large Language Models. Thus, also research groups with low computer science capacities are able to implement such a system for their experiments. For further guidance an overview of our server infrastructure is illustrated in Fig. S6.†
3 Conclusions
In summary, we have demonstrated a comprehensive data workflow interconnecting each data handling step, namely, the acquisition, analysis, visualization, and publication of data with a central relational database. Key improvements for every step to significantly reduce manual labor and ensure data quality are discussed. These include (i) directly linking rich metadata to result data during acquisition, (ii) comprehensible and shareable analysis procedures based on scripted routines including predefined quality control criteria, (iii) storing analysis parameters and result data in the database linked to originating experiments enabling data lineage tracking, (iv) straightforward and flexible visual comparison of experimental data to communicate results, (v) effortless uploading of experimental data to repositories in compliance with FAIR principles enabling reuse of the data by the research community.While we have highlighted the applicability in the field of electrochemistry, especially correlating other techniques to electrochemical experiments, this approach is generally applicable to other fields. Especially in light of limited expertise on IT infrastructure at research institutes, in our experience the presented workflow offers a system simple to implement. At the same time, it remains flexible to be customized for specific needs. Once implemented, also for users with little programming expertise, it is easy to adapt their data management to profit from a comprehensive workflow.
Finally, such a data management tool is, on the one hand, a key element to enable automation in materials research laboratories and the building of high-throughput experimental setups enabling the applicability of ML methods on a laboratory scale. On the other hand, increasing the amount and quality of openly published data will enhance big data analysis on an inter-laboratory scale.
Data availability
The data, showcasing the applicability of the research data management tool, is made available online, here https://zenodo.org/records/10417756. In specific, this is data visualized in Fig. 4 including experimental raw data, metadata, and analyzed data, as well as any analysis tools, routines to visualize the data, and the underlying database schema as MySQL file. The Jupyter Notebooks can be interactively executed with Binder, here https://mybinder.org/v2/zenodo/10.5281/zenodo.10417756/.Author contributions
Nico C. Röttcher: conceptualization: lead; data curation, formal analysis, investigation for SFC-ICPMS; methodology: lead; software: lead; writing – original draft: lead. Gun D. Akkoç: conceptualization: supporting; methodology: supporting; software: supporting; writing – review & editing: supporting. Selina Finger: data curation, formal analysis, investigation for eis writing – review & editing: supporting. Birk Fritsch: formal analysis for SFC-ICPMS peak extraction; software for EIS and SFC-ICPMS peak extraction; writing – review & editing: supporting. Jonas Möller: software for potentiostat – database communication writing – review & editing: supporting. Karl J. J. Mayrhofer: funding acquisition: lead; resources: lead; supervision: supporting; writing – review & editing: supporting. Dominik Dworschak: conceptualization: supporting; methodology: supporting; project administration: lead; supervision: lead; writing – review & editing: supporting.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Nico C. Röttcher, Gun D. Akkoc and Dominik Dworschak gratefully acknowledge the German Ministry of Education and Research (BMBF) for financial support within the project 03HY108A. Selina Finger and Birk Fritsch thank the German Ministry of Education and Research (BMBF) for financial support within the project 03HY103H.Notes and references
- N. Fujinuma and S. E. Lofland, Adv. Intell. Syst., 2023, 5, 2200290 CrossRef.
- K. J. Jenewein, G. D. Akkoc, A. Kormányos and S. Cherevko, Chem Catal., 2022, 2, 2778–2794 CrossRef CAS.
- C. Xiang, S. K. Suram, J. A. Haber, D. W. Guevarra, E. Soedarmadji, J. Jin and J. M. Gregoire, ACS Comb. Sci., 2014, 16, 47–52 CrossRef CAS PubMed.
- R. W. Epps, A. A. Volk, M. Y. S. Ibrahim and M. Abolhasani, Chem, 2021, 7, 2541–2545 CAS.
- S. Langner, F. Häse, J. D. Perea, T. Stubhan, J. Hauch, L. M. Roch, T. Heumueller, A. Aspuru-Guzik and C. J. Brabec, Adv. Mater., 2020, 32, 1907801 CrossRef CAS PubMed.
- B. P. MacLeod, F. G. L. Parlane, C. C. Rupnow, K. E. Dettelbach, M. S. Elliott, T. D. Morrissey, T. H. Haley, O. Proskurin, M. B. Rooney, N. Taherimakhsousi, D. J. Dvorak, H. N. Chiu, C. E. B. Waizenegger, K. Ocean, M. Mokhtari and C. P. Berlinguette, Nat. Commun., 2022, 13, 995 CrossRef CAS PubMed.
- B. Rohr, H. S. Stein, D. Guevarra, Y. Wang, J. A. Haber, M. Aykol, S. K. Suram and J. M. Gregoire, Chem. Sci., 2020, 11, 2696–2706 RSC.
- M. Kim, Y. Kim, M. Y. Ha, E. Shin, S. J. Kwak, M. Park, I.-D. Kim, W.-B. Jung, W. B. Lee, Y. Kim and H.-T. Jung, Adv. Mater., 2023, 35, 2211497 CrossRef CAS PubMed.
- C. C. Rupnow, B. P. MacLeod, M. Mokhtari, K. Ocean, K. E. Dettelbach, D. Lin, F. G. Parlane, H. N. Chiu, M. B. Rooney, C. E. Waizenegger, E. I. De Hoog, A. Soni and C. P. Berlinguette, Cell Rep. Phys. Sci., 2023, 4, 101411 CrossRef CAS.
- J. Noh, J. Kim, H. S. Stein, B. Sanchez-Lengeling, J. M. Gregoire, A. Aspuru-Guzik and Y. Jung, Matter, 2019, 1, 1370–1384 CrossRef.
- E. Kim, Z. Jensen, A. van Grootel, K. Huang, M. Staib, S. Mysore, H.-S. Chang, E. Strubell, A. McCallum, S. Jegelka and E. Olivetti, J. Chem. Inf. Model., 2020, 60, 1194–1201 CrossRef CAS PubMed.
- M. Vogler, J. Busk, H. Hajiyani, P. B. Jørgensen, N. Safaei, I. E. Castelli, F. F. Ramirez, J. Carlsson, G. Pizzi, S. Clark, F. Hanke, A. Bhowmik and H. S. Stein, Matter, 2023, 6, 2647–2665 CrossRef.
- D. Guevarra, K. Kan, Y. Lai, R. J. R. Jones, L. Zhou, P. Donnelly, M. Richter, H. S. Stein and J. M. Gregoire, Digital Discovery, 2023, 1806–1812 RSC.
- M. C. Swain and J. M. Cole, J. Chem. Inf. Model., 2016, 56, 1894–1904 CrossRef CAS PubMed.
- Z. Jensen, E. Kim, S. Kwon, T. Z. H. Gani, Y. Román-Leshkov, M. Moliner, A. Corma and E. Olivetti, ACS Cent. Sci., 2019, 5, 892–899 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Chem. Mater., 2017, 29, 9436–9444 CrossRef CAS.
- R. Ding, Y. Ding, H. Zhang, R. Wang, Z. Xu, Y. Liu, W. Yin, J. Wang, J. Li and J. Liu, J. Mater. Chem. A, 2021, 9, 6841–6850 RSC.
- R. Ding, X. Wang, A. Tan, J. Li and J. Liu, ACS Catal., 2023, 13267–13281 CrossRef CAS.
- X. Jia, A. Lynch, Y. Huang, M. Danielson, I. Lang’at, A. Milder, A. E. Ruby, H. Wang, S. A. Friedler, A. J. Norquist and J. Schrier, Nature, 2019, 573, 251–255 CrossRef CAS PubMed.
- P. Raccuglia, K. C. Elbert, P. D. F. Adler, C. Falk, M. B. Wenny, A. Mollo, M. Zeller, S. A. Friedler, J. Schrier and A. J. Norquist, Nature, 2016, 533, 73–76 CrossRef CAS PubMed.
- G. H. Gu, J. Noh, I. Kim and Y. Jung, J. Mater. Chem. A, 2019, 7, 17096–17117 RSC.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 160018 CrossRef.
- S.-A. Sansone, P. Rocca-Serra, D. Field, E. Maguire, C. Taylor, O. Hofmann, H. Fang, S. Neumann, W. Tong, L. Amaral-Zettler, K. Begley, T. Booth, L. Bougueleret, G. Burns, B. Chapman, T. Clark, L.-A. Coleman, J. Copeland, S. Das, A. de Daruvar, P. de Matos, I. Dix, S. Edmunds, C. T. Evelo, M. J. Forster, P. Gaudet, J. Gilbert, C. Goble, J. L. Griffin, D. Jacob, J. Kleinjans, L. Harland, K. Haug, H. Hermjakob, S. J. H. Sui, A. Laederach, S. Liang, S. Marshall, A. McGrath, E. Merrill, D. Reilly, M. Roux, C. E. Shamu, C. A. Shang, C. Steinbeck, A. Trefethen, B. Williams-Jones, K. Wolstencroft, I. Xenarios and W. Hide, Nat. Genet., 2012, 44, 121–126 CrossRef CAS PubMed.
- L. Harland, Knowl. Eng. Knowl. Manag., Berlin, Heidelberg, 2012, pp. 1–7 Search PubMed.
- C. Draxl and M. Scheffler, MRS Bull., 2018, 43, 676–682 CrossRef.
- I. E. Castelli, D. J. Arismendi-Arrieta, A. Bhowmik, I. Cekic-Laskovic, S. Clark, R. Dominko, E. Flores, J. Flowers, K. Ulvskov Frederiksen, J. Friis, A. Grimaud, K. V. Hansen, L. J. Hardwick, K. Hermansson, L. Königer, H. Lauritzen, F. Le Cras, H. Li, S. Lyonnard, H. Lorrmann, N. Marzari, L. Niedzicki, G. Pizzi, F. Rahmanian, H. Stein, M. Uhrin, W. Wenzel, M. Winter, C. Wölke and T. Vegge, Batteries Supercaps, 2021, 4, 1803–1812 CrossRef.
- S. Clark, F. L. Bleken, S. Stier, E. Flores, C. W. Andersen, M. Marcinek, A. Szczesna-Chrzan, M. Gaberscek, M. R. Palacin, M. Uhrin and J. Friis, Adv. Energy Mater., 2022, 12, 2102702 CrossRef CAS.
- A. Engstfeld and J. Rüth, Linuxrider and Nicohoermann, Echemdb/Echemdb: 0.6.0, Zenodo, 2023, https://zenodo.org/record/7834993 Search PubMed.
- J. K. Pedersen, C. M. Clausen, O. A. Krysiak, B. Xiao, T. A. A. Batchelor, T. Löffler, V. A. Mints, L. Banko, M. Arenz, A. Savan, W. Schuhmann, A. Ludwig and J. Rossmeisl, Angew. Chem., 2021, 133, 24346–24354 CrossRef.
- K. J. Jenewein, S. Thienhaus, A. Kormányos, A. Ludwig and S. Cherevko, Chem. Sci., 2022, 13, 13774–13781 RSC.
- E. B. Tetteh, D. Valavanis, E. Daviddi, X. Xu, C. Santana Santos, E. Ventosa, D. Martín-Yerga, W. Schuhmann and P. R. Unwin, Angew. Chem., Int. Ed., 2023, 62, e202214493 CrossRef CAS PubMed.
- J. A. Keith, J. R. McKone, J. D. Snyder and M. H. Tang, Curr. Opin. Chem. Eng., 2022, 36, 100824 CrossRef.
- G. Smith and E. J. F. Dickinson, Nat. Commun., 2022, 13, 6832 CrossRef CAS PubMed.
- C. Tenopir, S. Allard, K. Douglass, A. U. Aydinoglu, L. Wu, E. Read, M. Manoff and M. Frame, PLoS One, 2011, 6, e21101 CrossRef CAS PubMed.
- S. Zhu, K. Jiang, B. Chen and S. Zheng, J. Mater. Chem. A, 2023, 11, 3849–3870 RSC.
- ELIXIR Research Data Management Kit (RDMkit), What Is the Best Way to Name a File?, 2023, https://rdmkit.elixir-europe.org/data_organisation.html#what-is-the-best-way-to-name-a-file Search PubMed.
- FAIRmat, Guide to Writing a Research Data Management Plan, 2023, https://www.fairmat-nfdi.eu/uploads/documents/FAIRmat%20DMP%20guide%20March%202023.pdf Search PubMed.
- N. CARPi, A. Minges and M. Piel, J. Open Source Softw., 2017, 2, 146 CrossRef.
- N. Brandt, L. Griem, C. Herrmann, E. Schoof, G. Tosato, Y. Zhao, P. Zschumme and M. Selzer, Data Sci. J., 2021, 20, 8 CrossRef.
- R. Duke, V. Bhat and C. Risko, Chem. Sci., 2022, 13, 13646–13656 RSC.
- L. Banko and A. Ludwig, ACS Comb. Sci., 2020, 22, 401–409 CrossRef CAS PubMed.
- E. Soedarmadji, H. S. Stein, S. K. Suram, D. Guevarra and J. M. Gregoire, npj Comput. Mater., 2019, 5, 1–9 CrossRef.
- L. Fiedler, T.-C. Ma, B. Fritsch, J. H. Risse, M. Lechner, D. Dworschak, M. Merklein, K. J. J. Mayrhofer and A. Hutzler, ChemElectroChem, 2023, e202300373 CrossRef CAS.
- Python Software Foundation, Python 3.8.15 Documentation, 2019, https://docs.python.org/release/3.8.15/ Search PubMed.
- J. Reback, J. Brock Mendel, W. McKinney, J. V. den Bossche, M. Roeschke, T. Augspurger, S. Hawkins, P. Cloud, gfyoung, P. Hoefler, Sinhrks, A. Klein, T. Petersen, J. Tratner, C. She, W. Ayd, R. Shadrach, S. Naveh, M. Garcia, J. H. M. Darbyshire, J. Schendel, T. Wörtwein, A. Hayden, D. Saxton, M. E. Gorelli, F. Li, M. Zeitlin, V. Jancauskas, A. McMaster and T. Li, Pandas-Dev/Pandas: Pandas 1.4.4, Zenodo, 2022, https://zenodo.org/record/7037953 Search PubMed.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S. J. Van Der Walt, M. Brett, J. Wilson, K. J. Millman, N. Mayorov, A. R. J. Nelson, E. Jones, R. Kern, E. Larson, C. J. Carey, İ. Polat, Y. Feng, E. W. Moore, J. VanderPlas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E. A. Quintero, C. R. Harris, A. M. Archibald, A. H. Ribeiro, F. Pedregosa, P. Van Mulbregt, SciPy 1.0 Contributors, A. Vijaykumar, A. P. Bardelli, A. Rothberg, A. Hilboll, A. Kloeckner, A. Scopatz, A. Lee, A. Rokem, C. N. Woods, C. Fulton, C. Masson, C. Häggström, C. Fitzgerald, D. A. Nicholson, D. R. Hagen, D. V. Pasechnik, E. Olivetti, E. Martin, E. Wieser, F. Silva, F. Lenders, F. Wilhelm, G. Young, G. A. Price, G.-L. Ingold, G. E. Allen, G. R. Lee, H. Audren, I. Probst, J. P. Dietrich, J. Silterra, J. T. Webber, J. Slavič, J. Nothman, J. Buchner, J. Kulick, J. L. Schönberger, J. V. De Miranda Cardoso, J. Reimer, J. Harrington, J. L. C. Rodríguez, J. Nunez-Iglesias, J. Kuczynski, K. Tritz, M. Thoma, M. Newville, M. Kümmerer, M. Bolingbroke, M. Tartre, M. Pak, N. J. Smith, N. Nowaczyk, N. Shebanov, O. Pavlyk, P. A. Brodtkorb, P. Lee, R. T. McGibbon, R. Feldbauer, S. Lewis, S. Tygier, S. Sievert, S. Vigna, S. Peterson, S. More, T. Pudlik, T. Oshima, T. J. Pingel, T. P. Robitaille, T. Spura, T. R. Jones, T. Cera, T. Leslie, T. Zito, T. Krauss, U. Upadhyay, Y. O. Halchenko and Y. Vázquez-Baeza, Nat. Methods, 2020, 17, 261–272 CrossRef CAS PubMed.
- C. R. Harris, K. J. Millman, S. J. van der Walt, R. Gommers, P. Virtanen, D. Cournapeau, E. Wieser, J. Taylor, S. Berg, N. J. Smith, R. Kern, M. Picus, S. Hoyer, M. H. van Kerkwijk, M. Brett, A. Haldane, J. F. del Río, M. Wiebe, P. Peterson, P. Gérard-Marchant, K. Sheppard, T. Reddy, W. Weckesser, H. Abbasi, C. Gohlke and T. E. Oliphant, Nature, 2020, 585, 357–362 CrossRef CAS PubMed.
- T. A. Caswell, M. Droettboom, A. Lee, E. S. de Andrade, T. Hoffmann, J. Klymak, J. Hunter, E. Firing, D. Stansby, N. Varoquaux, J. H. Nielsen, B. Root, R. May, P. Elson, J. K. Seppänen, D. Dale, J.-J. Lee, D. McDougall, A. Straw, P. Hobson, hannah, C. Gohlke, A. F. Vincent, T. S. Yu, E. Ma, S. Silvester, C. Moad, N. Kniazev, E. Ernest and P. Ivanov, Matplotlib/Matplotlib: REL: V3.5.2, Zenodo, 2022, https://zenodo.org/record/6513224 Search PubMed.
- T. Kluyver, B. Ragan-Kelley, P. F. Rez, B. Granger, M. Bussonnier, J. Frederic, K. Kelley, J. Hamrick, J. Grout, S. Corlay, P. Ivanov, D. Avila, N. S. Abdalla, C. Willing and J. D. Team, in Positioning and Power in Academic Publishing: Players, Agents and Agendas, IOS Press, 2016, pp. 87–90 Search PubMed.
- E. F. Codd, Commun. ACM, 1970, 13, 377–387 CrossRef.
- Solid IT, DB-Engines Ranking, 2023, https://db-engines.com/en/ranking.
- W. Khan, T. Kumar, C. Zhang, K. Raj, A. M. Roy and B. Luo, Big Data Cogn. Comput., 2023, 7, 97 CrossRef.
- A. A. Topalov, I. Katsounaros, J. C. Meier, S. O. Klemm and K. J. J. Mayrhofer, Rev. Sci. Instrum., 2011, 82, 114103 CrossRef PubMed.
- M. M. Hielscher, M. Dörr, J. Schneider and S. R. Waldvogel, Chem.–Asian J., 2023, 18, e202300380 CrossRef CAS PubMed.
- T. Chen and C. Guestrin, Proc. 22nd ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., New York, NY, USA, 2016, pp. 785–794 Search PubMed.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Kopf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai and S. Chintala, 33rd Conf. Neural Inf. Process. Syst., Vancouver, Canada, 2019 Search PubMed.
- M. Abadi, P. Barham, J. Chen, Z. Chen, A. Davis, J. Dean, M. Devin, S. Ghemawat, G. Irving, M. Isard, M. Kudlur, J. Levenberg, R. Monga, S. Moore, D. G. Murray, B. Steiner, P. Tucker, V. Vasudevan, P. Warden, M. Wicke, Y. Yu and X. Zheng, Proc. 12th USENIX Symp. Oper. Syst. Des. Implement. OSDI ’16, Savannah, GA, USA, 2016, pp. 265–283 Search PubMed.
- S. Raschka, J. Patterson and C. Nolet, Information, 2020, 11, 193 CrossRef.
- H. S. Stein and J. M. Gregoire, Chem. Sci., 2019, 10, 9640–9649 RSC.
- H. S. Stein, A. Sanin, F. Rahmanian, B. Zhang, M. Vogler, J. K. Flowers, L. Fischer, S. Fuchs, N. Choudhary and L. Schroeder, Curr. Opin. Electrochem., 2022, 35, 101053 CrossRef CAS.
- R. Bose, Proc. 14th Int. Conf. Sci. Stat. Database Manag., 2002, 15–19 Search PubMed.
- A. A. Topalov, I. Katsounaros, M. Auinger, S. Cherevko, J. C. Meier, S. O. Klemm and K. J. J. Mayrhofer, Angew. Chem., Int. Ed., 2012, 51, 12613–12615 CrossRef CAS PubMed.
- N. C. Röttcher, Y.-P. Ku, M. Minichova, K. Ehelebe and S. Cherevko, J. Phys. Energy, 2023, 5, 024007 CrossRef.
- M. Murbach, B. Gerwe, N. Dawson-Elli and L.-k. Tsui, J. Open Source Softw., 2020, 5, 2349 CrossRef.
- R. Zhang, R. Black, D. Sur, P. Karimi, K. Li, B. DeCost, J. R. Scully and J. Hattrick-Simpers, J. Electrochem. Soc., 2023, 170, 086502 CrossRef.
- S. O. Klemm, A. A. Topalov, C. A. Laska and K. J. Mayrhofer, Electrochem. Commun., 2011, 13, 1533–1535 CrossRef CAS.
- J. Timoshenko and B. Roldan Cuenya, Chem. Rev., 2021, 121, 882–961 CrossRef CAS PubMed.
- Q. Lin, Curr. Opin. Electrochem., 2023, 37, 101201 CrossRef CAS.
- B. M. Weckhuysen, Chem. Commun., 2002, 97–110 RSC.
- Y. Ji, Z.-W. Yin, Z. Yang, Y.-P. Deng, H. Chen, C. Lin, L. Yang, K. Yang, M. Zhang, Q. Xiao, J.-T. Li, Z. Chen, S.-G. Sun and F. Pan, Chem. Soc. Rev., 2021, 50, 10743–10763 RSC.
- Y. Yang, Y. Xiong, R. Zeng, X. Lu, M. Krumov, X. Huang, W. Xu, H. Wang, F. J. DiSalvo, J. D. Brock, D. A. Muller and H. D. Abruña, ACS Catal., 2021, 11, 1136–1178 CrossRef CAS.
- B. Chen, H. Zhang, J. Xuan, G. J. Offer and H. Wang, Adv. Mater. Technol., 2020, 5, 2000555 CrossRef CAS.
- S. Zhou, K. Liu, Y. Ying, L. Chen, G. Meng, Q. Zheng, S.-G. Sun and H.-G. Liao, Curr. Opin. Electrochem., 2023, 41, 101374 CrossRef CAS.
- T.-H. Shen, R. Girod, J. Vavra and V. Tileli, J. Electrochem. Soc., 2023, 170, 056502 CrossRef CAS.
- Y. Yang, J. Feijóo, V. Briega-Martos, Q. Li, M. Krumov, S. Merkens, G. De Salvo, A. Chuvilin, J. Jin, H. Huang, C. J. Pollock, M. B. Salmeron, C. Wang, D. A. Muller, H. D. Abruña and P. Yang, Curr. Opin. Electrochem., 2023, 101403 CrossRef.
- J. C. Meier, I. Katsounaros, C. Galeano, H. J. Bongard, A. A. Topalov, A. Kostka, A. Karschin, F. Schüth and K. J. J. Mayrhofer, Energy Environ. Sci., 2012, 5, 9319–9330 RSC.
- A. Unwin, Harvard Data Sci. Rev., 2020, 2, 1–7 Search PubMed.
- P. Jupyter, M. Bussonnier, J. Forde, J. Freeman, B. Granger, T. Head, C. Holdgraf, K. Kelley, G. Nalvarte, A. Osheroff, M. Pacer, Y. Panda, F. Perez, B. Ragan-Kelley and C. Willing, Proc. 17th Python Sci. Conf., 2018, 113–120 Search PubMed.
- N. C. Röttcher, B. Fritsch and D. Dworschak, Repository for: Accelerating Materials Research with a Comprehensive Data Management Tool: A Case Study on an Electrochemical Laboratory, 2023, https://zenodo.org/records/10417756 Search PubMed.
- Binder Project Team, Binder Session for Zenodo Repository: 10.5281/Zenodo.10417756, 2023, https://mybinder.org/v2/zenodo/10.5281/zenodo.10417756/ Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ta06247c |
| This journal is © The Royal Society of Chemistry 2024 |