Designing nanodiscs as versatile platforms for on-demand therapy
Qianwen
Mu†
abc,
Haolan
Deng†
ab,
Xiaoyu
An
abc,
Gang
Liu
 bc and
Chao
Liu
bc and
Chao
Liu
 *ad
*ad
aState Key Laboratory of Stress Biology and Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Xiamen University, Xiamen 361102, China. E-mail: liuchao66888@xmu.edu.cn; Tel: +86 132-1502-1518
bState Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & Center for Molecular Imaging and Translational Medicine, School of Public Health, Xiamen University, Xiamen 361102, China
cState Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Biology, School of Life Sciences, Xiamen University, Xiamen 361102, China
dShenzhen Research Institute of Xiamen University, Shenzhen 518000, China
First published on 15th December 2023
Abstract
Nowadays, there has been an increasing utilization of nanomedicines for disease treatment. Nanodiscs (NDs) have emerged as a novel platform technology that garners significant attention in biomedical research and drug discovery. NDs are nanoscale phospholipid bilayer discs capable of incorporating membrane proteins and lipids within a native-like environment. They are assembled using amphiphilic biomacromolecular materials, such as apolipoprotein A1 or membrane scaffold proteins (MSPs), peptides, and styrene–maleic acid polymers (SMAs). NDs possess well-defined sizes and shapes, offering a stable, homogeneous, and biologically relevant environment for studying membrane proteins and lipids. Their unique properties have made them highly desirable for diverse applications, including cancer immunotherapy, vaccine development, antibacterial and antiviral therapy, and treating Alzheimer's disease (AD) and diabetes-related conditions. This review discusses the classifications, advantages, and applications of NDs in disease therapy.
1. Introduction
Nanomaterials have made a significant impact as drug delivery carriers, with nanodiscs (NDs) emerging as a forefront class of nanoparticles (NPs) for both treatment and diagnosis of various ailments. NDs are synthetic phospholipid particles with a unique morphology and size, which heightens their effectiveness in drug delivery. Pioneered by Sligar et al. in the early 2000s, these model membrane systems have dimensions of roughly 10 nm in diameter and a thickness ranging from 4.6 to 5.6 nm.1 NDs resemble high-density lipoprotein (HDL) in structure. HDL, with a diameter of approximately 10 nm, functions as a carrier for different types of lipids, such as cholesterol and triglycerides.2 HDL is made up of a variety of apolipoproteins, the most important of which is apolipoproteins A-1 (ApoA-1); HDL is a good model for loading poorly soluble small molecules. For addressing the solubility challenge, scientists have modified ApoA-1 proteins, leading to an array of membrane scaffold proteins (MSPs) and peptides, which can then be assembled into NDs of similar size.3A standout application of NDs is seen in EVOQ therapeutics’ nanodisc technology. This advanced system is designed to target disease-specific antigens directly to lymph nodes, presenting a promising avenue for autoimmune disease treatments. Notably, Amgen and subsequently Gilead Sciences have recognized the value of nanodiscs and invested $660 million on January 3, 2023, for the development of novel therapies targeting autoimmune diseases like systemic lupus erythematosus and rheumatoid arthritis; this demonstrates the significant potential of NDs in disease treatment. This review summarizes the classifications and benefits of NDs and introduces their applications in disease treatment.
2. Classification of NDs
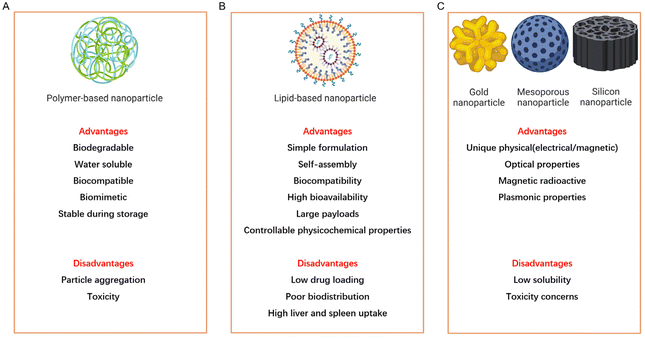
Since the FDA approved the first nanoparticles, known as INFeD (Sanofi Aventis, Allergan), for clinical use in 1974, there has been a steadily increasing interest in exploring nanomedicines as a potential treatment for various diseases.4 Research findings indicate that NPs can be broadly categorized into three main types: polymer-based nanoparticles, lipid-based nanoparticles, and inorganic particles (Fig. 1).5 Among these, lipid-based nanoparticles composed of lipids have gained significant attention for their ability to deliver hydrophobic drugs and nucleic acids, which results in prolonged circulation and enhanced drug delivery capabilities.6–8 Polymer-based nanoparticles, assembled using amphiphilic copolymers such as polymersomes, micelles, and dendrimers, which are typically formed with materials like polyethylene glycol (PEG) and polydimethylsiloxane (PDMS), offer improved stability and cargo-retention efficiency, making them well-suitable for drug delivery.9,10 Dendrimers, formed from charged polymers such as polyethylenimine (PEI) and polyamidoamine (PAMAM), have applications as theranostic and contrast agents.11,12 Inorganic particles, created from materials such as gold, iron, and silica, exhibit unique physical, magnetic, and optical properties and have been extensively studied for diagnostics, imaging, and photothermal therapy.13–16 Each type of NP possesses distinct advantages for disease therapy, but they also come with notable drawbacks.Nanoparticles typically range in size from 1 to 100 nm, making them susceptible to uptake by the liver and spleen. However, non-spherical NPs offer advantages over their spherical counterparts, including an extended blood circulation half-life and a higher cellular endocytosis rate. In recent years, significant attention has been directed toward the development of NDs, which are non-spherical NPs coated with amphiphilic molecules like proteins, synthetic polymers, or short-chain lipids. NDs draw inspiration from the transportation of lipid nanocarriers in the human bloodstream. Among these nanocarriers, high-density lipoprotein (HDL) plays a pivotal role in the transport and metabolism of lipids, particularly cholesterol and triglycerides.17 NDs can efficiently carry various substances, including drugs, nucleic acids, signal-emitting molecules, or dyes, making them highly effective nanocarriers for therapeutic applications or medical diagnostics.18 This versatility and capability make NDs promising tools in the field of nanomedicine.
NDs are considered ideal nanoformulations due to their enhanced anisotropic characteristics and multifunctional functionalization potential. Based on their composition (as illustrated in Fig. 2), NDs can be classified into four categories:19
(1) Lipid nanodiscs (LNDs). LDNs consist of amphiphilic biomacromolecules such as membrane scaffold proteins (MSPs),20 apolipoprotein A1 (ApoA-1),21 and peptides modified according to ApoA-1 (Fig. 2A).22
(2) Styrene–maleic acid (SMA) lipid nanoparticles (SMALPs) and novel SMALPs. SMALPs are formed through the copolymerization of styrene and maleic anhydride monomers. They are the hydrolyzed form of styrene–maleic anhydride (SMAnh) copolymers (Fig. 2B).23
(3) Polymer nanodiscs (PNDs). PNDs are assembled from MSPs or other biomacromolecules in the presence of amphiphilic block copolymers such as the bacterial ATP-binding cassette (ABC) transporter MsbA (Fig. 2C).24–26
(4) Hybrid nanodiscs (HNDs). HNDs encompass hybrid bicelles (HBs), edge-modified hybrid bicellars (EHBs), plane-modified hybrid bicellars (PHBs), and similar structures. HBs are an extension of bicelles and are composed of a long-chain cerasome-forming lipid and a short-chain phospholipid. The edge- and plane-modified hybrid bicellar nanodiscs are referred to as EHB and PHB, respectively (Fig. 2D).27
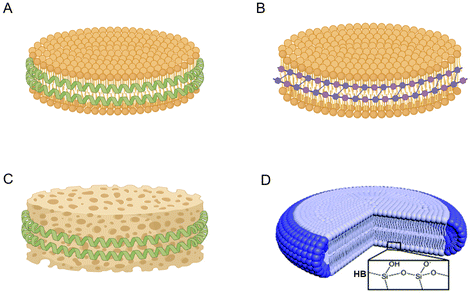 | ||
| Fig. 2 Different forms of NDs. (A) Lipid nanodiscs (LNDs) consist of amphiphilic biomacromolecules. (B) Styrene–maleic acid (SMA) lipid nanoparticles (SMALPs) formed through the copolymerization of styrene and maleic anhydride monomers. (C) Polymer nanodiscs (PNDs) assembled from MSPs or other biomacromolecules in the presence of amphiphilic block copolymers. (D) Hybrid nanodiscs (HNDs) composed of a long-chain cerasome-forming lipid and a short-chain phospholipid.27 | ||
3. Advantages of NDs
NDs present numerous advantages over other nanoparticle platforms, such as uncomplicated formulation, consistent reproducibility, enhanced stability, and superior biocompatibility. When employed in vivo, NDs demonstrate remarkable pharmacokinetics and biological distribution, allowing them to exhibit heightened specificity to target tissues and cellular receptors. This specificity enables more accurate drug release or lipid exchange. As a multifaceted platform, NDs have a broad spectrum of potential applications. To sum up, the benefits of NDs significantly overshadow those of traditional NPs.3.1 Size control and therapeutic targeting
One of the primary attributes of NDs is their uniform and adjustable size, which plays a crucial role in therapeutic targeting. NDs can be fabricated from diverse materials, resulting in varying sizes. For instance, NDs offer precise size control by leveraging specific membrane scaffold protein (MSP) structures, usually from 10 nm to 20 nm. In recent years, the development of cyclized MSP proteins (cMSP) has made it possible to obtain covalently circularized NDs (cNDs) with a size larger than 80 nm, which have better thermal stability and long-term stability; they can load drugs and membrane proteins more efficiently and achieve better targeted drug delivery, and achieve better applications in structural analysis.28–30 Assembled with styrene–maleic acid (SMA), NDs typically possess a size of around 20 nm—ideal for drug delivery and transporting small molecular probes across the blood–brain barrier (BBB).31,32 Additionally, strategic ligand modifications, such as planar and edge adjustments on NDs, can amplify the anisotropic effect, thereby bolstering targeting efficiency.33In recent studies, Zhang's team, in 2023, innovatively used bacterial outer membranes to design cell-based nanodiscs (OM-NDs) as an antibacterial vaccination platform. Using Pseudomonas aeruginosa as a model pathogen, they demonstrated that OM-NDs efficiently reached the lymph nodes in vivo due to their petite size. These NDs also interacted effectively with antigen-presenting cells (APCs), which sped up uptake and intensified immune responses. Such findings underscore NDs’ ideal sizing and adaptability for various material loadings.34 In 2017, Zhang Qiang's group delved into how nanomaterials’ biological attributes could be modulated by altering their shape and ligand modifications. They designed and engineered four types of nanoparticles, including hybrid nanospheres, NDs, and NDs modified with octahedral arginine or edge or planar modification (R8) sequences. The researchers discovered that the anisotropy induced by R8 modification significantly impacted endocytosis, intracellular transport, and even tissue penetration of nanoparticles. The maximum increase in cell uptake, comparing the planar modification to marginal modification of R8, reached up to 17-fold. The team also compared the cell uptake differences across six different cell lines. Their findings revealed that shape anisotropy and ligand alterations profoundly influence the endocytosis, internal transport, and tissue penetration capacities of nanoparticles.27 NDs, with their capacity for fine-tuned size adjustment and enhanced targeting, have emerged as potent assets in oncological research and treatments. These unique characteristics of NDs hold great promise for evolving imaging probes and delivery mechanisms for chemotherapeutics, vaccines, and anti-cancer agents.
3.2 Binding affinity and adhesion
Compared to spherical NPs, disk-shaped NDs offer distinctive perks. Their expansive surface area leads to prolonged circulation, augmented cellular intake, and refined biological distribution. Additionally, NDs’ flattened geometry instigates a torque within blood flow, propelling them to rotate and roll. This dynamism amplifies their proximity to vessel walls, reinforcing vascular adherence.35,36Recent advancements have highlighted challenges in systemically delivering STING agonists to tumors. However, a marked improvement in tumor penetration was observed when STING-activated cyclic dinucleotides (CDNs) were integrated into NDs for intravenous administration. This approach ensures that most tumor cells are exposed to STING agonists.37 Likewise, another investigation showed that disc-shaped nanocarriers decorated with N-methylated glutamate (CR(NMe)EKA) and loaded with Cys–Arg–Glu–Lys–Ala exhibited superior tumor targeting and penetration compared to spherical nanoparticles. This enhanced delivery system effectively transported tumor necrosis factor-associated apoptosis-inducing ligand (TRAIL) and paclitaxel (PTX) to lung melanoma tumor sites.38
3.3 Biocompatibility and biosafety
The foundation of NDs lies in predominantly biocompatible constituents. For example, the primary elements that comprise LNDs or PNDs are modifications of the human ApoA-1 protein, like membrane scaffold proteins (MSPs) or peptides. Studies have consistently vouched for the commendable pharmacokinetic traits and biocompatibility of LNDs and PNDs formulated with MSPs or ApoA-1 analogs.39,40 SMA, an amphiphilic biomacromolecule utilized in assembling SMALPs, has been extensively investigated for its biosafety. Furthermore, innovations are underway to create sturdier SMA copolymers that remain stable under acidic conditions or amid polyvalent ions.41 Silica, a key component in the construction of HNDs, is classified as “generally considered safe” by the US Food and Drug Administration (ID Code: 14808-60-7).33 These biosafe materials and their metabolic pathways contribute to the overall biocompatibility of NDs, making them a promising platform for various biomedical applications.The comprehensive benefits of NDs encompass size adaptability, targeting proficiency, biocompatibility, and seamless drug modification potential. Although considerable efforts have been made in preclinical research, a significant portion of ND-related studies remains in the validation phase, awaiting concrete clinical data. To harness NDs’ full potential in clinical contexts, continued research, validation, and clinical trials are imperative. As ND research continues to evolve, it is pivotal to rigorously assess NDs through clinical examinations to confirm their efficacy and safety, ensuring that they become a trustworthy instrument in clinical applications.
4. Application of NDs in disease therapy
NDs present a remarkably versatile molecular delivery platform, amenable to extensive modifications that enhance disease targeting and enable personalized tumor treatments. Numerous studies have underscored the utility of NDs in disease therapy, illuminating their remarkable versatility and untapped potential.42–47 The unique disc-shaped structure of NDs allows for efficient loading of poorly soluble drugs, thereby improving their therapeutic efficacy. Furthermore, NDs can be modified with vaccine adjuvants or anti-tumor drugs to facilitate tumor immunotherapy.19 Functional small molecules can also be loaded onto NDs to investigate transmembrane signaling pathways and employ visualization tracing techniques. In subsequent discussions, we will delve into the specific applications of NDs in disease therapy, providing comprehensive insights into their therapeutic potential and highlighting their contributions to advancing medical research and personalized medicine.4.1 Cancer immunotherapy
Cancer immunotherapy faces two formidable challenges: the insufficient infiltration of CD8+ T cells and the overexpression of programmed cell death protein 1 (PD-1) on the surfaces of activated T cells.48 Extensive research has focused on small-molecule drugs targeting the stimulator of interferon genes (STING), immune checkpoints, and toll-like receptors (TLR) as potential tumor therapy agents. Although these drugs exhibit promise against various cancers, their efficacy is hampered by the daunting barriers posed by tumor heterogeneity, the immunosuppressive tumor microenvironment (TME), and the blood–brain barrier.44,49,50 Thanks to their unique advantages, NDs have emerged as a promising platform for cancer immunotherapy. One approach involves utilizing NDs to deliver tumor antigens to dendritic cells, specialized immune cells that can activate T cells to recognize and eliminate cancer cells. NDs can be engineered with tumor antigens and adjuvants, such as toll-like receptor agonists, to enhance the immune response. By delivering these immunogenic components to dendritic cells, NDs facilitate the generation of a robust anti-tumor immune response.51–53 The advantages of employing NDs in cancer immunotherapy are manifold. Firstly, the disk-shaped structure of NDs provides a larger surface area for loading tumor antigens and adjuvants, enabling efficient delivery to dendritic cells. Secondly, the size and surface modification of NDs can be fine-tuned with precision to augment cellular uptake and activate immune cells. Thirdly, NDs can protect the encapsulated antigens and adjuvants from degradation and premature release, ensuring their effective delivery to the target cells. By harnessing NDs’ capabilities, researchers aim to overcome the limitations of current cancer immunotherapy approaches and achieve improved therapeutic outcomes. Ongoing research endeavors are imperative to optimize ND-based immunotherapeutic strategies and validate their efficacy in preclinical and clinical settings.Targeting the STING signaling pathway with innate immune stimuli has demonstrated potential in bolstering antitumor immunity. However, the effective delivery of STING agonists directly to tumors remains challenging. To tackle this, researchers have conjugated STING agonists, specifically cyclic dinucleotides (CDNs), with PEGylated lipids, creating CDN-PEG-lipids. When these are incorporated into LNDs, the resultant LNDs (as depicted in Fig. 3A) have shown improved tumor penetration upon intravenous administration. This led to improved co-localization of CDNs and tumor antigens in dendritic cells, increased uptake of LND-CDNs by cancer cells, and enhanced T cell activation.44,54 As carriers, LNDs ensure efficient compound delivery within solid tumors, reinforcing their potential to augment immunotherapy. Integrating chemotherapy drugs with immunotherapies has shown a promising synergy in immune activation. For instance, the commonly employed chemotherapy agent, adriamycin (DOX), has been channeled through NDs as delivery vehicles. This modality triggers immunogenic cell death in malignant cells, exerting an anti-tumor effect (Fig. 3B).46 Another notable study focused on NDs for chemoimmunotherapy targeting glioblastoma (GBM), wherein the vector, packed with docetaxel (DTX) and CpG (a toll-like receptor 9 agonist), efficiently delivered the chemotherapy drug to the GBM microenvironment, causing notable tumor regression (Fig. 3C).50
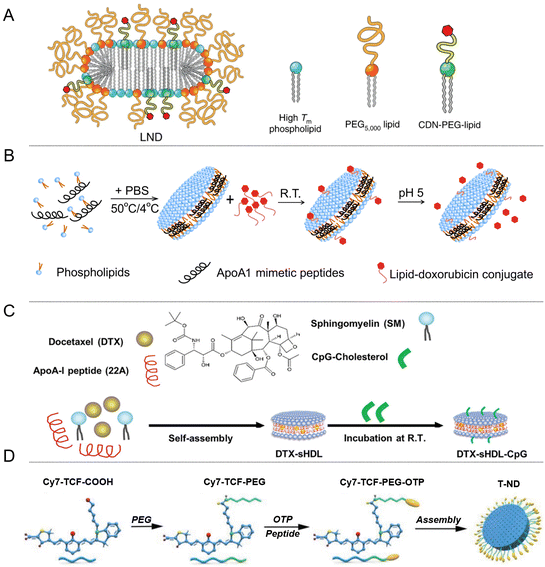 | ||
| Fig. 3 (A) LNDs conjugated with STING agonists, CDNs and PEGylated lipids to improve their delivery. (B) The chemotherapy drug DOX, which is widely used, delivered using NDs as carriers. (C) DTX-sHDL-CpG loaded with DTX and CpG, a toll-like receptor 9 (TLR9) agonist. (D) T-NDs modified with OTP and Cy7-TCF (2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran, TCF).44,46,50,55 | ||
Highly heterogeneous tumors represent a formidable obstacle to targeted therapies. Within this context, photothermal therapy (PTT) has emerged as a promising treatment modality. Ye's group utilized phage display technology to identify an osteosarcoma (OS) targeting peptide (OTP). Subsequently, they ingeniously integrated OTP and Cy7-TCF (2-dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran, TCF) into NDs, resulting in the creation of T-NDs (illustrated in Fig. 3D) tailored for heterogeneous tumors. Experimental results demonstrated that these specialized NDs effectively enhanced photothermal tumor ablation under near-infrared light and prolonged tumor retention.55
These studies highlight the potential of ND-based cancer therapy as a novel and adaptable approach for priming anti-tumor immunity and enhancing tumor sensitivity to immune checkpoint blockade. This provides a theoretical foundation for the precise treatment of highly heterogeneous tumors. By leveraging NDs’ unique targeting capabilities and photothermal properties, researchers aim to develop innovative strategies for combating complex and diverse tumor types. Further exploration and validation of these approaches are essential for advancing the field of precision medicine in the treatment of heterogeneous tumors.
4.2 Vaccine development
Based on soluble antigens paired with classic adjuvants, conventional cancer vaccines have demonstrated limited performance in clinical settings. This underscores the imperative need for innovative vaccine technologies to stimulate robust T cell responses for effective antitumor activity. To date, many reports have been published on using nanovesicles to integrate nanovaccines, which can achieve personalized tumor immunotherapy.56,57 Whether sourced from diverse membranes or loaded with antigenic peptides and tumor markers, NDs present a rich tapestry of antigenic features and inherent immunogenic attributes. They stand out as promising candidates for devising vaccines for tumor immunotherapy and countering infectious diseases.Extracting membrane proteins directly from the natural membrane environment to assemble NDs has great potential and can well retain the structure and activity of membrane proteins. In 2018, Tsyr-Yan Yu's group extracted bacteriorhodopsin from the natural membrane environment and assembled it into a covalently circularized nanodisc; the natural structure and activity of the protein are well retained.58 Zhang's group has successfully constructed a cellular ND, harnessing the bacterial outer membrane of Pseudomonas aeruginosa (OM-NDs) (Fig. 4A) as a platform for antimicrobial vaccine. Owing to their diminutive size, these NDs can be efficiently delivered to lymph nodes, forging effective interactions with antigen-presenting cells, resulting in accelerated uptake and improved immune stimulation. This approach has demonstrated the ability to elicit strong humoral and cellular immune responses against Pseudomonas aeruginosa, as confirmed in a mouse model of pneumonia.34 Furthermore, NDs offer the advantage of being loaded with antigenic peptides or specific tumor markers, enhancing their immunogenicity. Studies conducted in mice and nonhuman primates have validated the efficacy and safety of NDs loaded with antigenic peptides (Fig. 4B).49,59–61 Aldehyde dehydrogenase (ALDH) has been widely utilized as a marker for isolating cancer stem cells (CSCs), which proliferate extensively and drive tumor metastasis and recurrence and have been demonstrated to be present in more than 20 tumor types to date.62,63 James J. Moon's group developed synthetic NDs for vaccination against ALDHhigh CSCs. NDs facilitate antigen transport to lymph nodes and induce robust ALDH-specific T cell responses, presenting a renewed avenue for cancer immunotherapy aimed at CSCs (Fig. 4C).45
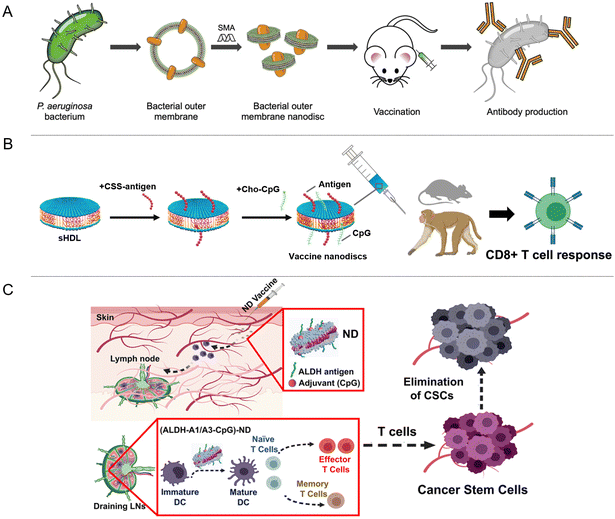 | ||
| Fig. 4 (A) OM-ND formulation used as a nanovaccine to protect against bacterial infection. (B) Vaccination of NDs loaded with antigenic peptides or specific tumor markers conducted in mice and nonhuman primates has validated the efficacy and safety. (C) Illustration of ND vaccination against ALDHhigh CSCs, ALDH-A1/A3-CpG-NDs facilitate antigen transport to lymph nodes and induce robust ALDH-specific T cell responses.34,45,60,61 | ||
Furthermore, there have been studies on loading the HIV envelope glycoprotein (Env) membrane protein onto NDs. These studies have successfully resolved the complex structure formed by Env and the membrane-proximal external region (MPER)-targeting antibody 10EB. Through the exploration of the molecular mechanism of the Env-MPER antibody complex, it was discovered that the tilt of Env plays a crucial role in the neutralization mechanism of MPER-targeted antibodies.64 A work studied the structure of MPER NDs combined with 4E10, and also found that membrane fusion was abolished by changing MPER dynamics and modifying average ectodomain tilt, providing information for future vaccine development.65 In another research study, MPER-TMD (MPER and the adjacent transmembrane domain) was assembled into NDs, and the structure of MPER-TMD and its mechanism of action combined with 10E8 were explored to provide a structural basis for vaccine design.66 These findings provide a solid structural foundation for developing vaccines based on the ND platform, with the potential to advance HIV immunotherapy.
4.3 Antibacterial and antiviral therapy
NDs have emerged as promising platforms for antimicrobial and antiviral therapies, aiming to enhance the targeting and eradication of bacterial and viral pathogens. The utilization of NDs for delivering antimicrobial peptides or drugs offers several benefits. Firstly, it protects antibacterial drugs from degradation, ensuring their stability and prolonging their effectiveness. Additionally, NDs facilitate the efficient uptake of these agents by pathogens, thereby improving their potency and selectivity. In the context of antiviral therapy, NDs can display viral or bacterial antigens, triggering robust and specific immune responses against targeted pathogens. This immune stimulation is critical in combating viral infections. To further optimize therapy, NDs can be functionalized with antigens, adjuvants, and targeting ligands such as antibodies or peptides. These modifications enhance the immunogenicity and specificity of the therapeutic approach, ultimately improving its overall efficacy.The misuse and overuse of antibiotics have contributed to the emergence of drug resistance during treating bacterial infections. Additionally, inflammation poses another significant challenge in antimicrobial therapy. Infections often trigger the recruitment of inflammatory cells, producing high levels of reactive oxygen species (ROS), which can cause oxidative damage to healthy tissues.67 Most antimicrobial agents currently available lack anti-inflammatory properties, highlighting the need to develop new therapeutic platforms capable of selectively targeting microbes while also possessing anti-inflammatory activity. Although non-antibiotic drugs, such as cationic peptides or metal-based nanomaterials, have been explored as antimicrobial agents, they often suffer from poor selectivity and may induce cytotoxicity in normal human somatic cells, leading to severe adverse effects.68 However, recent research has demonstrated the tremendous potential of Bi2Se3-NDs in treating bacterial infections. Bi2Se3-NDs exhibit several desirable properties. They effectively bind to lipoteichoic acid found in Gram-positive bacteria, physically disrupting the bacterial wall/membrane. Additionally, these nanosheets possess intrinsic antioxidant activity, allowing them to scavenge ROS generated by macrophages during bacterial infection. Importantly, Bi2Se3-NDs (Fig. 5A) demonstrate low off-target side effects and are effective against drug-resistant bacteria. They also exhibit high stability and biocompatibility, opening up new possibilities for developing non-antibiotic drugs.69 In addition, another study also reported that Bi2Se3-NDs can clear intestinal reactive oxygen and nitrogen species (RONS), effectively reduce intestinal inflammation, and have a good therapeutic effect in the treatment of inflammatory bowel disease.70
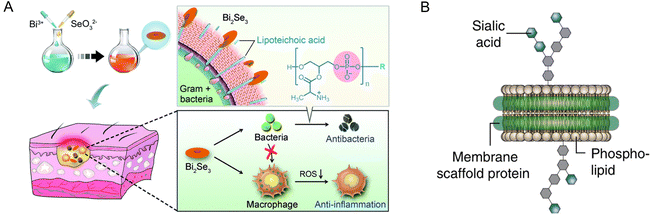 | ||
| Fig. 5 (A) Illustration of the preparation of Bi2Se3-NDs and the combined antibacterial and anti-inflammatory therapy of bacterial infections. (B) The total ganglioside-embedded nanodisc (NDTG) contains a decoy virus receptor to inhibit viral infections.69,74 | ||
Antiviral drugs that target the viral membrane offer the advantage of creating a barrier against drug resistance and exhibiting broad-spectrum activity across different viruses, irrespective of their specific viral membranes. However, there is a current lack of antiviral drugs that selectively target and destroy viral membranes, inhibiting a wide range of enveloped viruses.71–73 In a significant development, Dae-Hyuk Kweon has pioneered using NDs containing a decoy virus receptor to inhibit viral infections. The antiviral activity of these NDs is enhanced when a viral receptor, such as sialic acid, is incorporated into the nanoparticle. This incorporation allows the NDs to disrupt the viral envelope within endosomes, particularly at low pH levels. Consequently, the viral RNA becomes trapped within endolysosomes, undergoing enzymatic hydrolysis. In experimental mouse models, NDs effectively inhibit influenza virus infection, reducing morbidity and mortality without causing noticeable toxicity (Fig. 5B).74
Antimicrobial peptides (AMPs) play a crucial role in the innate immune system of animals, plants, fungi, and prokaryotes. These cationic and amphiphilic peptides directly target bacterial membranes, causing damage and compromising their integrity. AMPs have shown promising prospects in combating antibiotic-resistant infections. In a study conducted by Michael T. Marty et al., the interaction between different lipid bilayer NDs and AMP-lipids was investigated to gain insights into the potential mechanism of action of AMPs.75 Furthermore, Tomohiro Imura et al. isolated an antimicrobial peptide called PXT-5 and its modified version, modified PXT-5, from the skin of diploid Xenopus tropicalis. Through experiments, it was demonstrated that these antimicrobial peptides could be efficiently incorporated into NDs, highlighting the potential for their application in antimicrobial therapies.76
In conclusion, NDs offer substantial potential as versatile platforms for antibacterial and antiviral therapies. Their capability to deliver antimicrobial agents, induce specific immune responses, and be modified with various components significantly enhances the immunogenicity, specificity, and effectiveness of these therapeutic approaches. The studies mentioned above contribute to our understanding of the interaction between NDs and AMPs, furthering the development of innovative antimicrobial strategies.
4.4 Treatment of other diseases
In a study conducted by Gao's group in 2015, a nanostructure called monosialotetrahexosylganglioside (GM1)-modified recombinant high-density lipoprotein (GM1-rHDL) was reported (Fig. 6A). This nanostructure exhibited a high antibody-like affinity to Aβ and facilitated the degradation of Aβ by microglia and its efflux across the BBB. When administered intranasally, NDs demonstrated efficient distribution in the brain. The researchers loaded a neuroprotective peptide called NAP onto NDs, resulting in RNAP-GM1-rHDL. This formulation exhibited enhanced neuroprotection, reduced Aβ deposition, and improved neurological changes in AD model mice.31 In 2016, Gao's group developed a biomimetic nanocarrier called APOE-reconstituted high-density lipoprotein nanocarrier (ANC). Confocal microscopy studies demonstrated that ANC could cross cerebral blood vessels and accumulate around Aβ aggregates. Subsequently, the researchers loaded α-mangogenin (α-M) onto ANC, forming ANC-α-M (Fig. 6B). α-M is a polyphenolic drug that inhibits the formation of Aβ oligomers and fibers, and enhances the cellular degradation of Aβ. ANC-α-M exhibited a strong affinity for Aβ and effectively reduced amyloid deposition, decreased microglia proliferation, and improved memory deficits in SAMP8 mice.32 The BBB poses a challenge in delivering therapeutic and diagnostic drugs to the central nervous system (CNS). However, NDs with high BBB permeability and a solid binding affinity for Aβ have been developed, offering a promising platform for drug delivery in AD. These advancements hold the potential for effective treatment strategies for AD.
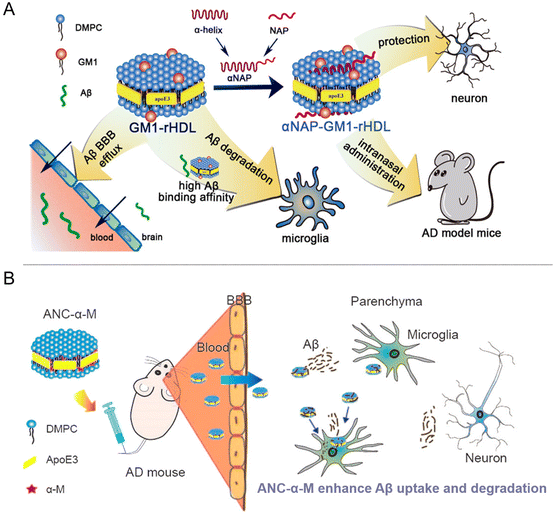 | ||
| Fig. 6 (A) The assembly process of GM1-rHDL, GM1-rHDL exhibited a high antibody-like affinity to Aβ and facilitated the degradation of Aβ. (B) The assembly process of ANC-α-M, which can enhance Aβ uptake and degradation in mouse models.31,32 | ||
Anna Schwendeman's research group has significantly advanced in developing a renal-targeted therapy strategy for DN. They have modified synthetic NDs by incorporating a renal targeting ligand called KT peptide and encapsulating LXR agonists within the NDs. This approach aims to specifically target the kidneys and deliver LXR agonists to address the underlying mechanisms of DN. The LXR agonists delivered by NDs play a crucial role in promoting the removal of excess lipids from mesangial cells, improving inflammation, and restoring normal kidney function. In experimental studies, the data demonstrated that the modified NDs, known as KT-sHDL/TO, effectively facilitated cholesterol efflux and inhibited mesangial cell proliferation. Moreover, these NDs exhibited significant therapeutic effects in a DN mouse model, showcasing their potential for future clinical translation in the treatment of DN. The research conducted by Anna Schwendeman's group provides promising insights into the development of targeted therapies for DN, offering new avenues for managing this prevalent complication of diabetes (Fig. 7A).102
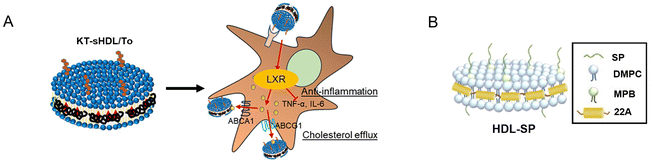 | ||
| Fig. 7 (A) KT-sHDL/TO loaded with a renal targeting ligand called KT peptide and LXR agonists, which can facilitate cholesterol efflux and inhibit mesangial cell proliferation. (B) Schematic illustration of HDL-SP NDs formulation, NDs extend the half-life of SP and improve the retention of SP in the bone marrow.102,106 | ||
Therapeutic approaches utilizing endogenous stem cell mobilizers have shown promise in providing effective cell-free therapy for various ischemic diseases. Substance P (SP), a peptide consisting of 11 sequences, has been identified as an effective endogenous mobilizer for mesenchymal stem cells (MSCs) and endothelial progenitor cells (EPCs), leading to wound healing and angiogenesis induction. It has been considered a potential treatment for severe ischemic diseases, including diabetic hind limb ischemia, ulcers, and wounds, without needing in vitro cell culture and expansion.103,104 However, the SP peptide has a short half-life and is rapidly degraded in the body. Diabetic patients, in particular, experience increased levels of neutral endopeptidase (NEP), an enzyme responsible for degrading SP. This results in a half-life of SP of less than 1 minute in diabetic mice, limiting its therapeutic potential.105 To overcome this challenge, Seung-Woo Cho's group has developed HDL-SP-simulated NDs. By combining SP with HDL NDs, the in vitro and in vivo half-life of SP is significantly extended, showing an over thousand-fold increase. This innovative approach improves the retention of SP in the bone marrow, leading to enhanced angiogenesis and restoration of blood perfusion in a diabetic limb ischemia model. As a result, limb retention is significantly improved, offering a new and promising strategy for treating diabetic ischemia (Fig. 7B).106
Indeed, studies have demonstrated that NDs are highly efficient drug carriers with several advantageous properties. One notable feature is their ability to enhance the stability and prolong the half-life of unstable polypeptide or protein drugs. Encapsulating these fragile drugs within NDs can reduce their susceptibility to degradation, thereby improving their therapeutic potential. One of the key advantages of NDs is their ultra-small size, typically around 10 nanometers. This nanoscale dimension allows them to traverse biological barriers and reach target tissues more effectively. Additionally, NDs exhibit high in vivo tolerance, meaning that they are well tolerated by the body and do not elicit significant adverse effects. This is crucial for their safe and efficient application as drug carriers. Furthermore, NDs possess a prolonged cycle half-life, allowing them to circulate in the body for an extended period. This extended circulation time increases the chances of drug delivery to target sites, enhancing the efficacy of the treatment.17,107 NDs have demonstrated remarkable potential as drug carriers due to their small size, biocompatibility, prolonged circulation, and high targeting capabilities. These properties make them efficient and versatile platforms for delivering various types of drugs to target tissues, thereby enhancing drug stability, improving pharmacokinetics, and optimizing therapeutic outcomes.
4.5 Novel biotechnology
Nanotechnology has been extensively explored for drug delivery and various emerging biological technologies.116–119 Yet, it is essential to note that traditional liposome-based drug delivery systems have intrinsic drawbacks, such as brief circulation duration, suboptimal drug encapsulation, and instability in biological media.120 NDs, have emerged as a solution, offering promise not only in disease treatment but also in a range of other innovative biological applications.121,1225. Conclusions
NDs are a versatile and innovative platform that can be used to deliver drugs or peptides for various disease treatments. NDs can protect the drugs or peptides from degradation, improve their pharmacokinetics and selectivity, and enhance their therapeutic efficacy. NDs can also be engineered to display or release enzymes, growth factors, or drugs that can promote tissue repair or regeneration, or to act as adhesives or scaffolds that can facilitate cell migration or attachment. NDs have been explored in various disease treatments, such as cancer immunotherapy, vaccine development, antibacterial and antiviral therapy, and treatment of diabetes mellitus and Alzheimer's disease. NDs have also shown promise as imaging agents and platforms for microsurgery.Despite the promising potential of NDs in various disease treatments, several challenges and limitations need to be addressed to translate them into clinical applications. First, the scalability and reproducibility of ND production and purification need to be improved to ensure the consistency and quality of NDs. Second, the safety and toxicity of NDs need to be thoroughly evaluated, especially in terms of their long-term effects on the immune system, the liver, and the kidney. Third, the pharmacokinetics and biodistribution of NDs need to be optimized to ensure the efficient delivery and clearance of drugs or peptides. Fourth, the targeting and specificity of NDs need to be enhanced to ensure the selective and effective accumulation of drugs or peptides in the diseased tissues or cells. Fifth, the cost-effectiveness and regulatory compliance of NDs need to be considered to ensure the affordability and accessibility of the therapy.
In the future, several strategies can be explored to overcome these challenges and improve the clinical potential of NDs. First, new MSP variants or alternative scaffold proteins can be developed to enhance the stability, homogeneity, and scalability of NDs. Second, new surface modifications or coatings can be introduced to enhance the biocompatibility, immunogenicity, and targeting of NDs. Third, new encapsulation or loading strategies can be developed to enhance the loading efficiency, stability, and specificity of drugs or peptides. Fourth, new imaging or diagnostic modalities can be developed to enhance the real-time monitoring and evaluation of the therapy. Fifth, new collaborations or partnerships can be established to facilitate the translation and commercialization of the technology. Overall, NDs hold great promise as a novel disease treatment platform and have the potential to revolutionize the field of medicine in the years to come.
Author contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was sponsored by the National Natural Science Foundation of China (81925019, U22A20333, 82272144) and the Shenzhen Science and Technology Innovation Commission (JCYJ20220530143411025, JCYJ20230807091404010).References
- A. Nath, W. M. Atkins and S. G. Sligar, Biochemistry, 2007, 46, 2059–2069 CrossRef CAS PubMed.
- S. Ben-Aicha, L. Badimon and G. Vilahur, Int. J. Mol. Sci., 2020, 21, 732 CrossRef CAS PubMed.
- M. C. Phillips, J. Lipid Res., 2013, 54, 2034–2048 CrossRef CAS.
- H. Park, A. Otte and K. Park, J. Controlled Release, 2022, 342, 53–65 CrossRef CAS PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- B. Fonseca-Santos, M. P. Gremião and M. Chorilli, Int. J. Nanomed., 2015, 10, 4981–5003 CrossRef CAS PubMed.
- R. Alyautdin, I. Khalin, M. I. Nafeeza, M. H. Haron and D. Kuznetsov, Int. J. Nanomed., 2014, 9, 795–811 CAS.
- I. Vhora, R. Lalani, P. Bhatt, S. Patil and A. Misra, Int. J. Pharm., 2019, 563, 324–336 CrossRef CAS PubMed.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- S. W. Lee, Y. M. Kim, C. H. Cho, Y. T. Kim, S. M. Kim, S. Y. Hur, J. H. Kim, B. G. Kim, S. C. Kim, H. S. Ryu and S. B. Kang, Cancer Res. Treat., 2018, 50, 195–203 CrossRef CAS.
- L. Palmerston Mendes, J. Pan and V. P. Torchilin, Molecules, 2017, 22, 1401 CrossRef PubMed.
- A. R. Menjoge, R. M. Kannan and D. A. Tomalia, Drug Discovery Today, 2010, 15, 171–185 CrossRef CAS PubMed.
- J. Wang, A. M. Potocny, J. Rosenthal and E. S. Day, ACS Omega, 2020, 5, 926–940 CrossRef CAS PubMed.
- L. S. Arias, J. P. Pessan, A. P. M. Vieira, T. M. T. Lima, A. C. B. Delbem and D. R. Monteiro, Antibiotics, 2018, 7, 46 CrossRef PubMed.
- K. W. Huang, F. F. Hsu, J. T. Qiu, G. J. Chern, Y. A. Lee, C. C. Chang, Y. T. Huang, Y. C. Sung, C. C. Chiang, R. L. Huang, C. C. Lin, T. K. Dinh, H. C. Huang, Y. C. Shih, D. Alson, C. Y. Lin, Y. C. Lin, P. C. Chang, S. Y. Lin and Y. Chen, Sci. Adv., 2020, 6, eaax5032 CrossRef CAS PubMed.
- C. Xu, J. Nam, H. Hong, Y. Xu and J. J. Moon, ACS Nano, 2019, 13, 12148–12161 CrossRef CAS PubMed.
- R. Kuai, D. Li, Y. E. Chen, J. J. Moon and A. Schwendeman, ACS Nano, 2016, 10, 3015–3041 CrossRef CAS PubMed.
- S. Ben-Aicha, L. Badimon and G. Vilahur, Int. J. Mol. Sci., 2020, 21, 732 CrossRef CAS PubMed.
- J. Bariwal, H. Ma, G. A. Altenberg and H. Liang, Chem. Soc. Rev., 2022, 51, 1702–1728 RSC.
- T. H. Bayburt and S. G. Sligar, FEBS Lett., 2010, 584, 1721–1727 CrossRef CAS.
- T. Nakayama, J. S. Butler, A. Sehgal, M. Severgnini, T. Racie, J. Sharman, F. Ding, S. S. Morskaya, J. Brodsky, L. Tchangov, V. Kosovrasti, M. Meys, L. Nechev, G. Wang, C. G. Peng, Y. Fang, M. Maier, K. G. Rajeev, R. Li, J. Hettinger, S. Barros, V. Clausen, X. Zhang, Q. Wang, R. Hutabarat, N. V. Dokholyan, C. Wolfrum, M. Manoharan, V. Kotelianski, M. Stoffel and D. W. Sah, Mol. Ther., 2012, 20, 1582–1589 CrossRef CAS PubMed.
- R. Kuai, L. J. Ochyl, K. S. Bahjat, A. Schwendeman and J. J. Moon, Nat. Mater., 2017, 16, 489–496 CrossRef CAS PubMed.
- J. M. Dörr, S. Scheidelaar, M. C. Koorengevel, J. J. Dominguez, M. Schäfer, C. A. van Walree and J. A. Killian, Eur. Biophys. J., 2016, 45, 3–21 CrossRef PubMed.
- M. C. Fiori, Y. Jiang, W. Zheng, M. Anzaldua, M. J. Borgnia, G. A. Altenberg and H. Liang, Sci. Rep., 2017, 7, 15227 CrossRef PubMed.
- M. E. Zoghbi, R. S. Cooper and G. A. Altenberg, J. Biol. Chem., 2016, 291, 4453–4461 CrossRef CAS PubMed.
- R. S. Cooper and G. A. Altenberg, J. Biol. Chem., 2013, 288, 20785–20796 CrossRef CAS PubMed.
- X. Y. Wang, L. Lin, R. F. Liu, M. Chen, B. L. Chen, B. He, B. He, X. L. Liang, W. B. Dai, H. Zhang, X. Q. Wang, Y. G. Wang, Z. F. Dai and Q. Zhang, Adv. Funct. Mater., 2017, 27, 1701027 CrossRef PubMed.
- M. L. Nasr, D. Baptista, M. Strauss, Z.-Y. J. Sun, S. Grigoriu, S. Huser, A. Plückthun, F. Hagn, T. Walz, J. M. Hogle and G. Wagner, Nat. Methods, 2017, 14, 49–52 CrossRef CAS PubMed.
- J. Miehling, D. Goricanec and F. Hagn, ChemBioChem, 2018, 19, 1927–1933 CrossRef CAS PubMed.
- Y. Yusuf, J. Massiot, Y.-T. Chang, P.-H. Wu, V. Yeh, P.-C. Kuo, J. Shiue and T.-Y. Yu, Langmuir, 2018, 34, 3525–3532 CrossRef CAS PubMed.
- M. Huang, M. Hu, Q. Song, H. Song, J. Huang, X. Gu, X. Wang, J. Chen, T. Kang, X. Feng, D. Jiang, G. Zheng, H. Chen and X. Gao, ACS Nano, 2015, 9, 10801–10816 CrossRef CAS PubMed.
- Q. Song, H. Song, J. Xu, J. Huang, M. Hu, X. Gu, J. Chen, G. Zheng, H. Chen and X. Gao, Mol. Pharm., 2016, 13, 3976–3987 CrossRef CAS PubMed.
- Q. Chen, G. Guan, F. Deng, D. Yang, P. Wu, S. Kang, R. Sun, X. Wang, D. Zhou, W. Dai, X. Wang, H. Zhang, B. He, D. Chen and Q. Zhang, Biomaterials, 2020, 251, 120008 CrossRef CAS PubMed.
- I. Noh, Z. Guo, J. Zhou, W. Gao, R. H. Fang and L. Zhang, ACS Nano, 2022, 2c08360, DOI:10.1021/acsnano.
- L. Yuan, Q. Chen, J. E. Riviere and Z. Lin, J. Drug Delivery Sci. Technol., 2023, 83, 104404 CrossRef CAS PubMed.
- G. Sharma, D. T. Valenta, Y. Altman, S. Harvey, H. Xie, S. Mitragotri and J. W. Smith, J. Controlled Release, 2010, 147, 408–412 CrossRef CAS PubMed.
- E. L. Dane, A. Belessiotis-Richards, C. Backlund, J. Wang, K. Hidaka, L. E. Milling, S. Bhagchandani, M. B. Melo, S. Wu, N. Li, N. Donahue, K. Ni, L. Ma, M. Okaniwa, M. M. Stevens, A. Alexander-Katz and D. J. Irvine, Nat. Mater., 2022, 21, 710–720 CrossRef CAS PubMed.
- S. Huang, L. Deng, H. Zhang, L. Wang, Y. Zhang, Q. Lin, T. Gong, X. Sun, Z. Zhang and L. Zhang, Nano Res., 2022, 15, 728–737 CrossRef CAS.
- A. Tahmasbi Rad, C. W. Chen, W. Aresh, Y. Xia, P. S. Lai and M. P. Nieh, ACS Appl. Mater. Interfaces, 2019, 11, 10505–10519 CrossRef CAS PubMed.
- H. Patel, B. Ding, K. Ernst, L. Shen, W. Yuan, J. Tang, L. R. Drake, J. Kang, Y. Li, Z. Chen and A. Schwendeman, Int. J. Nanomed., 2019, 14, 3069–3086 CrossRef CAS PubMed.
- M. C. Fiori, Y. Jiang, G. A. Altenberg and H. Liang, Sci. Rep., 2017, 7, 7432 CrossRef PubMed.
- L. Chen, C. Yu, W. Xu, Y. Xiong, P. Cheng, Z. Lin, Z. Zhang, L. Knoedler, A. C. Panayi, S. Knoedler, J. Wang, B. Mi and G. Liu, ACS Nano, 2023, 17, 3153–3167 CrossRef CAS PubMed.
- Q. Ren, S. Zhang and H. Bao, Commun. Biol., 2022, 5, 507 CrossRef CAS PubMed.
- E. L. Dane, A. Belessiotis-Richards, C. Backlund, J. Wang, K. Hidaka, L. E. Milling, S. Bhagchandani, M. B. Melo, S. Wu, N. Li, N. Donahue, K. Ni, L. Ma, M. Okaniwa, M. M. Stevens, A. Alexander-Katz and D. J. Irvine, Nat. Mater., 2022, 21, 710–720 CrossRef CAS PubMed.
- A. Hassani Najafabadi, J. Zhang, M. E. Aikins, Z. I. Najaf Abadiare, F. Liao, Y. Qin, E. B. Okeke, L. M. Scheetz, J. Nam, Y. Xu, D. Adams, P. Lester, T. Hetrick, A. Schwendeman, M. S. Wicha, A. E. Chang, Q. Li and J. J. Moon, Nano Lett., 2020, 20, 7783–7792 CrossRef CAS PubMed.
- R. Kuai, W. M. Yuan, S. Son, J. Nam, Y. Xu, Y. C. Fan, A. Schwendeman and J. J. Moon, Sci. Adv., 2018, 4, eaao1736 CrossRef PubMed.
- R. Patwa, N. Soundararajan, N. Mulchandani, S. M. Bhasney, M. Shah, S. Kumar, A. Kumar and V. Katiyar, Biopolymers, 2018, 109, e23231 CrossRef PubMed.
- P. S. Hegde and D. S. Chen, Immunity, 2020, 52, 17–35 CrossRef CAS PubMed.
- R. Kuai, X. Sun, W. Yuan, L. J. Ochyl, Y. Xu, A. Hassani Najafabadi, L. Scheetz, M. Z. Yu, I. Balwani, A. Schwendeman and J. J. Moon, J. Controlled Release, 2018, 282, 131–139 CrossRef CAS PubMed.
- P. Kadiyala, D. Li, F. M. Nuñez, D. Altshuler, R. Doherty, R. Kuai, M. Yu, N. Kamran, M. Edwards, J. J. Moon, P. R. Lowenstein, M. G. Castro and A. Schwendeman, ACS Nano, 2019, 13, 1365–1384 CAS.
- A. Harari, M. Graciotti, M. Bassani-Sternberg and L. E. Kandalaft, Nat. Rev. Drug Discovery, 2020, 19, 635–652 CrossRef CAS PubMed.
- W. Li, X. Zhang, C. Zhang, J. Yan, X. Hou, S. Du, C. Zeng, W. Zhao, B. Deng, D. W. McComb, Y. Zhang, D. D. Kang, J. Li, W. E. Carson 3rd and Y. Dong, Nat. Commun., 2021, 12, 7264 CrossRef CAS PubMed.
- J. Zhang, B. Fan, G. Cao, W. Huang, F. Jia, G. Nie and H. Wang, Adv. Mater., 2022, 34, e2205950 CrossRef PubMed.
- N. Gong and M. J. Mitchell, Nat. Mater., 2022, 21, 616–617 CrossRef CAS PubMed.
- P. Lin, Y. Xue, X. Mu, Y. Shao, Q. Lu, X. Jin, E. Yinwang, Z. Zhang, H. Zhou, W. Teng, H. Sun, W. Chen, W. Shi, C. Shi, X. Zhou, X. Jiang, X. Yu and Z. Ye, Small, 2022, 18, e2200179 CrossRef PubMed.
- X. Liu, C. Liu, Z. Zheng, S. Chen, X. Pang, X. Xiang, J. Tang, E. Ren, Y. Chen, M. You, X. Wang, X. Chen, W. Luo, G. Liu and N. Xia, Adv. Mater., 2019, 31, e1808294 CrossRef PubMed.
- C. Liu, X. Liu, X. Xiang, X. Pang, S. Chen, Y. Zhang, E. Ren, L. Zhang, X. Liu, P. Lv, X. Wang, W. Luo, N. Xia, X. Chen and G. Liu, Nat. Nanotechnol., 2022, 17, 531–540 CrossRef CAS PubMed.
- V. Yeh, T.-Y. Lee, C.-W. Chen, P.-C. Kuo, J. Shiue, L.-K. Chu and T.-Y. Yu, Sci. Rep., 2018, 8, 13501 CrossRef PubMed.
- R. Kuai, L. J. Ochyl, K. S. Bahjat, A. Schwendeman and J. J. Moon, Nat. Mater., 2017, 16, 489–496 CrossRef CAS PubMed.
- R. Kuai, X. Sun, W. Yuan, Y. Xu, A. Schwendeman and J. J. Moon, Bioconjug Chem., 2018, 29, 771–775 CrossRef CAS PubMed.
- A. H. Najafabadi, Z. I. N. Abadi, M. E. Aikins, K. E. Foulds, M. M. Donaldson, W. Yuan, E. B. Okeke, J. Nam, Y. Xu, P. Weerappuli, T. Hetrick, D. Adams, P. A. Lester, A. M. Salazar, D. H. Barouch, A. Schwendeman, R. A. Seder and J. J. Moon, J. Controlled Release, 2021, 337, 168–178 CrossRef CAS PubMed.
- P. Marcato, C. A. Dean, C. A. Giacomantonio and P. W. Lee, Cell Cycle, 2011, 10, 1378–1384 CrossRef CAS PubMed.
- C. Ginestier, M. H. Hur, E. Charafe-Jauffret, F. Monville, J. Dutcher, M. Brown, J. Jacquemier, P. Viens, C. G. Kleer, S. Liu, A. Schott, D. Hayes, D. Birnbaum, M. S. Wicha and G. Dontu, Cell Stem Cell, 2007, 1, 555–567 CrossRef CAS PubMed.
- K. Rantalainen, Z. T. Berndsen, A. Antanasijevic, T. Schiffner, X. Zhang, W. H. Lee, J. L. Torres, L. Zhang, A. Irimia, J. Copps, K. H. Zhou, Y. D. Kwon, W. H. Law, C. A. Schramm, R. Verardi, S. J. Krebs, P. D. Kwong, N. A. Doria-Rose, I. A. Wilson, M. B. Zwick, J. R. Yates 3rd, W. R. Schief and A. B. Ward, Cell Rep., 2020, 31, 107583 CrossRef CAS PubMed.
- S. Yang, G. Hiotis, Y. Wang, J. Chen, J. H. Wang, M. Kim, E. L. Reinherz and T. Walz, Nat. Commun., 2022, 13, 6393 CrossRef CAS PubMed.
- Y. Wang, P. Kaur, Z. J. Sun, M. A. Elbahnasawy, Z. Hayati, Z. S. Qiao, N. N. Bui, C. Chile, M. L. Nasr, G. Wagner, J. H. Wang, L. Song, E. L. Reinherz and M. Kim, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 22556–22566 CrossRef CAS PubMed.
- J. Wang, Y. Zhang, E. Archibong, F. S. Ligler and Z. Gu, Adv. Biosyst., 2017, 1, e1700084 CrossRef PubMed.
- Z. Xu, Z. Qiu, Q. Liu, Y. Huang, D. Li, X. Shen, K. Fan, J. Xi, Y. Gu, Y. Tang, J. Jiang, J. Xu, J. He, X. Gao, Y. Liu, H. Koo, X. Yan and L. Gao, Nat. Commun., 2018, 9, 3713 CrossRef CAS PubMed.
- J. Ouyang, M. Wen, W. Chen, Y. Tan, Z. Liu, Q. Xu, K. Zeng, L. Deng and Y. N. Liu, Chem. Commun., 2019, 55, 4877–4880 RSC.
- C. Zhang, Q. Li, J. Shan, J. Xing, X. Liu, Y. Ma, H. Qian, X. Chen, X. Wang, L. M. Wu and Y. Yu, Acta Biomater., 2023, 160, 252–264 CrossRef CAS PubMed.
- S. Speerstra, A. A. Chistov, G. V. Proskurin, A. V. Aralov, E. A. Ulashchik, P. P. Streshnev, V. V. Shmanai, V. A. Korshun and L. M. Schang, Antiviral Res., 2018, 149, 164–173 CrossRef CAS PubMed.
- M. C. Wolf, A. N. Freiberg, T. Zhang, Z. Akyol-Ataman, A. Grock, P. W. Hong, J. Li, N. F. Watson, A. Q. Fang, H. C. Aguilar, M. Porotto, A. N. Honko, R. Damoiseaux, J. P. Miller, S. E. Woodson, S. Chantasirivisal, V. Fontanes, O. A. Negrete, P. Krogstad, A. Dasgupta, A. Moscona, L. E. Hensley, S. P. Whelan, K. F. Faull, M. R. Holbrook, M. E. Jung and B. Lee, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 3157–3162 CrossRef CAS PubMed.
- S. Skalickova, Z. Heger, L. Krejcova, V. Pekarik, K. Bastl, J. Janda, F. Kostolansky, E. Vareckova, O. Zitka, V. Adam and R. Kizek, Viruses, 2015, 7, 5428–5442 CrossRef CAS PubMed.
- B. Kong, S. Moon, Y. Kim, P. Heo, Y. Jung, S. H. Yu, J. Chung, C. Ban, Y. H. Kim, P. Kim, B. J. Hwang, W. J. Chung, Y. K. Shin, B. L. Seong and D. H. Kweon, Nat. Commun., 2019, 10, 185 CrossRef PubMed.
- L. R. Walker and M. T. Marty, Biochemistry, 2020, 59, 2135–2142 CrossRef CAS PubMed.
- Y. Ikeda, T. Taira, K. Sakai, H. Sakai, Y. Shigeri and T. Imura, J. Oleo Sci., 2018, 67, 1035–1041 CrossRef CAS PubMed.
- V. W. Henderson and C. E. Finch, J. Neurosurg., 1989, 70, 335–353 CAS.
- R. Kandimalla, V. Thirumala and P. H. Reddy, Biochim. Biophys. Acta, Mol. Basis Dis., 2017, 1863, 1078–1089 CrossRef CAS PubMed.
- K. G. Mawuenyega, W. Sigurdson, V. Ovod, L. Munsell, T. Kasten, J. C. Morris, K. E. Yarasheski and R. J. Bateman, Science, 2010, 330, 1774 CrossRef CAS PubMed.
- J. M. Castellano, J. Kim, F. R. Stewart, H. Jiang, R. B. DeMattos, B. W. Patterson, A. M. Fagan, J. C. Morris, K. G. Mawuenyega, C. Cruchaga, A. M. Goate, K. R. Bales, S. M. Paul, R. J. Bateman and D. M. Holtzman, Sci. Transl. Med., 2011, 3, 89ra57 CAS.
- K. R. Wildsmith, M. Holley, J. C. Savage, R. Skerrett and G. E. Landreth, Alzheimers Res. Ther., 2013, 5, 33 CrossRef CAS PubMed.
- P. Zhang, Y. Kishimoto, I. Grammatikakis, K. Gottimukkala, R. G. Cutler, S. Zhang, K. Abdelmohsen, V. A. Bohr, J. Misra Sen, M. Gorospe and M. P. Mattson, Nat. Neurosci., 2019, 22, 719–728 CrossRef CAS PubMed.
- Y. Zhao, J. Cai, Z. Liu, Y. Li, C. Zheng, Y. Zheng, Q. Chen, H. Chen, F. Ma, Y. An, L. Xiao, C. Jiang, L. Shi, C. Kang and Y. Liu, Nano Lett., 2019, 19, 674–683 CrossRef CAS PubMed.
- T. H. Ferreira-Vieira, I. M. Guimaraes, F. R. Silva and F. M. Ribeiro, Curr. Neuropharmacol., 2016, 14, 101–115 CrossRef CAS PubMed.
- N. N. Nalivaeva and A. J. Turner, Chem.-Biol. Interact., 2016, 259, 301–306 CrossRef CAS PubMed.
- T. Wisniewski and F. Goñi, Neuron, 2015, 85, 1162–1176 CrossRef CAS PubMed.
- S. Mourtas, M. Canovi, C. Zona, D. Aurilia, A. Niarakis, B. La Ferla, M. Salmona, F. Nicotra, M. Gobbi and S. G. Antimisiaris, Biomaterials, 2011, 32, 1635–1645 CrossRef CAS PubMed.
- D. Brambilla, R. Verpillot, B. Le Droumaguet, J. Nicolas, M. Taverna, J. Kóňa, B. Lettiero, S. H. Hashemi, L. De Kimpe, M. Canovi, M. Gobbi, V. Nicolas, W. Scheper, S. M. Moghimi, I. Tvaroška, P. Couvreur and K. Andrieux, ACS Nano, 2012, 6, 5897–5908 CrossRef CAS PubMed.
- M. Gobbi, F. Re, M. Canovi, M. Beeg, M. Gregori, S. Sesana, S. Sonnino, D. Brogioli, C. Musicanti, P. Gasco, M. Salmona and M. E. Masserini, Biomaterials, 2010, 31, 6519–6529 CrossRef CAS PubMed.
- J. G. Tsun, S. Yung, M. K. Chau, S. W. Shiu, T. M. Chan and K. C. Tan, PLoS One, 2014, 9, e105787 CrossRef PubMed.
- G. Russo, P. Piscitelli, A. Giandalia, F. Viazzi, R. Pontremoli, P. Fioretto and S. De Cosmo, J. Nephrol., 2020, 33, 1001–1008 CrossRef CAS PubMed.
- T. Toyama, M. Shimizu, K. Furuichi, S. Kaneko and T. Wada, Clin. Exp. Nephrol., 2014, 18, 201–205 CrossRef CAS PubMed.
- G. M. Ducasa, A. Mitrofanova and A. Fornoni, Curr. Diab. Rep., 2019, 19, 144 CrossRef CAS PubMed.
- P. Liu, L. Peng, H. Zhang, P. M. Tang, T. Zhao, M. Yan, H. Zhao, X. Huang, H. Lan and P. Li, Front. Physiol., 2018, 9, 343 CrossRef PubMed.
- E. E. Morin, Y. Guo, H. He, W. Yuan, W. N. Souery, M. V. Fawaz, Y. E. Chen and A. Schwendeman, Front. Pharmacol., 2020, 11, 513031 CrossRef CAS PubMed.
- E. Kiss, B. Kränzlin, K. Wagenblaβ, M. Bonrouhi, J. Thiery, E. Gröne, V. Nordström, D. Teupser, N. Gretz, E. Malle and H. J. Gröne, Am. J. Pathol., 2013, 182, 727–741 CrossRef CAS PubMed.
- Y. Wang, S. Xiao, S. Zhou, R. Zhang, H. Liu, Y. Lin and P. Yu, Front. Physiol., 2020, 11, 552483 CrossRef PubMed.
- K. Feng, C. Ma, Y. Liu, X. Yang, Z. Yang, Y. Chen, T. Xu, C. Yang, S. Zhang, Q. Li, Z. Wei, D. Zhao, P. Zeng, J. Han, J. Gao, Y. Chen and Y. Duan, Theranostics, 2021, 11, 2634–2654 CrossRef CAS PubMed.
- F. Oroojalian, F. Charbgoo, M. Hashemi, A. Amani, R. Yazdian-Robati, A. Mokhtarzadeh, M. Ramezani and M. R. Hamblin, J. Controlled Release, 2020, 321, 442–462 CrossRef CAS PubMed.
- G. Wang, Q. Li, D. Chen, B. Wu, Y. Wu, W. Tong and P. Huang, Theranostics, 2019, 9, 6191–6208 CrossRef CAS PubMed.
- H. He, R. Adili, L. Liu, K. Hong, M. Holinstat and A. Schwendeman, Sci. Adv., 2020, 6, eabd0130 CrossRef CAS PubMed.
- H. He, T. A. Halseth, L. Mei, C. Shen, L. Liu and A. Schwendeman, J. Controlled Release, 2022, 348, 1016–1027 CrossRef CAS PubMed.
- S. Amadesi, C. Reni, R. Katare, M. Meloni, A. Oikawa, A. P. Beltrami, E. Avolio, D. Cesselli, O. Fortunato, G. Spinetti, R. Ascione, E. Cangiano, M. Valgimigli, S. P. Hunt, C. Emanueli and P. Madeddu, Circulation, 2012, 125, 1774–1786 CrossRef CAS PubMed.
- H. S. Hong, S. Kim, S. Nam, J. Um, Y. H. Kim and Y. Son, Wound Repair Regen., 2015, 23, 268–277 CrossRef PubMed.
- J. J. Bowden, A. M. Garland, P. Baluk, P. Lefevre, E. F. Grady, S. R. Vigna, N. W. Bunnett and D. M. McDonald, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 8964–8968 CrossRef CAS PubMed.
- H. J. Park, R. Kuai, E. J. Jeon, Y. Seo, Y. Jung, J. J. Moon, A. Schwendeman and S. W. Cho, Biomaterials, 2018, 161, 69–80 CrossRef CAS PubMed.
- M. Hoekstra, Atherosclerosis, 2017, 258, 153–161 CrossRef CAS PubMed.
- W. L. Wong, X. Su, X. Li, C. M. Cheung, R. Klein, C. Y. Cheng and T. Y. Wong, Lancet Global Health, 2014, 2, e106–e116 CrossRef PubMed.
- E. Loane, J. M. Nolan and S. Beatty, Invest. Ophthalmol. Vis. Sci., 2010, 51, 5897–5905 CrossRef PubMed.
- Y. Wang, S. L. Connor, W. Wang, E. J. Johnson and W. E. Connor, Exp. Eye Res., 2007, 84, 591–598 CrossRef CAS PubMed.
- M. N. Oda, P. L. Hargreaves, J. A. Beckstead, K. A. Redmond, R. van Antwerpen and R. O. Ryan, J. Lipid Res., 2006, 47, 260–267 CrossRef CAS PubMed.
- M. Ghosh, A. T. Singh, W. Xu, T. Sulchek, L. I. Gordon and R. O. Ryan, Nanomedicine, 2011, 7, 162–167 CrossRef CAS PubMed.
- A. Moschetti, L. N. Vine, K. Lethcoe, R. K. Dagda, P. Ellison and R. O. Ryan, Lipids, 2020, 55, 141–149 CrossRef CAS PubMed.
- A. Moschetti, C. A. Fox, S. McGowen and R. O. Ryan, Front. Nanotechnol., 2022, 4, 955022 CrossRef PubMed.
- B. Park, Y. K. Cha, J. Kwak, K. S. Hwang, H. S. Kim, S. Park, Y. Pak, T. H. Park, H. S. Song and J. H. Kim, Nano Lett., 2022, 22, 6825–6832 CrossRef CAS PubMed.
- T. Murakami, Biotechnol. J., 2012, 7, 762–767 CrossRef CAS PubMed.
- M. Niu, Y. Lu, L. Hovgaard and W. Wu, Int. J. Nanomed., 2011, 6, 1155–1166 CAS.
- N. T. Huynh, C. Passirani, P. Saulnier and J. P. Benoit, Int. J. Pharm., 2009, 379, 201–209 CrossRef CAS PubMed.
- S. Krol, R. Ellis-Behnke and P. Marchetti, Maturitas, 2012, 73, 61–67 CrossRef CAS PubMed.
- A. K. Agrawal, H. Harde, K. Thanki and S. Jain, Biomacromolecules, 2014, 15, 350–360 CrossRef CAS PubMed.
- A. Nath, W. M. Atkins and S. G. Sligar, Biochemistry, 2007, 46, 2059–2069 CrossRef CAS PubMed.
- A. Nath, A. J. Trexler, P. Koo, A. D. Miranker, W. M. Atkins and E. Rhoades, Methods Enzymol., 2010, 472, 89–117 CAS.
- D. C. Carter and J. X. Ho, Adv. Protein. Chem., 1994, 45, 153–203 CrossRef CAS PubMed.
- J. S. Mandeville and H. A. Tajmir-Riahi, Biomacromolecules, 2010, 11, 465–472 CrossRef CAS PubMed.
- S. Damiati, A. Scheberl, S. Zayni, S. A. Damiati, B. Schuster and U. B. Kompella, Biophys. Chem., 2019, 251, 106178 CrossRef CAS PubMed.
- X. Jia, Y. K. Y. Chin, A. H. Zhang, T. Crawford, Y. Zhu, N. L. Fletcher, Z. Zhou, B. R. Hamilton, M. Stroet, K. J. Thurecht and M. Mobli, Commun. Chem., 2023, 6, 48 CrossRef PubMed.
- J. Ferlay, E. Steliarova-Foucher, J. Lortet-Tieulent, S. Rosso, J. W. Coebergh, H. Comber, D. Forman and F. Bray, Eur. J. Cancer, 2013, 49, 1374–1403 CrossRef CAS PubMed.
- G. C. Barnett, C. M. West, A. M. Dunning, R. M. Elliott, C. E. Coles, P. D. Pharoah and N. G. Burnet, Nat. Rev. Cancer, 2009, 9, 134–142 CrossRef CAS PubMed.
- T. N. Zamay, V. S. Prokopenko, S. S. Zamay, K. A. Lukyanenko, O. S. Kolovskaya, V. A. Orlov, G. S. Zamay, R. G. Galeev, A. A. Narodov and A. S. Kichkailo, Nanomaterials, 2021, 11, 1459 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |