Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sunlight-mediated inactivation of health-relevant microorganisms in water: a review of mechanisms and modeling approaches
Kara L.
Nelson
 *a,
Alexandria B.
Boehm
*a,
Alexandria B.
Boehm
 b,
Robert J.
Davies-Colley
c,
Michael C.
Dodd
d,
Tamar
Kohn
e,
Karl. G.
Linden
f,
Yuanyuan
Liu
g,
Peter A.
Maraccini
b,
Kristopher
McNeill
b,
Robert J.
Davies-Colley
c,
Michael C.
Dodd
d,
Tamar
Kohn
e,
Karl. G.
Linden
f,
Yuanyuan
Liu
g,
Peter A.
Maraccini
b,
Kristopher
McNeill
 h,
William A.
Mitch†
h,
William A.
Mitch†
 b,
Thanh H.
Nguyen
i,
Kimberly M.
Parker
b,
Thanh H.
Nguyen
i,
Kimberly M.
Parker
 j,
Roberto A.
Rodriguez
k,
Lauren M.
Sassoubre
l,
Andrea I.
Silverman
j,
Roberto A.
Rodriguez
k,
Lauren M.
Sassoubre
l,
Andrea I.
Silverman
 m,
Krista R.
Wigginton
n and
Richard G.
Zepp
o
m,
Krista R.
Wigginton
n and
Richard G.
Zepp
o
aCivil and Environmental Engineering, University of California, Berkeley, CA, USA. E-mail: karanelson@berkeley.edu; Tel: +1 510 643 5023
bCivil and Environmental Engineering, Stanford University, Stanford, CA, USA
cNational Institute of Water and Atmospheric Research Ltd., Hamilton, New Zealand
dCivil and Environmental Engineering, University of Washington, Seattle, WA, USA
eCivil and Environmental Engineering, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
fCivil, Environmental, and Architectural Engineering, University of Colorado, Boulder, CO, USA
gSchool of Earth Sciences and Engineering, Nanjing University, Nanjing, China
hInstitute for Biogeochemistry and Pollutant Dynamics, ETH, Zurich, Switzerland
iCivil and Environmental Engineering, University of Illinois, Urbana-Champaign, Urbana, IL, USA
jDepartment of Energy, Environmental and Chemical Engineering, Washington University, St. Louis, MO, USA
kEpidemiology, Human Genetics and Environmental Sciences, University of Texas Health Science Center, El Paso, TX, USA
lCivil, Structural, and Environmental Engineering, University at Buffalo, NY, USA
mCivil and Urban Engineering, New York University, NY, USA
nCivil and Environmental Engineering, University of Michigan, Ann Arbor, MI, USA
oNational Exposure Research Laboratory, US Environmental Protection Agency, Athens, GA, USA
First published on 26th July 2018
Abstract
Health-relevant microorganisms present in natural surface waters and engineered treatment systems that are exposed to sunlight can be inactivated by a complex set of interacting mechanisms. The net impact of sunlight depends on the solar spectral irradiance, the susceptibility of the specific microorganism to each mechanism, and the water quality; inactivation rates can vary by orders of magnitude depending on the organism and environmental conditions. Natural organic matter (NOM) has a large influence, as it can attenuate radiation and thus decrease inactivation by endogenous mechanisms. Simultaneously NOM sensitizes the formation of reactive intermediates that can damage microorganisms via exogenous mechanisms. To accurately predict inactivation and design engineered systems that enhance solar inactivation, it is necessary to model these processes, although some details are not yet sufficiently well understood. In this critical review, we summarize the photo-physics, -chemistry, and -biology that underpin sunlight-mediated inactivation, as well as the targets of damage and cellular responses to sunlight exposure. Viruses that are not susceptible to exogenous inactivation are only inactivated if UVB wavelengths (280–320 nm) are present, such as in very clear, open waters or in containers that are transparent to UVB. Bacteria are susceptible to slightly longer wavelengths. Some viruses and bacteria (especially Gram-positive) are susceptible to exogenous inactivation, which can be initiated by visible as well as UV wavelengths. We review approaches to model sunlight-mediated inactivation and illustrate how the environmental conditions can dramatically shift the inactivation rate of organisms. The implications of this mechanistic understanding of solar inactivation are discussed for a range of applications, including recreational water quality, natural treatment systems, solar disinfection of drinking water (SODIS), and enhanced inactivation via the use of sensitizers and photocatalysts. Finally, priorities for future research are identified that will further our understanding of the key role that sunlight disinfection plays in natural systems and the potential to enhance this process in engineered systems.
Environmental significanceThe manuscript provides a comprehensive synthesis of the current understanding of the mechanisms by which sunlight causes damage to microorganisms, ultimately leading to inactivation. This topic is important for understanding the fate and transport of microbiological contaminants in all sunlit surface waters, including fresh and marine ecosystems, as well as engineered treatment systems. |
1. Introduction
Sunlight has long been recognized as a disinfectant. The sunlight-mediated inactivation of microorganisms is relevant in many types of applications and to many aquatic environments. In both fresh and marine surface waters, sunlight-mediated damage influences microbial ecology, with implications for microbial food webs and microbially mediated biogeochemical processes.1 It also strongly influences the persistence of human pathogens and indicator organisms in contaminated waters (e.g., sunlight is a major determinant of swimming beach water quality).2 Sunlight is the key factor contributing to inactivation of indicator organisms and pathogens in engineered natural systems like wastewater treatment ponds (WTP)3 and open-water wetlands for treatment of wastewater and stormwater.4 Solar disinfection of drinking water (SODIS) is promoted around the world as a low-cost method for household water treatment.5,6 The goal of this paper is to review the tremendous progress that has been made in the last several decades in understanding the mechanisms by which sunlight damages health-relevant microorganisms in water. Based on this understanding, we present a mechanistic approach for modeling inactivation, discuss the implications of sunlight-mediated inactivation for common applications in the field of water quality, and identify knowledge gaps and research priorities. The review focuses on mechanisms that occur in both viruses and bacteria, including indicator organisms and human pathogens, because sunlight inactivation is most relevant and best understood for these two classes of microorganisms. Short sections review sunlight inactivation of protozoan cysts and antibiotic resistance genes.2. Conceptual model of sunlight inactivation
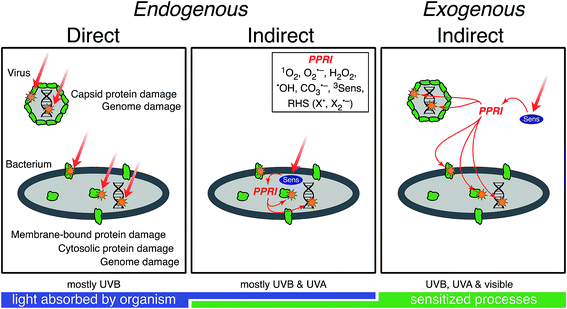
Sunlight-mediated inactivation is a type of photoinactivation, a term that also includes disinfection by artificial radiation sources whose spectral irradiance typically differs appreciably from that of sunlight. Although the emphasis of this review is natural sunlight, the discussion of mechanisms is also relevant to artificial radiation sources. Indeed, much of the information on solar inactivation comes from experiments with artificial sources. A conceptual model of photoinactivation mechanisms of viruses and bacteria is shown in Fig. 1. This conceptual model provides a framework for discussing the underlying principles and mechanisms in more detail in subsequent sections. Direct photoinactivation occurs when a chromophore endogenous to the microorganism (e.g., nucleic acids, proteins, or other macromolecules that occur in microorganisms) absorbs a photon resulting in changes to the chemical structure of the chromophore. Indirect photoinactivation occurs when an endogenous (a constituent of the microorganism) or exogenous (not a constituent of the microorganism) chromophore absorbs a photon and sensitizes the production of photo-produced reactive intermediates (PPRI) that, in turn, damage virus or cell components. Chromophores that produce PPRI are called sensitizers. As indicated in Fig. 1, viruses are primarily damaged through endogenous direct and exogenous indirect mechanisms, whereas all three mechanisms may contribute to bacterial inactivation. Although the three mechanisms are described separately, they likely occur simultaneously and interact, especially in bacteria. For example, direct damage to a bacterial enzyme, such as catalase, could exacerbate indirect inactivation by causing higher levels of photo-chemically produced hydrogen peroxide to persist within a bacterial cell.3. Solar irradiance and water optics
Different regions of the solar spectrum contribute to the three main mechanisms of damage, as shown in Fig. 1. Endogenous direct damage is primarily initiated by photons in the UVB range (280–320 nm) whereas endogenous indirect damage can involve photons in the UVB and UVA (320–400 nm) ranges. Photons in the UVB, UVA, and visible (400–700 nm) light regions can contribute to exogenous damage. The main reason for this dependence on wavelength is that different chromophores are involved, with different absorption spectra and quantum yields, as reviewed in Section 4. An implication of this dependence on wavelength is that because sunlight can vary appreciably in spectral quality, particularly in the UV range and underwater, the mechanisms contributing to solar inactivation of microorganisms may vary with solar zenith angle (a function of latitude, time of year, and time of day), atmospheric conditions, water quality, and depth in the water column.In Fig. 2, we provide examples of spectral irradiance of sunlight for different zenith angles, total atmospheric ozone concentrations, and for an overcast sky. The spectral quality of solar irradiance is fairly consistent throughout the visible and UVA range despite major changes in the magnitude of solar irradiance. UVB wavelengths, however, are preferentially absorbed by atmospheric ozone. Differential absorption of the sunlight spectrum is exacerbated when the sun is lower in the sky due to the longer path through the atmosphere (i.e., larger air mass). For example, while UVA and visible light vary seasonally in irradiance by about a factor of two between summer and winter at mid-latitudes, UVB varies by a factor of four (Table 1). A similar effect occurs over the course of a day. During the equinox, at mid-latitudes, the UVA and visible light intensities reach 50% of their maximum value about four hours before solar noon, while UVB reaches the 50% mark almost a full hour later (the UVB “sunrise” and “sunset” lag and precede visible sunrise and sunset7). Due to these large differences in irradiance, we can expect the sunlight-mediated inactivation rate to vary by several orders of magnitude as a function of location, season, time of day, and weather conditions.
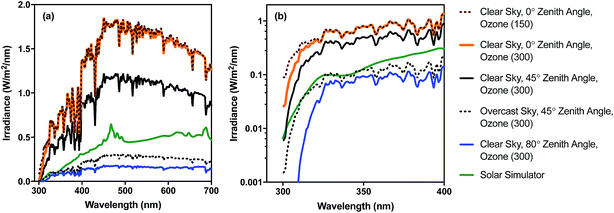 | ||
| Fig. 2 Spectral irradiance of sunlight under different conditions for (a) UV-visible range (300–700 nm) and (b) UV range (300–400 nm) shown using log scale. Sunlight spectra were generated with the RADTRANX‡ routine in Hydrolight5 (ref. 8) for varying ozone concentration (300 ppb is approx. average in the stratosphere), solar altitudes (zenith angle), and overcast versus clear sun. The solar simulator spectrum is for a 1000 W Oriel simulator with airmass and atmospheric attenuation filters, measured using a Stellarnet spectroradiometer, as reported in Silverman and Nelson (2016). | ||
| Radiation type | Wavelength range (nm) | Irradiance, E (μmol photons per m2 per s) | E summer/Ewinter | |
|---|---|---|---|---|
| Dec 21 | Jun 21 | |||
| a SMARTS was used for generating these values (reference atmosphere), because it can account for wavelengths down to 280 nm. A limitation of SMARTS is that it does not account for the impact of cloud cover. Ozone concentration = 300 Dobson units. | ||||
| UVB | 280–320 | 2.0 | 8.4 | 4.2 |
| UVA | 320–400 | 76.2 | 174 | 2.3 |
| Visible | 400–700 | 1010 | 2050 | 2.0 |
As solar radiation penetrates waters it undergoes further spectral shifts due to wavelength-dependent irradiance attenuation (spectral filtering) by water; as a result, the water quality and water depth also exert significant influence over the sunlight-mediated inactivation rates. The transmission of irradiance over a depth interval in the water column can be described as:10,11
| Ed(z, λ) = Ed(0, λ)e−Kd(λ)z | (1) |
In very clear natural waters, the absorption spectrum (and attenuation spectrum) is dominated by water, such that spectral irradiance tends to be concentrated in the blue-visible window near the attenuation minimum (Fig. 3), giving such waters their blue color. However, most other constituents of natural waters result in preferential attenuation of shorter wavelengths. The main dissolved substance in natural waters that attenuates radiation is natural organic matter (NOM), specifically the colored dissolved organic matter (CDOM), whose absorbance increases exponentially with declining wavelength, resulting in yellow or orange (visible) light penetrating most strongly. Thus, UV wavelengths are strongly attenuated by CDOM in most natural waters. For example, irradiance attenuation in the solar UV is nearly 1000-fold higher in water from the humic-stained Lake Hochstetter, New Zealand, than in the very clearest natural waters (Fig. 3); this effect has been observed in many natural waters.12 Consequently, enhancements in CDOM concentrations caused by runoff tend to reduce inactivation by UVB whereas droughts that reduce runoff result in deeper UVB penetration that enhances inactivation.12
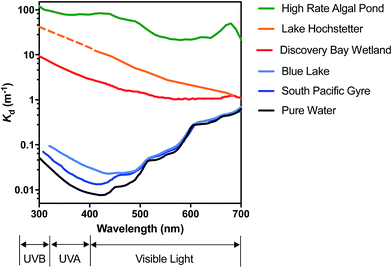 | ||
| Fig. 3 Spectral irradiance attenuation in contrasting waters: pure water, the clearest known seawater on earth (S. Pacific Gyre near Easter Island14), a clear lake water (Blue Lake, NZ (ref. 15)), a humic-stained lake (Lake Hochstetter, NZ;16 UV data are extrapolated), a constructed wetland for polishing wastewater (Discover Bay wetland, CA17); and a ‘super-eutrophic’ water laden with phytoplankton (a high-rate algal pond treating wastewater18). | ||
Suspended particles exacerbate this bias against shorter wavelengths in colored waters, since short wavelengths are more efficiently scattered by particles, which increases their average pathlength and therefore their absorption. For example, suspended sediments in turbid waters such as the Mississippi River can dominate attenuation of solar UV radiation.13 In eutrophic waters, with high phytoplankton concentrations, irradiance attenuation has appreciable spectral structure owing to light absorption by chlorophyll-a (with two absorption peaks at about 440 nm and 676 nm) and accessory photosynthetic pigments, resulting in green (visible) light penetrating deepest. Solar UV is also strongly attenuated in these eutrophic waters. The spectral irradiance attenuation in a high rate algal pond (common wastewater treatment technology; Fig. 3) has broadly similar spectral shape to eutrophic waters generally, and with attenuation 3000-fold greater than in pure water in the solar UVB range.
Due to the greater attenuation of UVB, the relative importance of UVA and visible light compared to UVB increases with depth. A key implication for sunlight inactivation is that exogenous processes become relatively more important with increasing light attenuation (or depth in the water column). Because microorganisms have differing susceptibility to endogenous and exogenous mechanisms, this spectral filtering can lead to large shifts in the relative photoinactivation rates between organisms with water depth (see Section 8).
4. Photochemistry and photobiology fundamentals
4.1. Chromophores and sensitizers
The first step in photoinactivation is absorption of a photon by a chromophore (Fig. 1), but the chromophores involved in endogenous and exogenous processes are markedly different. In viruses, chromophores involved in the endogenous direct and indirect inactivation are limited to amino acids (tryptophan, tyrosine, cysteine disulfide) and nucleic acid bases that primarily absorb light in the UVB range.19 In bacteria, chromophores also include coenzymes, vitamins and metalloproteins (see Table 2); therefore the range of light absorption is wider and encompasses the UVB, UVA, and visible light ranges (Table 2). While there is documented evidence that some chromophores undergo direct damage (e.g., nucleotide bases) and others act as sensitizers (e.g., riboflavin), it is likely that most chromophores experience both direct damage and initiate sensitized reactions (i.e., most chromophores are also sensitizers).| Compound | Absorbance wavelength range | ||
|---|---|---|---|
| UVB (280–320 nm) | UVA (320–400 nm) | Visible (400–700 nm) | |
| Endogenous chromophores in viruses and bacteria | |||
| DNA | + | − | − |
| RNA | + | − | − |
| Proteins (Trp, Tyr, CysS) | + | − | − |
| 4-Thiouracil | + | + | − |
| NADH | + | + | − |
| Flavins (e.g., riboflavin) | + | + | + |
| Porphyrins (e.g., cytochromes) | + | + | + |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Exogenous chromophores in natural waters | |||
| CDOM | ++ | + | + |
| Nitrate | + | − | − |
| Nitrite | + | − | − |
| Metal complexes | + | + | + |
Exogenous sensitizers are derived from the environment, with organic matter being the most important class. CDOM absorbs light over the UVB, UVA and visible range, though the absorption decreases exponentially with increasing wavelength (Table 2). This exponential decrease in absorption can be characterized as
aCDOM,λ = aCDOM,λ0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e−S(λ − λ0) e−S(λ − λ0) | (2) |
4.2. Photochemical reactions of chromophores (direct and indirect)
A chromophore (CHROM) that absorbs a photon is promoted to an excited singlet state (1CHROM*; Fig. 4). 1CHROM* are short-lived (nanosecond lifetimes) and generally return to their ground states, emitting heat or light (fluorescence), although some undergo intersystem crossing (ISC) to longer-lived (microsecond lifetimes) excited triplet states (3CHROM*).291CHROM* or 3CHROM* may directly undergo photochemical transformation, resulting in endogenous direct inactivation. The best-studied chemical structures in biomolecules that promote direct photoreactions are adjacent pyrimidine nucleobases (C, T or U), which can dimerize upon irradiation;30 pyrimidine hydrates can also be formed.31 Double-stranded nucleic acids are generally less photoreactive than single-stranded nucleic acids. In RNA, uracil dimer reactions have lower quantum yields than the corresponding thymine dimer reactions in DNA.27,32–34 In contrast, hydrate pyrimidine products form to a greater extent in RNA than DNA due to the low quantum yields of the thymidine hydrate reactions compared to uracil hydrate reactions.35 The extent of nucleic acid photoproduct formation is dependent on solution pH36 and ionic strength.37 Nucleic acids sequence and structure also have significant impacts on base photoreactivity.38 Although most research on the direct photolysis of nucleic acids have focused on UVC wavelengths, the pyrimidine products can also form by UVA and UVB.30,39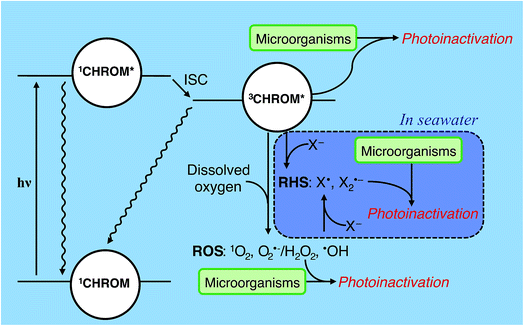 | ||
| Fig. 4 Indirect photoinactivation sensitizers and intermediates. CHROM refers to both endogenous chromophores and exogenous chromophores (see Table 2). Reactive oxygen species (ROS) can be formed by all sensitizers when oxygen is present. Carbonate radicals (not shown) may affect exogenous photoinactivation under some conditions. Reactive halogen species (RHS) may contribute to exogenous photoinactivation, particularly in seawater, but this has yet to be confirmed experimentally. ISC = intersystem crossing; X = halide. | ||
Besides direct photoreactions, 3CHROM* can furthermore promote reactions of biomolecules through sensitized processes, resulting in endogenous or exogenous indirect inactivation. Sensitized photooxidations include 3CHROM* acting directly as an oxidant, or acting as a sensitizer and promoting the formation of PPRI, such as reactive oxygen species (ROS).23,40–42 ROS include: (1) singlet oxygen (1O2) formed by energy transfer to dissolved oxygen, (2) superoxide (O2˙−) and hydrogen peroxide (H2O2) formed by electron and proton transfer to dissolved oxygen, and (3) hydroxyl radical (˙OH) formed by processes involving 3CHROM*, but also including other processes, such as the photolysis of nitrate or nitrite and Fenton reactions involving dissolved iron and H2O2.21,40–42 In some waters, other intermediates such as carbonate radical (CO3˙−)43 or reactive halogen species (RHS; X˙, X2˙−)44 might contribute to photoreactions; see Section 4.5.
The concentrations of individual PPRI can vary by orders of magnitude, depending on the water composition.42,45 Typical concentration ranges of some exogenous PPRI in sunlit surface waters are 10−17 to 10−15 M for hydroxyl radical, 10−14 to 10−12 M for singlet oxygen and carbonate radical, and 10−12 to 10−10 M for superoxide.46
Most PPRI selectively react with electron-rich sites on biomolecules. In nucleic acids, PPRI most readily oxidize guanine (G), producing 7,8-dihydro-8-oxoguanine (8-oxo-G) and other products.47 In proteins, PPRI mostly target the electron-rich amino acid side chains of tryptophan, tyrosine, histidine, methionine, cysteine, and cystine.48–51 Hydroxyl radical, which is a highly reactive and nonselective oxidant, can, in principle react with all of the amino acid side chains and backbones. Nevertheless, ˙OH has been found to preferentially oxidize the so-called RKPT amino acids (arginine (R), lysine (K), proline (P), and threonine (T)), leading to formation of carbonyl-containing derivatives.52 In addition, ˙OH hydroxylates aromatic amino acids.50
While the potential photochemical reactions of PPRI with individual biomolecules are fairly well understood, the reactions occurring with whole bacterial cells or virus particles are less well-understood, and may differ substantially. Due to the organisms' higher order structure, additional damage may occur (e.g., via radical chain reactions to adjacent molecules), or damage may be mitigated or attenuated (e.g., due to poor accessibility of PPRI to reactive sites, or quenching of PPRI). Furthermore, modification of a site within an organism does not necessarily result in inactivation, due to repair mechanisms and to the high redundancy of protein and membrane components. Thus, the relevant sites of photochemical damage are difficult to predict based on the known photochemistry of free biomolecules alone. In Sections 5, 6, and 7, we review what is known about types of damage and causes of inactivation in microorganisms, as well as other pathways to damage in bacteria involving oxidative stress and internal Fenton chemistry.
4.3. Action spectra for endogenous inactivation
In the UVC range (100–280 nm), beyond the range of the solar spectrum at the Earth's surface, action spectra (relative inactivation as a function of wavelength) for viruses and bacteria closely match the absorption spectra of nucleic acids (maxima around 260 nm), indicating that direct damage to nucleic acids is the primary mechanism of damage.53 As summarized in recent reviews, the wavelengths present in sunlight incident on the Earth's surface (>280 nm) cause less inactivation of viruses and bacteria with increasing wavelength.54,55 In bacteria irradiated under aerobic conditions, the action spectrum deviates strongly from the absorption spectra of endogenous chromophores, due to the complex pathways involved in indirect endogenous damage.53 Thus, empirical relationships are needed to describe the wavelength-dependence of endogenous inactivation. Two main approaches have been used to develop quantitative relationships – either exposing microorganisms to narrow bands of radiation, or broadband exposure (polychromatic) modified with cutoff filters.1 Cullen proposed that only the former be called “action spectra” and that the latter be called “biological weighting functions”.56 Recent research on sunlight inactivation has not adhered to this distinction, but it is important to note that the former approach does not capture interactions between different wavelengths, nor photorepair, and these phenomena are believed to be particularly important for bacteria exposed to sunlight.57 A further disadvantage of using narrow bands is that to generate inactivation data in a reasonable timeframe, the irradiances are often much higher than in natural sunlight. An outstanding challenge with developing action spectra is the choice of a functional form (e.g., algebraic function or look-up table);57 to date, there is no consensus for the most useful functional form for waterborne indicator organisms and pathogens.58 Action spectra are discussed further in the mechanism and modeling sections.4.4. Interaction of exogenous sensitizers with microorganisms
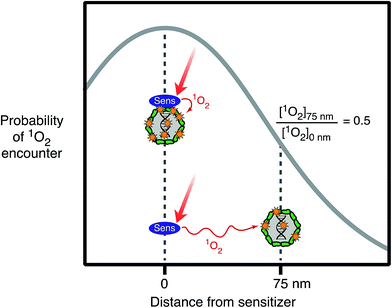
For exogenous sensitizers, the properties of the sensitizer itself, in particular its ability to associate with the organism, can affect the efficiency of exogenous inactivation. Natural organic matter exists as a mixture with components that are supramolecular, colloidal and particulate,59 and these assemblies may sorb to viruses and bacteria. Viruses and bacteria with sorbed DOM may experience enhanced photoreactivity, because they are bound to the sources of the PPRI. For example, singlet oxygen's short lifetime (3.6 μs (ref. 60)) means that it can only take part in reactions within a small sphere of diffusion from where it was generated. This phenomenon is illustrated in Fig. 5, which shows that the probability of an encounter with photochemically produced 1O2 decreases by 50% if the sensitizer is separated by a distance of 75 nm from the virus, compared to if the sensitizer is sorbed to the virus, due to quenching of 1O2 as it diffuses away from the sensitizer.61 Higher rates of inactivation due to sorbed organic matter have been demonstrated for some viruses62,63 and Ent. faecalis.64Relative to photoinactivation, there has been more work on the effects of sensitizer association on the photodegradation of small molecules. For example, studies have shown that the interaction with DOM enhanced the photodegradation of mirex,65,66 organic probe compounds of singlet oxygen,61,67 histidine,68 and also mercury(0).69 Compared to free molecules, it has proven difficult to experimentally demonstrate enhanced photoreactions for organic matter-bound microorganisms. Nevertheless, it is clear that viruses and bacteria will associate with organic matter-rich (micro)phases62,70 and the likelihood of enhanced exogenous photoinactivation in such cases deserves further study.
Association between organic matter and viruses is governed by interactions with the outer surface of the protein capsid, and is influenced by electrostatic, steric, and hydrophobic interactions, and cation bridging between carboxylate groups.71 Preferential adsorption of the hydrophobic, higher molecular weight fractions of an aquatic fulvic acid to Bacillus subtilis was reported.70 The interaction of microorganisms with organic matter can be enhanced by ionic strength and divalent cations (see Sections 5 and 6). However, our current understanding is inadequate to predict the association between organic matter and microorganisms in real water matrices, and the subsequent influence on photoinactivation.
4.5. Photoinactivation in seawater
A number of studies have shown that photoinactivation occurs more quickly in marine versus fresh waters for both bacteria and viruses.72–78 To date, these studies have primarily been observational and a complete mechanistic understanding for the salinity and other water quality effects is lacking. Salinity can potentially influence both endogenous and exogenous mechanisms. In isolated DNA, ionic strength enhanced the quantum yield of pyrimidine dimer formation due to the impact ionic strength has on nucleic acid configuration;37 this effect could potentially be relevant for non-enveloped viruses, but has not been studied directly. In Gram-negative bacteria, enhanced inactivation in seawater was attributed to greater loss of internal cell integrity when cytoplasmic membranes were damaged by sunlight (through either endogenous or exogenous mechanisms).79,80With respect to exogenous mechanisms, the higher ionic strength in seawater may enhance organism–sensitizer interactions.62,81 In addition, high ionic strength can influence the concentration and relative distribution of PPRI by a variety of mechanisms. First, ionic strength has been shown to decrease the loss rate of excited triplet state chromophores (3CHROM*) via electron transfer interactions with solution constituents, including other DOM moieties.82 However, ionic strength did not affect 3CHROM* formation rates or loss rates by energy transfer to other solution components. The net result was a near doubling in the steady-state (ss) 3CHROM* concentration, [3CHROM*]SS. 3CHROM* is the precursor for 1O2 formation, and some studies report higher [1O2]SS in seawater compared to freshwater.83 Overall, the skewing of 3CHROM* away from electron transfer processes may be expected to impact indirect exogenous inactivation processes. Furthermore, halides, particularly Br−, are the predominant ˙OH scavengers in seawater,40,84 leading to decreased [˙OH]SS in halide-rich waters. Halide scavenging of ˙OH40 or halide oxidation by excited state ketones85 forms halogen radicals of the form X˙ or X2˙−.44,86 Modeling indicates that halogen radicals promote the formation of carbonate radical, and that concentrations of halogen and carbonate radicals exceed that of ˙OH by several orders of magnitude in sunlit seawater.87,88 The conversion of ˙OH to these more selective radical oxidants is anticipated to focus the oxidizing power of the system on electron-rich functional group targets.44,87,89
5. Virus mechanisms
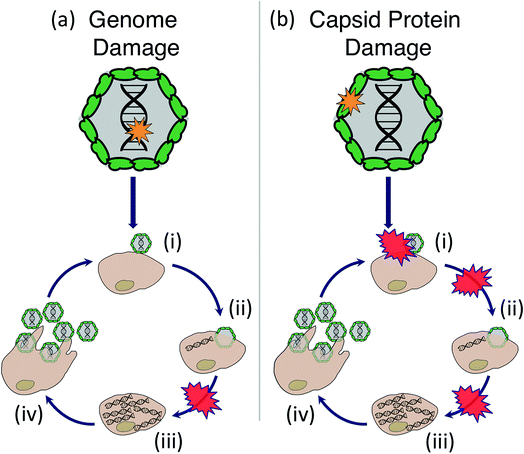
Sunlight disinfection mechanisms for viruses were first investigated by Davies-Colley et al.90 DNA F+ coliphages were only susceptible to endogenous direct inactivation, whereas RNA F+ coliphages could also undergo exogenous indirect inactivation. Since this first study, sunlight disinfection rates have been assessed for various other human and bacterial viruses, and more insight has been gained into their susceptibility to the different inactivation mechanisms. The main findings are reviewed in this section. To assist with our discussion of mechanisms, the potential stages of the virus life cycle that could be disrupted due to endogenous or exogenous damage to viral nucleic acids or capsids are illustrated in Fig. 6. Many knowledge gaps remain in terms of identifying specific sites of damage and which life cycle stages are impacted.5.1. Endogenous mechanisms
Under full-spectrum sunlight, all viruses investigated to date have been found to undergo endogenous inactivation.91–95 Among the viruses studied, human adenovirus (HAdV) and MS2 appear to be the most resistant whereas poliovirus and somatic phages are particularly sensitive.91,93,96 Even for the relatively resistant viruses, however, sunlight inactivation via endogenous mechanisms was found to be the main inactivation process in clear natural waters.93It is likely that endogenous inactivation of viruses mainly occurs by the direct mechanism, though indirect processes have been documented. One example of endogenous indirect inactivation (photosensitization) was identified in MS2 illuminated with (UVC at 254 nm UV254), resulting in an RNA-sensitized cleavage of the capsid protein backbone.97 However, this mechanism was found to be of minor importance compared to overall inactivation.98 The negligible contribution of endogenous indirect inactivation can be explained by the simple structure of many viruses, which consist of a genome surrounded by a protein capsid, and lack intrinsic biochemistry. As a result of this simple structure, viruses contain few internal sensitizers that absorb light in the solar wavelength range; consequently, endogenous indirect inactivation is typically not an efficient inactivation mechanism, and occurs at a much slower rate than endogenous direct inactivation (e.g., Love et al.93). However, experimentally, direct and indirect endogenous mechanisms are difficult to separate, and it is often more appropriate to group them together under the category “endogenous inactivation”.
Only one study to date has investigated the impact of endogenous sunlight damage on the virus life cycle. Sunlight was found to inhibit viral RNA synthesis (Fig. 6a(iii)) of rotavirus, which could explain about half of the inactivation that was observed; the remaining inactivation was attributed to post-translational steps.99 As discussed in Section 4, nucleic acid and protein monomers are susceptible to direct endogenous reactions. It is therefore likely that these reactions play a role in virus inactivation. At this time, no studies have identified the specific sites of virus genome and protein damage that are targeted in endogenous sunlight reactions. Qiao and Wigginton monitored the reactions in viral RNA oligomers exposed to simulated sunlight, but detected no decay with either mass spectrometry or RT-qPCR after 5100 J m−2 UVB.31 Nonetheless, insight into the expected molecular-level modifications induced by sunlight can be obtained from laboratory studies using UVC radiation.
RNA coliphage inactivation studies using a low-pressure UVC lamp (emitting at 254 nm) showed that different regions in the RNA genome exhibited varied susceptibility to UVC irradiation,100,101 and that each genome lesion caused inactivation. Earlier work on ssRNA Tobacco Mosaic Virus found that under some conditions, not all RNA lesions caused inactivation. A complicating factor in studying viral genome reactivity is that commonly employed methods do not detect potentially important reactions in the nucleic acids. For example, reverse transcriptase-based methods (like RT-qPCR) do not detect the same UV-induced RNA reactions as mass spectrometry methods.31 In addition, UVC-induced protein damage, specifically cysteine oxidation followed by backbone cleavage, was reported for selected phages,97,100,102 and was associated with the coliphage's inability to inject its genome into the host cell (Fig. 6b(ii)). Compared to low-pressure UV, the broad spectrum radiation of medium-pressure UV lamps (emitting down to 200 nm) led to a more significant contribution of protein damage in human adenovirus.103 It is therefore reasonable to expect that endogenous inactivation induced by sunlight causes damage to both genomes and protein capsids. The ability of some viruses, notably adenovirus104 and several bacteriophages,105 to hijack their host cells machinery and repair DNA damaged by UV254 has been reported. Similarly, repair of sunlight-induced damage has been reported.106
As might be expected based on viral chromophores, the action spectra of sunlight inactivation closely follow the absorption spectra of the nucleic acids and proteins, as shown by Lytle and Sagripanti, who developed a composite action spectrum for viruses by compiling inactivation data for both RNA and DNA viruses at different wavelengths in the solar spectrum.107 Although considerable work has been conducted to develop action spectra for several viruses in the UVC/low UVB range (e.g., 107–110) the only publications to date for the sunlight spectrum are for MS2 and PRD1.111
5.2. Exogenous mechanisms
In waters containing external sensitizers at concentrations that can occur in natural waters, inactivation rates by full spectrum sunlight of HAdV, human rotavirus, PRD1, and MS2 were faster than endogenous inactivation rates (after correcting for light attenuation),92,112–114 demonstrating that these viruses are susceptible to exogenous indirect inactivation. As discussed previously, exogenous inactivation of MS2 was greater with increasing association of the virus with the sensitizers.114 In contrast, inactivation of poliovirus,92 porcine rotavirus95,113 as well as other F+ DNA coliphages90 did not increase markedly in the presence of exogenous sensitizers and at environmentally relevant temperatures, indicating that the rate of any exogenous inactivation is too low to detect in the presence of endogenous inactivation. Results for phiX174 have ranged from a small91 to a significant contribution from exogenous sensitizers.115Several studies have investigated sunlight-mediated inactivation in the absence of UVB, to study the contribution of different PPRI to exogenous inactivation without the confounding effects of endogenous inactivation (Table 2; light absorption by endogenous chromophores in viruses is limited to the UVB region). Results indicate that 1O2 is an important contributor to overall indirect inactivation of MS2,114,116,117 phiX174 and human adenovirus91 in natural waters. In contrast, 1O2 produced by NOM was not important for the inactivation of porcine rotavirus.95 Several other PPRI can inactivate viruses, including hydroxyl radicals,91,113,118,119 triplet state organic matter,116 and carbonate radicals.91 Although each of these species can inactivate viruses in isolation, their relative importance also depends on solution characteristics and the contribution of endogenous inactivation. In particular the concentration of NOM, which both produces and quenches reactive species and attenuates light, can be expected to play an important role, as explored in Section 8.
Damage induced by PPRI has been most thoroughly investigated for 1O2. Exposure to 1O2 inhibited MS2 genome replication and reduced the virus's ability to bind to its E. coli host.98 The binding inhibition was due to chemical modifications in the virus assembly protein (Fig. 6b(i)). Specifically, damage to MS2 capsids as a result of 1O2 included oxidation of protein side chains,97 in particular of solvent-exposed methionine residues.100 RNA oligomers are reactive with 1O2, with purine bases being more reactive than pyrimidine bases; however the detected modifications in RNA oligomers have yet to be linked to inactivation of intact viruses.31 Protein damage (crosslinking) was also reported upon exposure to 1O2 produced by functionalized fullerenes.120 For adenovirus, both genome damage and significant protein damage by 1O2 was detected. Protein damage likely led to a loss in binding ability or a disruption of early infection processes within the host cell.94 Damage induced by environmentally relevant PPRI besides 1O2 have not been adequately examined.
5.3. Virus characteristics governing susceptibility to sunlight inactivation
If the factors that govern virus susceptibility to sunlight are understood, it may be possible to predict inactivation for viruses that are difficult to culture (and for which it is therefore difficult to quantify inactivation rates). For endogenous inactivation, some efforts have been made to establish generally applicable concepts of virus susceptibility. For example, Lytle and Sagripanti (2005)107 showed that endogenous inactivation by radiation in the UVC/B range depends on the size and type of the viral genome; when normalized by genome size, the inactivation of viruses of the same family, and to a lesser extent of the same genome type, can be estimated reasonably well. They generally found that double-stranded (ds) DNA viruses were the most resistant to UVC light, followed by dsRNA viruses, single-stranded (ss) RNA viruses, and finally ssDNA viruses; these findings are consistent with previous work by Rauth.109 The difference in the susceptibility of ds and ssDNA viruses was attributed to two main factors: the redundancy of the genetic information encoded in dsDNA, and their ability to undergo repair in the host cell.121 The difference between ssRNA and ssDNA viruses was explained by the greater photochemical reactivity of DNA compared to RNA. Very little work of this kind has been done to understand viral photoinactivation due to sunlight, however. One effort used a similar approach to relate virus susceptibility to genome size,93 and showed that within the somatic DNA coliphages isolated from a polluted shallow coastal water, a positive correlation was found between genome size and endogenous inactivation rate constants for sunlight. However, the relationship between genome length and inactivation is not linear. For example, the endogenous inactivation rate constant of poliovirus was five times greater than that of MS2, whereas the length of the genome of polio is about twice that of MS2.93 Similarly, Reovirus was found to be more sensitive to UVC than expected based on its genome size.109 Two explanations were offered for the observed discrepancies: first, virus morphology affects genome packaging and may thereby influence its susceptibility to radiation damage; and second, the presence of light-sensitive proteins likely contributes to inactivation.The characteristics that render a virus susceptible to exogenous indirect inactivation have proven difficult to assess. Generally, it appears that exogenous inactivation is only relevant for viruses that are relatively resistant to endogenous inactivation, and in waters that produce appreciable concentrations of PPRI. For viruses that are readily inactivated by endogenous inactivation, the exogenous contribution to inactivation may be too small to detect.91,92 In addition to the susceptibility of the protein capsid and genome to damage by PPRI, the association between viruses and sensitizers is expected to play a role, as discussed in Section 4.4 and Fig. 5. Recent advances have been made in understanding the interactions between viruses and DOM71 that could be insightful for explaining differential responses of viruses to sensitizers.
Few studies to date have attempted to pinpoint the virus characteristics that render a virus susceptible to inactivation by PPRI.100 The presence of oxidizable protein side chains has been suggested as a cause for a virus' susceptibility to transient species.122 However, the presence of such side chains is not sufficient to explain inactivation: first, not all side chains are accessible to transient species,100 and second, protein oxidation is not always causal to inactivation.98 In fact, for MS2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 98 as well as for HAdV,94 the major contribution to inactivation by singlet oxygen was found to arise from genome damage rather than protein oxidation. Within a closely related group of F+ RNA coliphages, strong similarities in 1O2-mediated inactivation kinetics were observed, and small differences could be explained based on the length and composition of the genome, in particular the number of the most easily oxidizable nucleobase guanine.100 Similar to endogenous inactivation, exogenous indirect inactivation may thus be governed by the genome composition, length and type. To further test this hypothesis, however, information is needed for a broad variety of viruses to assess the effects of PPRI on viral genome and proteins, and to determine the effect of this damage on virus infectivity.
98 as well as for HAdV,94 the major contribution to inactivation by singlet oxygen was found to arise from genome damage rather than protein oxidation. Within a closely related group of F+ RNA coliphages, strong similarities in 1O2-mediated inactivation kinetics were observed, and small differences could be explained based on the length and composition of the genome, in particular the number of the most easily oxidizable nucleobase guanine.100 Similar to endogenous inactivation, exogenous indirect inactivation may thus be governed by the genome composition, length and type. To further test this hypothesis, however, information is needed for a broad variety of viruses to assess the effects of PPRI on viral genome and proteins, and to determine the effect of this damage on virus infectivity.
6. Bacterial mechanisms
The ability of sunlight to inactivate bacteria has been recognized for a long time. Compared to viruses, bacterial cells are vastly more complex, with more potential targets of photochemical damage and molecules that can serve as sensitizers. Furthermore, bacteria have an adaptive regulatory response to sunlight, which induces several stress responses that help to protect against or repair damage.1,53,123,124 The general picture that emerges is that at some point during sunlight exposure, the protective and repair strategies become overwhelmed, leading to irreversible cell death (inactivation); for bacteria derived from batch cultures, these cellular processes are manifested as a lag phase that often precedes measureable inactivation. Thus, it is difficult to characterize the mechanisms that definitively lead to inactivation in bacteria, as many types of stress and damage may occur simultaneously, and it is challenging to identify which particular mechanism or combination of mechanisms leads to irreversible damage. Furthermore, the dominant mechanisms may be different for different environmental conditions, depending on changes in the solar spectrum, depth in the water column, and the water quality (type and concentration of sensitizers, oxygen, pH, salinity). The following sections summarize what is known about mechanisms, without attempting to rate their importance.6.1. Endogenous mechanisms
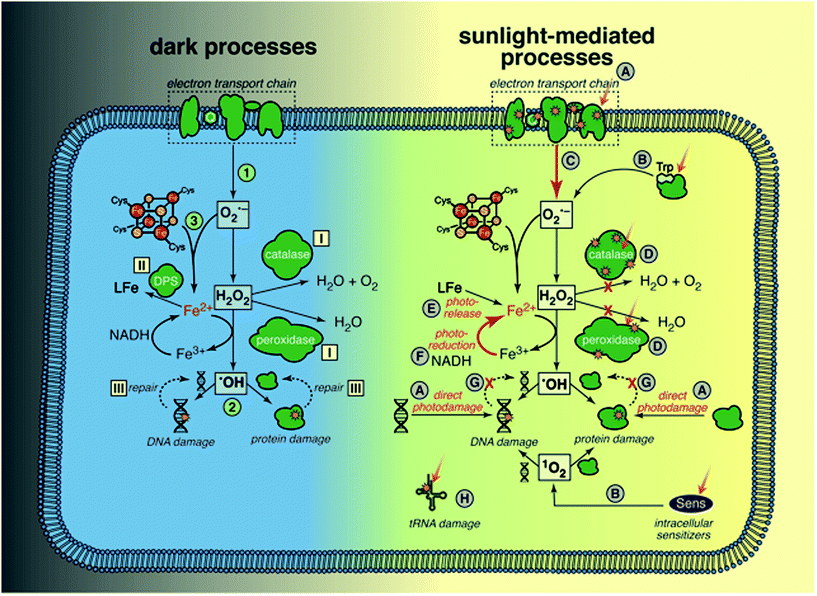
Sunlight is known to cause direct damage to bacterial DNA, via similar mechanisms reviewed above for dsDNA viruses, resulting in various photoproducts including dimers and single-strand breaks.53 Studies of the wavelength effects on bacterial inactivation provided early evidence for the importance of the indirect endogenous mechanism in bacteria; inactivation of E. coli K12 by UV wavelengths up to 313 nm was independent of the oxygen concentration in solution (interpreted as direct endogenous inactivation), whereas inactivation above 313 nm was strongly dependent on oxygen (interpreted as indirect endogenous inactivation).125 Inactivation of Ent. faecalis and Staphylococcus aureus by natural sunlight was also shown to be much faster under oxic conditions than anoxic conditions.126,127 Most mechanistic studies have been carried out only under aerobic conditions, under which it is difficult to separate direct and indirect endogenous mechanisms. For this reason, and because the mechanisms clearly interact, they are discussed together.To provide a framework for understanding many of the ways through which sunlight can cause cellular damage, we first review oxidative stress, summarizing from several recent reviews.128,129 Oxidative stress may affect any bacterial cell in an aerobic environment, and does not require exposure to radiation; these cells must constantly manage oxidative stress to survive. Dark oxidative processes are summarized in the left side of Fig. 7; we will return to a discussion of sunlight-mediated processes (right side of Fig. 7) in several paragraphs. The specific processes illustrated in Fig. 7 are referenced in the text with the corresponding number or letter. (1) Cytoplasmic O2˙− is produced when dissolved oxygen oxidizes reduced enzyme moieties and electron shuttles, such as in flavoenzymes and quinones, either in the electron transport chain or the cytosol. H2O2 is produced by a second electron transfer at the redox site of the enzyme, or by spontaneous or enzymatic dismutation in the cytoplasm. Neither H2O2 nor O2˙− is directly reactive with most organic biomolecules, including nucleotides, amino acids, and unsaturated lipids (unlike in eukaryotic cells, lipid peroxidation by O2˙− is not believed to be significant, due to the lack of polyunsaturated lipids in bacterial membranes). (2) A more important pathway of damage is the production of ˙OH/Fe(IV) by H2O2 and ferrous iron via the Fenton reaction,130 making any biomolecule containing or associated with solvent-accessible reduced iron susceptible (with reactivity depending on the ligand). Because ˙OH is so reactive and non-specific, the reaction products are diverse. Iron has an affinity for nucleic acids and thus DNA is a key target of intracellular Fenton reactions, leading to strand cleavage and formation of adducts. A wide range of ferroenzymes can also be damaged, with ˙OH initiating a chain reaction in some cases, and resulting in a wide range of products, including carbonyls. (3) O2˙− can exacerbate Fenton damage by releasing iron from enzymes, and reducing ferric to ferrous iron.
Cellular defense mechanisms to cope with oxidative stress (even in the absence of radiation) include: (I) enzymes to reduce the intracellular concentrations of ROS (superoxide dismutases (SOD) for O2˙−, and catalases and peroxidases for H2O2), (II) extremely tight controls on the levels of intracellular iron (controlling import, and sequestering cytoplasmic iron in ferritins and other iron binding proteins like Dps), and (III) repair of damaged proteins and DNA. Some of these defense mechanisms are regulated by inducible stress responses that are activated by ROS (e.g., OxyR protein system, which among other things increases levels of alkylhydroperoxidase (Ahp) and catalase as well as iron sequestration by Dps, and SoxRS protein system, which increases levels of SOD).
We now return to the possible ways in which sunlight might contribute to endogenous damage, with reference to right side of Fig. 7. Sunlight wavelengths in the UVB range may cause direct damage to DNA and proteins (A);53 several specific examples are also mentioned below (D and H). There are multiple lines of evidence that sunlight increases ROS levels in cells: accumulation of ROS in E. coli exposed to sunlight as measured using a fluorescent probe;131 increased expression or levels of ROS scavenging enzymes in E. coli123 and Ent. faecalis exposed to sunlight wavelengths;127 the finding that E. coli mutants lacking genes that regulate production of ROS scavengers and iron levels are more sensitive to sunlight wavelengths;132,133 and increased survival of E. coli and Ent. faecalis under simulated sunlight by addition of histidine, a membrane-permeable 1O2 quencher.134 One source of ROS is photoproduction by endogenous sensitizers (B), which is typically described as the classic indirect endogenous mechanism of sunlight inactivation. Examples include the production of 1O2 by flavoenzymes53 and H2O2 by tryptophan.135 Porphyrins are also endogenous sensitizers in E. coli, but it is not clear whether the porphyrins themselves are damaged, disrupting the electron transport chain, or if they sensitize production of ROS.55,136 Others have suggested that once the respiratory chain is damaged by sunlight, ROS production by adventitious reduction of oxygen increases because the regular electron transfer pathway is disrupted (C).137 Another mechanism through which ROS levels may increase is via sunlight damage to scavenging enzymes (D), such as direct photolysis of catalase and Ahp.133,138,139
Given the toxicity of iron in aerobic cells, reactive iron levels are tightly regulated in bacteria.140 There is evidence that sunlight increases the pool of reactive, reduced iron,133 for example via photoreduction (F) and release of iron from the siderophore enterobactin (E).141 The accumulation of reducing equivalents after the respiratory chain is damaged could also increase the rate at which Fe(III) is reduced to Fe(II), accelerating damage by the Fenton reaction (F).137 Another possibility is that DNA and protein repair processes that require energy are reduced, due to damage to the electron transport chain (G).
An intriguing possibility that is distinct from the mechanisms that exacerbate oxidative stress is that UVA wavelengths cause direct damage to transfer RNA, due to the chromophore 4-thiouridine, which causes a growth delay in E. coli, and has been suggested to offer a protective effect against UVA exposure by retarding protein expression (H).142–144
Another line of research has been to characterize loss of function or activity in E. coli exposed to sunlight wavelengths. There is evidence that bacterial cell membranes are damaged during sunlight exposure.137,145–147 More specifically, several key membrane functions related to the electron transport chain were reduced at low light fluences, including loss of proton motive force, which reduced efflux pump activity and ATP synthesis.147 Specific functions of the respiratory chain were also affected at low light fluences, including NADH oxidase, succinate oxidase, and lactate oxidase,137 followed by reduction in ATPase activity (for oxidative phosphorylation). The complete loss of membrane potential and glucose uptake did not occur until similar light fluence as loss of culturability. The membrane became permeable only at fluences higher than those that caused loss of culturability;147 similar results have been found for Ent. faecalis and Staphylococcus.126,127 Based on the prior discussion of mechanisms, loss of these membrane functions is likely a result of damage to components of the electron transport chain and other transmembrane proteins that contain either chromophores or accessible iron. Simultaneous with membrane damage, damage to cytoplasmic proteins has also been documented.148
Various studies have documented wavelength effects on E. coli, indicating that endogenous damage decreases steeply as wavelength increases in the solar range, with wavelengths above 400 nm having minimal effect (in the absence of sensitizers).125,135,149–151 Three lab strains and three environmental isolates of E. coli were found to have similar wavelength dependence based on filter cut-off experiments conducted with a solar simulator.150 The susceptibility of Ent. faecalis extends to longer wavelengths than E. coli (up through 500 nm), although UV is still more potent than visible light.151,152 Action spectra for solar wavelengths have recently been produced, using quasi-monochromatic LEDs for E. coli, and using polychromatic simulated sunlight for E. coli and enterococci cultured in the laboratory as well as those concentrated from treated wastewater.151,153
Although much less is known about sunlight inactivation mechanisms in other bacteria of concern for water quality, especially pathogens, some inferences can be made based on an understanding of their physiology. For example, all bacteria likely contain porphyrins that may serve as endogenous sensitizers.55 Also, it is likely that all bacteria have peroxidases or catalases to scavenge endogenous H2O2;128Pseudomonas aeruginosa was found to be protected from UVA irradiation by catalase.154 However, there is evidence that oxidative stress responses are complex and diverse. The oxyR gene was identified in protecting Salmonella typhimurium,128,138 whereas the sodA gene was identified in Ent. faecalis,124,127 and the msrA gene in S. aureus.126 On the other hand, the obligate anaerobe Bacteroides thetaiotaomicron was inactivated faster than a suite of seven other Gram-positive and Gram-negative bacteria under oxic and anoxic conditions; although it possesses oxidative stress response genes, they may not have been activated in this study when grown under anoxic conditions.155 Evidence of damage to lipids and proteins was found in a study of Acinetobacter and Pseudomonas exposed to UVB and UVA wavelengths.156
6.2. Exogenous mechanisms
Enterococci appear to be susceptible to exogenous inactivation, but E. coli are not noticeably susceptible except at high pH, which can occur (temporarily) in highly eutrophic waters such as wastewater treatment ponds or open water wetlands, due to the high photosynthetic rate of algae,157 or at high salinity, such as in seawater.79,80 Evidence that enterococci are susceptible to inactivation by exogenous sensitizers was provided by experiments in WTP effluent using both simulated and natural sunlight; the inactivation rate of enterococci was higher in WTP effluent than in buffered, sensitizer-free water,134,158 indicating that the sensitizing effects of chromophores in the water outweighed light-attenuation in the shallow reactors used. A study of eight health-relevant bacteria (Bacteroides thetaiotaomicron, Campylobacter jejuni, Ent. faecalis, E. coli K12, E. coli O157:H7, Salmonella enterica serovarTyphimurium LT2, Staphylococcus aureus, and Streptococcus bovis) to exogenous inactivation by synthetic and natural sensitizers confirmed that the Gram-positive bacteria were more susceptible to exogenous inactivation than the Gram-negative bacteria.155,159 When UVB wavelengths were present, all of the Gram-positive bacteria experienced faster inactivation in the presence of at least one synthetic sensitizer. However, the natural sensitizers only increased the inactivation rate when the UVB wavelengths were not present. Interestingly, the natural sensitizers (Suwanee River NOM and DOM isolated from a constructed wetland) also increased inactivation rates (via the exogenous mechanism) of E. coli K12 and S. enterica when the UVB wavelengths were not present. Recent results indicate that DOM isolated from wastewater and constructed wetlands adsorbs to Ent. faecalis cells, and that sunlight inactivation rate increased with the mass of adsorbed DOM.64 Synthetic sensitizers are also known to be more effective when associated with, or taken up by bacteria.155,160The reactive species responsible for exogenous mechanisms have not been well characterized. Kadir and Nelson (2014) found that polyhistidine, which is too large to be transported across the cell wall, decreased the inactivation rate of Ent. faecalis in WTP water, implicating 1O2 produced exogenously; consistent with this interpretation, quenchers of ˙OH, O2−, and H2O2 did not reduce the inactivation rate.152 Singlet oxygen is also known to be an effective reactive species for photodynamic therapy, with Gram-positive bacteria being more susceptible than Gram-negative bacteria; one possible explanation is that Gram-negative bacteria are protected by their outer membrane, whereas 1O2 can diffuse through the peptidoglycan layer of Gram-positive bacteria and damage the cytoplasmic membrane.161 Under conditions that compromise the outer membrane of Gram-negative bacteria, however, they appear to become more susceptible.138,158
Overall, there are a complex set of factors that influence whether exogenous mechanisms are relevant under specific conditions. These factors include: bacterial species and physiological state, the wavelengths of light, the characteristics of the sensitizer and its association with the bacterium.
6.3. Interactions between mechanisms
The three mechanisms of sunlight damage may interact for bacteria. For example, catalase may be damaged directly,133,138,139 which then increases indirect endogenous and exogenous damage. Similarly, a DNA repair enzyme may be damaged by an indirect endogenous mechanism, which then increases net direct DNA damage. As a final example, hydrogen peroxide produced exogenously may diffuse across cell membranes to cause indirect endogenous damage.1626.4. Pigmentation
Enterococci that contain carotenoids are less susceptible to both endogenous and exogenous sunlight inactivation, presumably due to the ability of the pigments to scavenge singlet oxygen and other reactive intermediates.161,163,164 As a result, pigmented strains become dominant with prolonged sunlight exposure.164,165 This difference in susceptibility complicates the use of enterococci as fecal indicator bacteria, as the fraction of pigmented and non-pigmented strains may vary with time and in different waters. Pigmentation may also protect some pathogenic bacteria from sunlight inactivation, such as Staphylococcus aureus.161,166 Fortunately, the pigmented S. aureus was found to be inactivated at higher rates than the non-pigmented Ent. faecalis,126 suggesting that non-pigmented enterococci may still be a conservative indicator of pathogenic bacteria.6.5. Damage versus inactivation
Because bacteria have multiple strategies to repair sunlight damage, there is a possibility that sub-lethal injury could lead to recovery and regrowth. Nonetheless, multiple studies have demonstrated that regrowth is uncommon for a range of conditions. In laboratory experiments with E. coli simulating disinfection with SODIS and photo-Fenton (see Section 10.3), no recovery or regrowth was observed, although cells retained culturability longer on less selective media.167 Using microcosm experiments, Ent. faecalis appeared to become permanently inactivated by sunlight in clear seawater and not to experience repairable injuries within 48 h,127 similar to findings of others on Salmonella and Shigella.168 Davies-Colley et al. held pond samples in the dark after they were exposed to sunlight, and found that enterococci counts continued to decrease over the 24 h holding period, although E. coli showed some increase – presumably due to repair mechanisms.1586.6. Effects of bacterial physiology
The susceptibility of bacteria to sunlight has been shown to be affected by the prior growth conditions,2 which has implications for the design of laboratory experiments, comparing results for different conditions, and relating experiments with lab cultures to environmentally acclimated bacteria. With respect to the latter, several studies have found bacteria sourced from wastewater to be less susceptible to sunlight than laboratory-grown organisms,150,151,164 although another study found that differences depended on the water quality.169 A faster growth rate during culturing was reported to increase the susceptibility of both E. coli170 and Ent. faecalis cells to sunlight wavelengths, and stationary phase Ent. faecalis were more resistant than cells harvested in exponential phase.171E. coli grown under aerobic conditions were more susceptible to sunlight than cells grown under anaerobic conditions; furthermore, after sunlight exposure, cell counts were higher when plated in the presence of ROS scavengers (pyruvate or catalase).167,172E. coli grown in a low-iron media were inactivated more slowly by sunlight than cells grown on iron-rich media.133 Finally, prior exposure of E. coli cells to a sub-lethal UVA fluence rate increased survival to a lethal fluence rate of UVA.123 Thus, the life history of bacteria may also affect their susceptibility to sunlight. Based on the current understanding of sunlight inactivation mechanisms, it is likely that physiological differences influence susceptibility to sunlight because of varying sources of, or responses to, oxidative stress.7. Other organisms and biomolecules
7.1. Sunlight-mediated inactivation of protozoan cysts
The sunlight-mediated inactivation mechanisms of Cryptosporidium parvum oocysts have been explored by Liu et al. (2015).173 Inactivation rates (as determined by in vitro cell culture) were faster in the presence of UVB light compared to when the UVB wavelengths were blocked. Direct damage to DNA was implicated as the dominant mechanism by UVB, whereas indirect endogenous mechanisms were implicated when only UVA and visible wavelengths were present. Inactivation by UVA-induced endogenous radical damage was higher at 40 °C than 25 °C, whereas inactivation by UVA-induced genome damage was not sensitive to temperature. Natural organic matter (Suwannee River NOM and wastewater effluent NOM) did not enhance inactivation, likely due to a thick oocyst wall, which renders oocysts resistant to exogenous inactivation. Studies focusing on mechanisms of damage have illustrated damage to the oocyst wall after 10 h of exposure to UVA/visible light,174 and interference with sporozoite exocytosis, which is a fundamental cellular process required for sporozoites to attach to and invade host cells.175Most other research investigating the effects of sunlight on waterborne protozoan pathogens has been conducted in the context of solar disinfection of drinking water (SODIS; see Section 9.3), and has focused on the effects of reactor geometry, water turbidity, and temperature. A number of studies on container effects have found that containers that transmit more or shorter-wavelength UV light achieved faster inactivation rates of C. parvum oocysts,174,176–181 consistent with the findings on mechanisms above. In general, other protozoan cysts, including Entamoeba histolytica/dispar, Naegleria gruberi, and Giardia lamblia/muris/duodenalis have been found to be susceptible to photoinactivation.5,174,182–185 The cysts of Acanthamoeba polyphaga/castellanii appear to be an exception,182 and were only detectably inactivated by sunlight at elevated temperatures (>45 °C)183 or in the presence of riboflavin.186 No studies have directly compared the inactivation rates of protozoan cysts to those of bacteria or viruses (with the exception of Acanthamoeba). Although it is difficult to compare rates from different studies given the differences in light spectra and irradiance, results to date suggest that protozoan cysts may be generally more resistant to sunlight than viruses and bacteria. This trend is different from that for inactivation by UV254,187 to which protozoan cysts have similar susceptibility as bacteria, and are more susceptible than most viruses, and underscores that there are differences in the principal inactivation mechanisms of UV254 and sunlight.
A particular challenge with interpreting some of the published research on protozoan cysts is the use of different assays to measure inactivation. Dye permeability and in vitro excystation were found to underestimate oocyst inactivation compared to animal infectivity tests (Swiss CD-1 suckling mice).177 Another challenge is different sources of oocysts; because oocysts cannot be propagated in vitro, propagation through animals such as calves and mice is required. Also oocysts for experiments are usually purified from feces of infected animals; however, purified oocysts have been shown to lose infectivity within 24 weeks during storage at 4 °C in autoclaved water.188 We recommend that future studies of oocyst inactivation should document the source of oocysts, the storage conditions of oocysts, and should quantify response by either in vitro cell culture189 or animal infectivity.
7.2. Sunlight-mediated degradation of antibiotic resistance genes
An emerging concern that is relevant to the transmission of bacterial pathogens via sunlit waters is the fate of antibiotic resistance genes (ARGs).190 ARGs are now recognized as widespread contaminants of aquatic systems,191–193 leading to concerns that their presence may contribute to the dissemination of antibiotic resistance traits amongst bacterial populations via horizontal gene transfer (HGT) processes (including conjugation, transduction, and natural transformation).194 ARGs are present as intracellular genomic and plasmid DNA in viable antibiotic resistant bacteria (ARB), and as extracellular (i.e., free) DNA protected within cell debris, phage capsids, extracellular polymeric substances, or on clay mineral surfaces. Even extracellular ARGs may be capable of transferring their encoded resistance traits to non-resistant bacterial populations by means of transduction or natural transformation.195 Thus, it is desirable to examine not only if solar radiation will yield inactivation of viable ARB cells, but also whether or not it is likely to render ARGs incapable of conferring resistance traits by any of the three means of HGT.Although very little information is currently available regarding the effects of UVB, UVA, or broadband sunlight in ARGs, substantial past work illustrates that monochromatic UVC radiation (UV254) can eliminate the ability of various ARGs to transform competent non-resistant recipient bacteria to the corresponding resistance phenotypes, whether such ARGs are contained in intracellular or extracellular DNA.196–199 Studies in which qPCR was utilized to quantify residual copy number of ARGs contained in extracellular and intracellular DNA from several genera of ARB during UVC irradiation are generally consistent with these findings.199–201 However, ∼2- to 10-fold higher fluences are required to achieve >2-log degradation of ARGs than to yield comparable ARB cell inactivation (i.e., ARGs are degraded more slowly than ARB cells are inactivated).199–201 One complicating factor of ARG fate is that regions in the DNA outside of the ARG sequence are necessary for transformation; thus measuring the decrease in portions or all of the resistance gene with qPCR following UV treatment is a conservative measurement of transformation potential.199 For extracellular plasmids carrying ARGs, plasmid nicking was not a major reaction pathway at UV254 fluences used for water treatment.
In light of the above, it is also reasonable to anticipate some degradation of ARGs (intracellular or extracellular) during solar irradiation, given the susceptibility of DNA to direct and indirect damage by sunlight, as described in previous sections. In general, it can also be expected that extracellular ARGs will undergo more rapid sunlight-driven degradation than intracellular ARGs, as many bacterial species are capable of DNA photorepair under solar illumination, as well as dark repair.202,203 Furthermore, extracellular DNA is likely to be susceptible to both exogenous direct and indirect mechanisms of damage.
In one series of studies, accelerated decay of several intracellular resistance genes (as monitored by qPCR) was observed in micro- and mesocosms that were seeded with wastewater and irradiated with simulated and/or natural sunlight, relative to dark controls.204–206 However, considering that wastewater matrixes were used to seed the meso-/microcosms – it remains unclear whether these observations were due specifically to sunlight-mediated DNA damage or to unidentified alterations in the microbial ecology of the investigated systems upon exposure to sunlight.
It has also been reported that the ability of a plasmid-borne cat gene to transform recipient bacterial cells to chloramphenicol resistance can be gradually eliminated during exposure of extracellular preparations of the host plasmid in TE buffer (pH 8) to artificial UVC, UVB, and UVA light, as well as natural sunlight.207 Fluence requirements to yield comparable levels of deactivation were ∼10-fold higher for irradiation by UVA compared to by UVB, and also ∼10-fold higher for UVB compared to UVC. Loss of activity correlated well with induction of cyclobutane pyrimidine photodimers by artificial UV ranges and natural sunlight, suggesting that photodimer formation represents the primary mechanism of ARG deactivation.207 Although not specific to ARGs, several recent studies also illustrate that qPCR signals for the 23S rRNA gene contained within extracellular or intracellular genomic DNA of Enterococcus spp. can persist even at solar fluence values several times those sufficient to yield 5-log inactivation of the bacterial cells themselves.127,208
Taken together, results to date suggest that sunlight-driven degradation of ARGs will likely proceed with markedly slower kinetics than ARB cell inactivation, in analogy with observations for UVC irradiation. Recent findings also suggest that ARB cells themselves may be more resistant to inactivation during solar irradiation than cells of non-resistant strains, possibly due to upregulation in expression of a wider array of stress response and repair genes in ARB than in non-resistant strains.209 Even if ARB cells are effectively inactivated by solar irradiation, their ARG-containing DNA may remain intact and capable of transferring resistance traits to non-resistant bacteria. A challenge with assessing this risk is that ARGs detected by qPCR may no longer be capable of transferring resistance via transformation. Considering the potential public health and ecological implications of ARGs persisting during transit through natural surface waters, further research on this topic is highly desirable.
8. Modeling of inactivation rates
Models of sunlight-mediated inactivation are needed for use in the design of treatment processes that rely on solar disinfection, to quantify the fate of pathogens and indicators in recreational waters, and more generally as components of ecological models for surface waters. Although the impact of sunlight has been incorporated into models for these different applications, in most cases inactivation is modeled as a simple first-order process with time. Reported sunlight inactivation rate constants vary over several orders of magnitude even for the same microorganism. One large source of variability is that these “overall” rate constants do not separate out the key parameters that are now known to influence inactivation rates based on the growing mechanistic understanding reviewed in the previous sections. To improve the predictive ability of models, and to better understand the sources of variability, in this section we review more detailed approaches that have been applied to model photoinactivation of viruses and bacteria. We do not consider the effects of other potential loss processes that may occur simultaneously with photoinactivation (e.g., physical removal, die-off due to unfavorable environmental conditions, predation) or transport processes. Thus, we analyze a volume element (batch) of water, which can be modeled assuming that it is either stratified or well-mixed. The approaches discussed here can be used to model laboratory experiments conducted with artificial light sources (e.g., UVB, UVA, visible light, or solar simulators) or natural sunlight, as well as to model surface water bodies exposed to natural sunlight. Of course, other die-off mechanisms and transport processes must also be accounted for in surface waters.There are two main steps in modeling photo-inactivation (Fig. 8): (1) estimating the irradiance spectrum to which the organisms are exposed, and (2) predicting the inactivation that occurs as a result of the irradiance spectrum. The first step can be further broken down into: (1a) characterizing the radiation spectrum incident upon the water body of interest, and (1b) accounting for differential transmission across the UV-visible spectrum within the water column. The second step can be further broken down into: (2a) predicting inactivation due to endogenous inactivation and (2b) due to exogenous inactivation. Because exogenous inactivation results from absorption of photons by chromophores in the water, this step involves (2b1) predicting the concentration of reactive intermediates in the sunlit water, and (2b2) predicting the inactivation caused by the reactive intermediates.
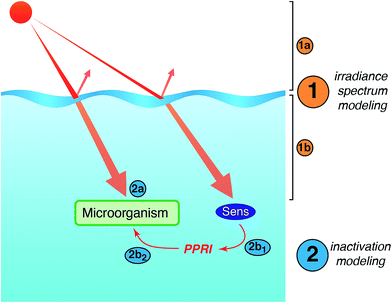 | ||
| Fig. 8 Main steps involved in modeling sunlight inactivation of microorganisms in sunlit surface waters. | ||
8.1. Measuring or predicting the incident irradiance (step 1a)
Step 1a involves either empirical measurement of the irradiance spectrum for a specific radiation source or sunlight condition, or prediction of the solar irradiance based on (assumed or measured) meteorological and atmospheric conditions. Because of the marked wavelength-dependence of photoinactivation, it is necessary to characterize the irradiance spectrum over the entire (UV-visible) wavelength range that may contribute to inactivation (i.e., the curves shown in Fig. 2). The main option for empirical measurement of the irradiance incident upon a water body (e.g., an open reactor or a natural water body) is a spectroradiometer, which measures the wavelength-specific irradiance over the desired range. Additional discussion of approaches for measuring irradiance is provided in Section 9.Alternatively, several models exist for predicting the incident irradiance from natural sunlight, which is necessary when modeling photoinactivation for light conditions that cannot be measured directly (e.g., different locations or times). To date, sunlight inactivation models have used the Simple Model of the Atmospheric Radiative Transfer of Sunshine (SMARTS)9 and the Tropospheric Ultraviolet and Visible Radiation Model (TUV).43,210 However, a unique challenge with predicting photoinactivation compared to other sunlight processes is that rates are very sensitive to UVB wavelengths, which are highly variable and represent only a minor fraction of the total irradiance; neither measurement instruments nor atmospheric models have been tailored to provide accurate results in the UVB region.211 To improve the accuracy of photoinactivation models, it will be necessary to develop more accurate methods for both measuring and predicting sunlight in the UVB range. A further challenge with predictive modeling is the difficulty of accounting for the effects of cloud cover on the sunlight spectrum.
8.2. Accounting for spectral transmission into water (step 1b)
Models of sunlight inactivation have employed the same approaches developed for aquatic photochemistry to account for differing light transmission through water across the UV-visible solar spectrum. To determine the rate of photoinactivation at a given depth in the water column, the irradiance at that depth must be determined by correcting the incident irradiance (e.g., Fig. 2) for light attenuation within the water column (e.g., Fig. 3). Alternatively, the depth-average irradiance can be determined if the water body is well-mixed. For laboratory experiments with collimated light sources, it is common to estimate sunlight attenuation due to absorption using Beer's Law, assuming that reflection at the surface and scattering by particles suspended in the water are negligible.117 For natural water bodies exposed to sunlight, the diffuse downwelling attenuation coefficient is used (eqn (1)), which can either be measured directly or modeled using fundamental optical measurements (e.g. Kirk 2011,10 or using an optical model like Hydrolight8). In some strongly coloured waters, scattering effects are relatively minor and can be neglected (e.g., wastewater polishing wetland211) whereas in most waters scattering significantly increases attenuation above that due to absorption alone (e.g., high rate algal pond, Fig. 3 (ref. 212)). The models ideally should account for a weak influence of solar zenith angle.17 To date, more sophisticated radiative transfer models that account for sunlight penetration into water bodies, such as HydroLight,213 have not been used for modeling sunlight inactivation, and we emphasize that this is an important direction for future research.8.3. Modeling inactivation rate constants (step 2)
Although synergies between sunlight inactivation mechanisms likely exist, the current understanding is insufficient to include such interactions in modeling, and so mechanisms are assumed to be independent and additive. Thus, the total inactivation rate constant (ktot) is represented as the sum of the rate constant for endogenous mechanisms (kendo), exogenous mechanisms (kexo), and any light-independent (dark) mechanisms (kdark).| ktot = kendo +kexo + kdark | (3) |
Current mechanistic models do not explicitly account for other factors that may influence sunlight inactivation rates, including dissolved oxygen, pH, the physiological state of bacteria, the extent of aggregation or particle-association, or repair processes (occurring at temperature-dependent rates). An approach to account for the synergistic effect of temperature (30–55 °C) on endogenous inactivation was recently developed and is discussed briefly in the next section.214
 | (4) |
In the second approach, Mattle et al. (2015) modeled P(λ) for three viruses (MS2, PhiX174, and HAdV) as the product of a constant apparent quantum yield for the virus (Φ; units = number of viruses inactivated per photon absorbed53) and the viral extinction coefficient (εvirus(λ); virus−1 cm−1).91 This approach assumes that all photons absorbed by virus components have equal likelihood to cause inactivation, independent of their wavelength (i.e., the action spectrum is proportional to the absorption spectrum of the virus). The number of photons absorbed by virus components is a function of their nucleic acid and endogenous protein chromophores, is a small fraction of the photons incident upon a virus, and is restricted to UVB wavelengths (Section 5.1). To generate similar modeling values for viruses other than those studied by Mattle et al., two pieces of experimental information are needed: the absorption spectrum of the virus, and its inactivation rate under a known light spectrum. Alternatively, it has been proposed that the parameters for additional viruses can be estimated without further experimentation, if the genome size and type are known, by assuming that the quantum yield as well as the extinction coefficient scales with genome type and size.107 This approach is not applicable to bacteria, for which the absorption spectrum does not match the action spectrum.125 Instead, Zepp58 proposed an approach for determining wavelength-specific quantum yields that could be applied to bacteria using cutoff filter experiments and a fitting function as described by Rundel,57 but this has yet to be applied in practice.
The third approach is similar to that proposed by Zepp58 (using experiments with cut-off filters), but directly yields values for P(λ). Fisher et al. used an empirical approach to determine action spectra and values for P(λ) based on cutoff filter experiments for the viruses MS2 and PRD1.215 This approach assumes that photons of different wavelengths may have different contributions to inactivation, but cannot differentiate whether the effect is due to differences in absorption by viral chromophores or differences in the damage caused by photons of different wavelength. Using this approach, Nguyen et al. captured seasonal effects (summer vs. winter sunlight) on the inactivation rate of MS2 in clear water, and the effect of light attenuation by strongly humic-coloured wetland water using simulated sunlight.211 However, successful prediction of inactivation rates was hampered by difficulty in accurately determining UVB irradiance, a problem noted above in Section 8.1. The third approach has also been applied to bacteria; Silverman et al. (2016) developed P(λ) functions for E. coli and enterococci grown in the laboratory and concentrated from wastewater.151 The wastewater bacteria P(λ) functions were used to predict inactivation rates in wetland water and clear seawater, and it was noted that further work is needed to account for the increased bacterial inactivation that occurs with high dissolved oxygen and pH in algal laden waters, which are important for E. coli.158,216 Mostafa et al. (2016) used an action spectrum for Ent. faecalis to predict the effect of light attenuation on its endogenous inactivation rate in water containing different types of organic matter.217 Finally, Roser et al. (2016) recently published action spectra for E. coli and enterococci based on experiments with monochromatic LEDs that cover the sunlight range; the results appear roughly consistent with other work conducted with polychromatic light.153
The main difference in these three approaches is their response to changes in the spectral irradiance. If the shape of the light spectrum does not change (i.e., all wavelengths increase or decrease directly proportional to total irradiance), the three approaches predict the same relative changes in kendo. However, as discussed in Section 3, the relative proportion of shorter (i.e., UVB) to longer wavelengths changes due to zenith angle and atmospheric conditions (Fig. 2), as well as wavelength-dependent attenuation in the water column (Fig. 3).
Another approach has been developed for modeling the indirect endogenous inactivation of E. coli that explicitly accounts for damage by ROS (e.g., superoxide, hydroxyl radical).139 Steady-state concentrations of intracellular ROS species are determined as a function of their production and loss processes (Fig. 7), and inactivation results from second-order reactions between ROS and bacteria. A strength of this approach is that it provides more insight into the specific endogenous reactions, which allows for the manipulation of a wide range of factors that influence kendo. For example, the authors expanded the model to account for the impact of temperature on the individual reactions, as well as the independent and synergistic effects of temperature in the range 30–55 °C;214 the authors have also explored the effect of different catalase loss rates (due to photoinactivation and thermal degradation). A disadvantage of this modeling approach is that it does not account for direct endogenous inactivation, nor wavelength-specific effects.
 | (5) |
Measuring PPRI concentrations in situ is experimentally challenging. However, models have been developed that predict [PPRI]ss as a function of water depth (or depth-averaged values) based on easy-to-determine water composition (e.g., the APEX model, Bodrato and Vione 2014; GCSOLAR, U.S. EPA).45,218 Such models take into account the dominant formation and quenching processes of each reactive species, as well as the changes in irradiance spectrum throughout the water column.
Second-order rate constants have been measured in laboratory experiments for MS2, PhiX174, HAdV, and rotavirus. Values of kPPRI for all four viruses are close to the diffusion limit for hydroxyl radical (∼1010 M−1 s−1)91,118,119 and 1–2 orders of magnitude lower for singlet oxygen, 3DOM* or carbonate radicals.91,114,116 Furthermore, the measured kPPRI for a given reactive species were typically within one order of magnitude for the different viruses (MS2, PhiX174, HAdV), except for 3DOM*, for which slightly larger differences between viruses were observed.91 It should be noted that the rate constants for singlet oxygen and 3DOM* were determined using model sensitizers (Rose Bengal and anthraquinone-2-sulfonate, respectively), and that association between the sensitizers and viruses cannot be ruled out. Therefore, the apparent rate constants may vary depending on the degree of association.219
While relatively straightforward in its application, this model of exogenous virus inactivation has several shortcomings. First, it does not take into consideration synergies between different reactive species or between reactive species and UVB light. Second, the model does not capture virus–sensitizer interactions, which may enhance inactivation, as discussed in Section 4.4.
The approach of Silverman et al. attempts to capture both of these aspects, but is more empirical.17 Recognizing that 1O2 was found to be the most important contributor to exogenous inactivation of MS2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91,114 and a likely contributor to inactivation of Ent. faecalis,134 exogenous inactivation was modeled as an apparent second order reaction with 1O2 as the only reactive species, with the assumption that other reactive species scale proportionally with 1O2, which is an oversimplification. This approach, in which the apparent k1O2 values for each organism were first determined experimentally in water from a constructed wetland, was used to model MS2
91,114 and a likely contributor to inactivation of Ent. faecalis,134 exogenous inactivation was modeled as an apparent second order reaction with 1O2 as the only reactive species, with the assumption that other reactive species scale proportionally with 1O2, which is an oversimplification. This approach, in which the apparent k1O2 values for each organism were first determined experimentally in water from a constructed wetland, was used to model MS2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 and Ent. faecalis64kexo in the same wetland. The value for kMS2–1O2 of 1.1 × 1013 M−1 h−1, is about ten times greater than that measured by Mattle et al.91 using Rose Bengal (RB). This difference may be due to two factors: (1) the apparent kMS2–1O2 accounts for the contribution of other reactive species and possibly their interactions; and (2) the sensitizers in the wetland may have had greater association with MS2 than RB, such that MS2 was exposed to higher concentration of ROS than the measured bulk-phase concentration. A strength of this approach is that the apparent k1O2 values account for other reactive species as well as association between the sensitizer and organisms. However, this is also a drawback, because the apparent k1O2 must be measured specifically for each water of interest, as the value has been observed to vary among waters.17,96
17 and Ent. faecalis64kexo in the same wetland. The value for kMS2–1O2 of 1.1 × 1013 M−1 h−1, is about ten times greater than that measured by Mattle et al.91 using Rose Bengal (RB). This difference may be due to two factors: (1) the apparent kMS2–1O2 accounts for the contribution of other reactive species and possibly their interactions; and (2) the sensitizers in the wetland may have had greater association with MS2 than RB, such that MS2 was exposed to higher concentration of ROS than the measured bulk-phase concentration. A strength of this approach is that the apparent k1O2 values account for other reactive species as well as association between the sensitizer and organisms. However, this is also a drawback, because the apparent k1O2 must be measured specifically for each water of interest, as the value has been observed to vary among waters.17,96
8.4. Putting it all together
Combining the approaches described above, the overall inactivation rate constant due to sunlight (eqn (3)) can be determined. If the interest is in inactivation at a specific depth, ktot(z) is calculated using E(λ, z) and [PPRI]ss,z. If the interest is a well-mixed water column, a depth-averaged 〈ktot〉 is calculated using depth-averaged irradiance 〈E(λ, z)〉 and 〈[PPRI]ss〉.17,169 Applications include modeling the effects of changes in the irradiance spectrum and/or changes in water quality on the sunlight-mediated inactivation of viruses and bacteria.In Fig. 9 and 10, we illustrate several insights from this modeling approach. The strong influence of spectral sunlight attenuation on the endogenous inactivation rate constant (kendo) is shown in Fig. 9, by comparing the different waters from Fig. 2. We used a water depth of 20 cm and assumed the water was well mixed. The first step was thus to calculate the average irradiance spectrum transmitted through each water (Fig 9, panel (b)), using an incident irradiance spectrum for June in Northern California, as reported in Silverman et al. 2015.17
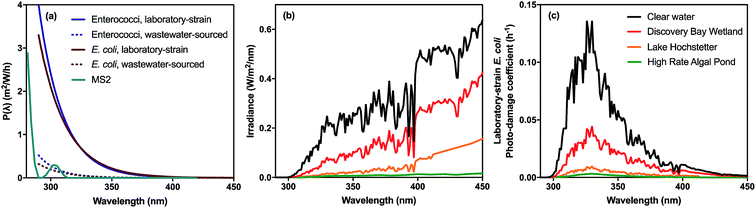 | ||
| Fig. 9 Impact of action spectrum and sunlight attenuation in the water column on the endogenous inactivation rate constant for diverse waters. The action spectra for different microorganisms are shown in panel (a). In panel (b) the irradiance spectrum averaged over 20 cm depth is shown for different waters (a subset of those in Fig. 3). In panel (c) we show the photodamage coefficient for lab-strain E. coli, which is the product of the action spectrum and the irradiance spectrum averaged over a 20 cm depth. The area under the photodamage spectrum is kendo. Irradiance spectra were determined for a single atmospheric condition (as reported in Silverman et al.,17 for a summer day at 38° latitude), accounting for attenuation in the water column for the waters shown in Fig. 3. Resulting values for kendo for the different waters are: clear water (5.2 h−1), Discovery Bay Wetland (1.9 h−1), Lake Hochstetter (0.41 h−1), and High Rate Algal Pond (0.12 h−1). Biological weighting functions for bacteria are from Silverman et al. (2016) and for MS2 was modified from Fisher et al. (2011).111,151 | ||
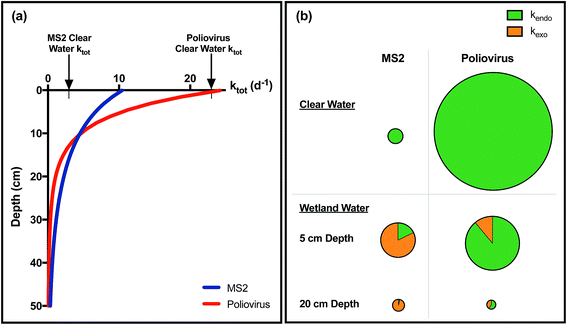 | ||
Fig. 10 Effect of wetland sensitizers and water depth on the sunlight-mediated inactivation rate constant, ktot, of MS2 and poliovirus. (A) ktot in wetland water (red and blue curves) as a function of depth (ktot in shallow clear water is shown at depth zero with an arrow). (B) Contribution of endogenous and exogenous mechanisms to ktot in clear water and two different depths of wetland water. The area of the circle is proportional to the value of ktot. Modeling parameters from Silverman et al.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96 96 | ||
The second step was to incorporate the action spectra (i.e., P(λ)) to calculate values for kendo; P(λ) spectra for E. coli, enterococci, and MS2 are shown in Fig. 9 panel (a). In panel (c), the product of the irradiance spectrum and the action spectrum – referred to as the photodamage spectrum for endogenous inactivation – is shown for laboratory strain E. coli for the different waters. The area under each curve is kendo. The modeled endogenous inactivation rate is reduced from 5.2 h−1 in clear water to 0.12 h−1 in the algal pond water, due to strong attenuation of sunlight by chromophores in the water. Another insight provided by the photodamage spectra is the dominant wavelengths contributing to inactivation. The peak wavelengths contributing to E. coli (panel c) and enterococci (not shown) inactivation occur around 330 nm. Note that a similar approach was used by Mbonimpa et al. (2012) to model inactivation of E. coli by sunlight; however, a standard DNA action spectrum was used which does not account for other mechanisms of sunlight damage.220
Note that the curves in Fig. 9 do not account for the contribution of exogenous mechanisms to inactivation, which can be initiated by longer wavelengths (Table 2). In Fig. 10 we account for both endogenous and exogenous inactivation rates, and illustrate how dramatically the rate constants can vary for MS2 and poliovirus due to their different susceptibilities to endogenous and exogenous mechanisms, using the model parameters from Silverman et al., also for the month of June in Northern California.17 In clear water (only endogenous inactivation, minimal light absorption by water column), ktot for poliovirus is ∼8 times greater than MS2. However, in wetland water (containing sensitizers), ktot for poliovirus is only ∼2.5 times greater at the water surface, because MS2 is much more susceptible to exogenous inactivation. Even at 5 cm depth in wetland water, ktot for MS2 is still greater than it is in clear water because of the large contribution by exogenous inactivation. In contrast, ktot for poliovirus is smaller at 5 cm depth in the wetland relative to clear water, because attenuation of sunlight by the organic matter decreases the endogenous rate more than the sensitizing effect contributes to exogenous inactivation. At depths greater than 11 cm, ktot for MS2 is actually greater than ktot for poliovirus, because kexo decreases less rapidly with depth than kendo, due to the dependence of kendo on shorter wavelengths, which are attenuated more efficiently by organic matter, as discussed in Section 3. If the water column is well-mixed, ktot for MS2 and poliovirus are equal if the water column is about 50 cm deep, and the inactivation rate constant for MS2 is still greater in this case than it is in shallow clear water.
Next, we describe several applications of the mechanistic modeling approach. Silverman et al. (2015)17 and Nguyen et al. (2015)64 modeled the sunlight-mediated and dark inactivation of MS2 and fecal indicator bacteria (E. coli and enterococci), respectively, in a pilot-scale, open water unit process wetland operating for one year. The sunlight spectrum was predicted using SMARTS, 1O2 was used as a surrogate for all PPRI, and 1O2 concentrations were estimated as a function of the organic matter concentration.221 The wetland hydraulics were modeled using simple two-parameter model, with the dispersion coefficient determined from a tracer test. There was surprisingly good agreement between modeled MS2 and measured F+ coliphage removals throughout the year (MS2 is one member of the F+ coliphage family). The agreement was not as good for the indicator bacteria, as the monitoring data were highly variable. Exogenous inactivation mechanisms were predicted to dominate inactivation of MS2 and also contributed significantly to the inactivation of non-pigmented enterococci. One challenge is that E. coli and enterococci concentrated from wastewater were more resistant to sunlight than lab strains and isolates cultured in the lab; therefore, a correction factor was developed to convert rates measured in the lab to those in field.
Kohn et al.43 modeled the inactivation rates of phages MS2 and phiX174 in two different water matrices, a WTP and a natural surface water, for different sunlight conditions (season and latitudes). The endogenous inactivation rate was determined using eqn (4), in which P(λ) was determined from each viruses' molar extinction coefficient and quantum yield.91 The exogenous inactivation rate constant was determined using eqn (5), and the second-order rate constants reported in Mattle et al.91 and steady-state PPRI concentrations determined using the APEX model. As expected, the contribution of the exogenous mechanism in WTP water was significant for MS2 (>50% for water depths > 1 m), and dominated by 1O2 with small contributions from 3CDOM* and OH˙. Inactivation of phiX174 was dominated by endogenous mechanisms. Because longer wavelengths contribute to exogenous mechanisms, the inactivation rate of MS2 was less sensitive than that of phiX174 to changes in solar irradiance due to season and latitude. Despite some differences in the modeling approaches used by Kohn et al. and Silverman et al., the Kohn et al. model was able to predict with fairly good agreement the experimentally measured rate of inactivation of F+ coliphage reported by Silverman et al.17
Other modeling efforts have incorporated some aspects of this mechanistic approach. Williamson et al. predicted that climate change is reducing solar disinfection of surface waters due to higher CDOM concentrations from runoff; the influence of wavelength on endogenous photoinactivation of Cryptosporidium oocysts was accounted for using a photoaction spectrum modified from the DNA absorption spectrum.12 Empirical approaches have also been used to account for the dual role of natural organic matter (attenuation vs. photosensitization) in the inactivation of the bacteriophage phiX174![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 115 and Ent. faecalis.217
115 and Ent. faecalis.217
The results in this section illustrate how models can be used to capture the sensitivity of sunlight inactivation rate constants to the magnitude and spectral qualities of sunlight irradiance, as well as the impacts of water quality on attenuating sunlight and producing PPRI. These environmental factors result in inactivation rate constants that can vary over several orders of magnitude for the same organism. Importantly, it was also shown that specific viruses and bacteria respond differently to these environmental conditions, leading to large changes in their relative inactivation rate constants.
9. Standardizing methods for photoinactivation experiments
The previous sections illustrate that a large number of factors influence photoinactivation rates. Thus, experiments to investigate the mechanisms or kinetics of sunlight-mediated inactivation of microorganisms should be conducted and reported in a manner to facilitate inter-laboratory comparisons. The light source is one of the most difficult aspects to characterize with sufficient detail, and is therefore one of the greatest sources of variability between field sites and laboratories.211,222 Given the wavelength-specificity of inactivation, the full UV-visible irradiance spectrum should be reported for both artificial sources and natural sunlight. For example, a graph should be provided of the spectral irradiance (280–700 nm; Fig. 2b), complemented by values of the integrated spectral irradiance for UVB, UVA, and visible radiation bands. A logarithmic scale (e.g., Fig. 2) often clarifies differences in the UVB region where spectral irradiances are much lower than elsewhere in the solar spectrum and change rapidly with wavelength. Alternatively, an inset of the UVB region can be provided.A spectroradiometer is the preferred physical instrument for measuring the spectrum of incident irradiance over the entire UV-visible range. However, spectroradiometers for the solar spectrum have not been designed to measure UVB with much accuracy, particularly <300 nm. Because these wavelengths contribute disproportionally to inactivation, especially for viruses, the difficulty in measuring these wavelengths accurately is a major source of uncertainty (see Nguyen et al. 2014 for further discussion211). The sunlight fluence (the product of irradiance and time) can be calculated for the wavelength range(s) of interest; fluence is useful for normalizing results to allow comparison between results from different radiation exposures. If a spectroradiometer is not available, a radiometer can be used to measure irradiance over a defined spectrum, and the wavelength range should be reported along with the total irradiance. Any physical device should be recently calibrated to NIST or other comparable international standards across the wavelength range of interest.
Spectral filtering of incident light by the water column can be modeled based on laboratory measurements of the UV-visible attenuation spectrum (see Section 8.2). Alternatively, the downwelling irradiance can be measured in situ in the water body of interest, using submersible instruments. In either case, it is important to provide a clear description of any assumptions that are made. In the solar UV range, light scattering can sometimes be neglected such that the vertical attenuation coefficient, Kd(λ) can be approximated with the absorption coefficient, a(λ) – notably in strongly colored wetland waters of relatively low scattering. To determine whether scattering is significant, the absorption spectrum of a filtered and unfiltered sample should be compared. If necessary, spectral scattering coefficients can be measured directly using modified spectrophotometers (e.g., with integrating sphere) or field measurements with suitable instruments.14,15 If the experiments are conducted in closed containers (i.e., the light passes through the wall of container to reach the sample), then absorption of light by the containers must be characterized.222
Chemical actinometry complements spectroradiometry by providing a measure of the average fluence received by cells or virions in a closed reactor. In particular, actinometry is a useful tool for normalizing fluence between different experimental conditions (e.g., variable irradiance or attenuation).127,223,224 For closed experimental reactors (no open water surface, e.g., merry-go-round reactor), it is difficult to measure the irradiance spectrum in the reactor with physical instruments, so chemical actinometry is preferred. The main drawback to using actinometry for characterizing photoinactivation is that the absorption spectra of existing chemical actinometers differ from the action spectrum for photoinactivation, requiring the use of cut-off filters or bandpass filters to isolate spectral regions of interest. The development of new chemical actinometers and methods to facilitate wavelength-specific actinometry would be a useful contribution to photoinactivation research.
Other water quality parameters should be reported to characterize potential sources of PPRI and any impacts of water quality on inactivation. Both dissolved and particulate sensitizers are potentially relevant. Dissolved organic sensitizers can be approximated as CDOM, which is quantified as the absorption coefficient of a filtered sample at standard wavelengths such as 440 nm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 or 340 nm
10 or 340 nm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 or other UV wavelengths (e.g. 280 nm, 254 nm; see eqn (2)). Characterizing absorption by particles (e.g., wastewater particles, algae) is more difficult because particles can both absorb and scatter light (see previous page). In addition to scattering coefficients, bulk parameters such as total suspended solids, volatile suspended solids, chlorophyll a, or particulate organic carbon should be reported for samples with high particulate matter. Other parameters that can influence PPRI include NO3−, NO2−, and iron (sources of ˙OH). Also, the steady-state concentrations of key PPRI such as 1O2, ˙OH, and H2O2 can be measured. Other relevant parameters that help capture effects of water quality include pH, salinity or ionic strength, divalent cations, dissolved oxygen, and temperature.
11 or other UV wavelengths (e.g. 280 nm, 254 nm; see eqn (2)). Characterizing absorption by particles (e.g., wastewater particles, algae) is more difficult because particles can both absorb and scatter light (see previous page). In addition to scattering coefficients, bulk parameters such as total suspended solids, volatile suspended solids, chlorophyll a, or particulate organic carbon should be reported for samples with high particulate matter. Other parameters that can influence PPRI include NO3−, NO2−, and iron (sources of ˙OH). Also, the steady-state concentrations of key PPRI such as 1O2, ˙OH, and H2O2 can be measured. Other relevant parameters that help capture effects of water quality include pH, salinity or ionic strength, divalent cations, dissolved oxygen, and temperature.
Experiments using organisms cultured in the laboratory must explain in detail the preparation [source of stock culture, growth media, host cells (viruses and phage only), culture conditions, purification steps] and enumeration methods (e.g., selective vs. non-selective media, additives such as pyruvate or antibiotics, and incubation conditions, such as aerobic vs. anaerobic). The extent of the enrichment or purification of the tested stock organism should be clearly described, as residual material from the cultivation (e.g., bacterial debris from bacteriophage propagation) may attenuate light or act as sensitizers. Experiments with indigenous organisms from the environment, which respond to sunlight differently than lab-derived organisms, should provide comprehensive information on the sources of the organisms, concentration methods, and specificity of the enumeration methods used.
Microbial concentration data should be presented as either the log10 or Naperian (natural) logarithm of normalized and unnormalized concentrations, with clear indication of which is used. The Naperian (natural) logarithm of concentration values should always be used for calculation of first-order decay rates. When a shoulder or tail is present, more complex decay models may be necessary.171,222,225 To compare rates between different light conditions (due to differences in irradiance, spectra, or light attenuation), it is necessary to normalize decay rates based on light incident on the target microorganisms (i.e., light transmitted through the water column), which can be accomplished using the fluence,127,226,227 employing a screening factor,117 or calculating rates as a function of the photon flux.91,114
10. Applications of sunlight disinfection
The growing understanding of sunlight-mediated inactivation mechanisms, and knowledge about which actual pathogens are susceptible to which mechanisms, provides opportunities to improve our understanding of the fate of microorganisms in engineered systems and natural surface waters exposed to sunlight, as well as optimize the design of engineered systems to enhance disinfection. There are still significant challenges that need to be overcome, however, to translate the mechanistic modeling approaches described in Section 8 into useful models for real-world application. In addition, accounting for other loss processes (not related to sunlight) and accurately characterizing transport in real water bodies are major challenges to modeling natural systems.10.1. Recreational waters and shellfish waters
Recreational waters are monitored for fecal indicator bacteria (FIB; total coliforms, fecal coliforms, E. coli, and enterococci) around the world to characterize risk for bathers.228 If local standards are exceeded, then bathing waters are considered unfit for swimming. Sunlight has been shown to affect concentrations of FIB in recreational waters, both seawater2,229,230 and freshwater.231,232 This means that water sampled in the afternoon, after several hours of sunlight exposure, may have lower FIB levels and comply with water quality standards, while water sampled at night or in the early morning may not. Understanding when sunlight will be important in reducing bacterial concentrations can help guide the design of water quality monitoring to protect public health. Models of natural surface water quality that account for sunlight effects often assume either a constant sunlight inactivation rate, or a rate that varies as a function of sunlight irradiance.231,233 To date, the more complete approach outlined in Section 8 to account for sunlight inactivation has not been integrated into models predicting the concentrations of indicator organisms in recreational waters.Microbial water quality is also a concern in surface waters used for shellfish harvesting and bivalve aquaculture, as pathogens can be concentrated in the flesh of filter-feeding shellfish. Sunlight is recognized as a key variable that can affect the concentrations of microbial pollutants in shellfisheries.234 Norovirus is a particular concern, and its inactivation by simulated sunlight has been studied with the aim of informing shellfish protection.235
Microbial community analysis has also been proposed to identify sources of microbial pollution by matching microbial communities between surface water samples and potential sources.236,237 However, the differential effect of sunlight on microbial species in surface waters over time complicates this potential source tracking method.238
Recreational waters' bacterial standards were set using epidemiology studies that relate indicator microorganism concentrations to health risk. It is presumed in these studies that an indicator-pathogen relationship exists that gives rise to an indicator-health relationship.239 Epidemiology studies have not specifically considered the effect of sunlight, although a recent study found that risks were reduced when swimming on very sunny versus less sunny days.240 If sunlight differentially affects indicator bacteria and the actual pathogens causing recreational waterborne illness, then the indicator-health relationship may be different under high sunlight versus low sunlight conditions. Thus, research that aims to understand the differential response of indicators and pathogens to sunlight is a priority for improving the management of recreational water quality.
10.2. Natural treatment systems for wastewater and stormwater
One of the oldest and most widespread applications of sunlight-mediated inactivation is the disinfection of wastewater in natural treatment systems, such as wastewater treatment ponds (WTP).241 A typical WTP system has an overall hydraulic retention time of weeks to months, and is comprised of a series of ponds: primary (anaerobic or facultative), secondary (facultative), and maturation. The conditions are particularly conducive to photoinactivation in maturation ponds, due to their shallow depths (typically 0.5 m) and high concentrations of planktonic algae, which give rise to supersaturated dissolved oxygen and elevated pH during sunlit hours when photosynthetic rates are high.3 In such ponds, the rate of inactivation of indicator bacteria and viruses by sunlight-mediated processes has been shown to be much greater than the rate of removal by dark processes (such as sedimentation of particle-associated organisms and predation).212,242Because algal pond waters have very high attenuation in the UV range (Fig. 3), and this attenuation is strongly wavelength-dependent, longer wavelengths are relatively more important to overall photoinactivation. Thus, direct inactivation by UVB may be low or negligible, whereas exogenous processes may be very important for organisms that are susceptible. Both dissolved organic matter158,243 and particulate organic matter114,152,158 have been shown to contribute to inactivation. Because direct inactivation may be minimal, a potential concern is that viruses that are not susceptible to exogenous inactivation may be removed less efficiently.18 For E. coli, inactivation was shown to be enhanced by high dissolved oxygen and high pH in microcosm experiments with algal pond water,216 although it is unclear whether the increased inactivation is due to more efficient endogenous indirect mechanisms or that the cells become susceptible to exogenous mechanisms,158 and it is difficult to isolate the effect of these mechanisms in full-scale ponds.212,244
The open water unit-process wetland discussed in Section 8 appears to be a promising design for maximizing sunlight-mediated removal of indicator bacteria and viruses in a natural treatment system. In this pilot-scale system (20 cm water depth), the removals were estimated to be dominated by exogenous mechanisms for F+ RNA coliphage, endogenous mechanisms for E. coli and poliovirus, and Ent. faecalis was removed by both endogenous and exogenous mechanisms. Modeling suggested that wetland cells up to 40 cm may achieve even higher removals, unless deeper water causes a shift in the algal population from diatoms that accumulate in the benthic sediment layer to planktonic algae, in which case the light attenuation increase could bias model predictions.
High-rate ponds (HRPs), which employ a low-power paddle-wheel to provide mixing and circulate the water in a raceway configuration, are a proven design for achieving high levels of sunlight-mediated disinfection212 as well as improved nutrient and oxygen-demand removal. HRPs have the major advantage over conventional ponds of preventing short-circuiting,11 which can dramatically compromise overall disinfection efficiency.4 A 30 cm and 45 cm deep HRP with the same hydraulic retention time were found to provide very similar removal of E. coli.11 Operating ponds in semi-batch (draw and fill) mode is another option for improving hydraulic efficiency and increasing sunlight-mediated inactivation.242 A direct comparison of the disinfection efficiency of open water wetlands (algae predominately in biomat at bottom) and HRPs (algae predominately suspended throughout water column), and the advantages and disadvantages of each design, would be a valuable contribution to guide improvements in disinfection in natural treatment systems.
One application of sunlight-mediated inactivation in natural treatment systems that is relatively unexplored is stormwater treatment. Many stormwater management approaches involve retaining water in ponds or wetlands with substantial open-water areas. The dominant sunlight inactivation mechanisms in stormwater ponds may be expected to vary depending on the concentration of organic matter (light attenuation, sensitizers) and the water depth.
10.3. Solar disinfection of drinking water (SODIS)
Thorough reviews on the theory and practice of SODIS have been published by Reed (2004)6 and more recently updated by McGuigan et al. (2012).5 The most common approach involves filling plastic beverage bottles (e.g., 1.5 L polyethylene terepthlalate (PET)) with water to be treated and exposing them to sunlight for one day; if the weather is cloudy the recommended exposure time is 2 d.5 The main photochemical mechanism through which bacteria are inactivated during conventional SODIS is via endogenous indirect inactivation; direct inactivation is likely minimal because PET bottles do not transmit UVB light,222 and exogenous inactivation is likely minimal in most waters used for drinking because few exogenous sensitizers are present. In the absence of high temperatures, inactivation of most viruses during conventional SODIS is likely to be poor,245 particularly in waters with low photoreactivity,222 given that sunlight-mediated inactivation of viruses occurs via direct and exogenous mechanisms. The use of container materials that are more transparent to sunlight, in particular UVB wavelengths, can increase photoinactivation of indicator bacteria and viruses by SODIS.222During SODIS there is the potential for the temperature of the water to increase significantly during sunlight exposure, which increases the effectiveness of photoinactivation.5 Synergistic temperature effects are notable above 30 °C;5,214,246 above ∼70 °C thermal inactivation (pasteurization) becomes faster than photoinactivation. Various modifications to SODIS containers have been reported to enhance temperature effects, including painting the bottom of the container black, placing the bottles on a black surface, placing the bottles in a solar box oven, and concentrating the sunlight through the use of mirrors, in particular Compound Parabolic Collectors.5,176,247 Mechanistic modeling has provided valuable insights into the effects of key parameters, such as clear versus cloudy skies, turbidity, and container material on the inactivation of E. coli in SODIS bottles and parabolic reactors.246
Considerable research efforts have been directed toward the development of practical approaches for enhancing sunlight-driven disinfection. One of the simplest such approaches involves the addition of H2O2 into solution during solar irradiation to accelerate inactivation of certain viruses, bacteria, and fungi relative to conventional SODIS processes.222,248–252 The observed benefits of H2O2 addition can been attributed to its participation in endogenous or exogenous photo-Fenton processes involving Fe(II) or Fe(III) associated with biomolecules present in the target microbial agents,222,248,253,254 or naturally-occurring Fe(III) associated with Fe-(hydr)oxide complexes, Fe-organic ligand complexes (formed through interactions with acidic groups in NOM), and/or solid Fe oxides present in the water to be treated. Photo-Fenton processes are in each case expected to lead to production of such oxidants as ˙OH or Fe(IV), depending on solution pH.254–256 Accordingly, inactivation appears to be particularly effective when waters dosed with H2O2 already also contain significant iron in the bulk solution,252,254,257 or are amended with copper in the presence of ascorbic acid (likely due to the ability of copper to participate in Fenton-like reactions).222,248
For waters that are not naturally enriched in Fe, the effectiveness of SODIS processes can also be improved by addition of Fe (with or without addition of H2O2) to drive exogenous photo-Fenton processes.254,258,259 Fe addition may also yield the benefit of enhanced endogenous photo-Fenton chemistry within bacterial cells due to siderophore-mediated intracellular accumulation of added Fe.260 Amending waters with an organic acid such as citrate can serve to yield further improvements in oxidant yields, by improving Fe solubility through formation of stable metal–ligand complexes, which may themselves participate in ligand-to-metal charge transfer upon solar irradiation.254 The use of innocuous, naturally-occurring, and widely-accessible reagents (Fe, H2O2, and/or organic acids) is an added benefit of this approach to SODIS enhancement. Applications of photo-Fenton processes to enhance microbial inactivation kinetics have been successfully demonstrated in a variety of natural water matrixes under field conditions at scales ranging from 1 L water bottles up to 50 L.254
An alternative approach to SODIS enhancement is the solar “photoactivation” of free available chlorine (HOCl/OCl−) to yield ˙OH, RHS, and O3.261 The production of ˙OH and O3 during exposure of chlorine-containing solutions to natural sunlight under conditions typical of SODIS processes (e.g., pH 8, T = 33 °C) can accelerate inactivation of highly chlorine-resistant B. subtilis endospores and Cryptosporidium parvum oocysts by more than 200% compared to either chlorine-containing dark controls or light controls in the absence of chlorine.24,25 This approach could present interesting opportunities for improving the effectiveness of either chlorine-based disinfection or SODIS.261,262 It is important to note that photoactivation of free available chlorine by UVB and UVA wavelengths may lead to elevated levels of disinfection byproducts.263 Thus, the use of this approach would require careful selection of treatment conditions to minimize risks of byproduct exposure while maximizing inactivation of recalcitrant pathogens.
The effectiveness of solar disinfection may also be enhanced by the addition of exogenous photosensitizer compounds to increase PPRI, although the practicality of this approach has not been demonstrated.264 Organic sensitizers include flavins and psoralens (furocoumarins). Riboflavin has been shown to accelerate the rates with which various viral, bacterial, and protozoan pathogens can be inactivated during exposure to simulated sunlight,186,265 presumably due to interactions of the pathogens with either 1O2 or the excited triplet-state of the riboflavin itself.266 Significant enhancements of viral and bacterial inactivation rates have also been observed in UVA or natural sunlight-irradiated solutions dosed with pure synthetic 5-methoxypsoralen (MOP), as well as lime juice, likely due to attack of DNA in the target organisms by photoexcited MOP and psoralens in the lime fruit (which is higher than in lemons).267–269
Alternatively, heterogeneous photocatalysts have been used for production of PPRI, such as TiO2 mineral phases.264,270 Undoped TiO2 phases (e.g., anatase, Degussa P25) may be excited by absorption of UVA light, resulting in the coupled reduction of O2 and oxidation of H2O at the photocatalyst surface, in turn leading to the formation of such ROS as O2˙−, H2O2, and ˙OH.271,272 Certain doped TiO2 phases are also known to exhibit similar photoactivity over the visible region of the solar spectrum.272 TiO2 photocatalysts are generally used either in suspension273 or in coatings applied to the inner surfaces of plastic or glass reactors.272 The use of these materials during SODIS processes has been shown to yield improved inactivation of a wide variety of microorganisms and has been demonstrated to function under both small- and large-scale conditions.182,258,259,272 A number of recent studies have also evaluated the use of C60 fullerenes functionalized with hydrophilic surface groups (e.g., –OH, –NH2, etc.) as photocatalysts in solar disinfection. Such materials are reported to generate 1O2 and/or ˙OH upon photoexcitation with UVA radiation, and have been shown to accelerate inactivation of various viruses, bacteria, and fungi in aqueous solution.122,274–276 Other nanomaterial-based catalysts are being explored, such as vertically aligned MoS2 nanofilms.277 However, the practicality of applying such approaches under field conditions may ultimately be limited, as heterogeneous photocatalysis approaches require the use of either suspensions of photocatalyst – which must be removed prior to water consumption, or coated reactor surfaces – which suffer from mass transfer constraints on exposure of organisms to ROS generated at the catalyst–water interface. Despite decades of intensive investigation in the laboratory, practical field designs employing photocatalysts have not emerged.278
10.4. Solar radiation and microbial ecology
Sunlight strongly influences the bacterial and viral assemblages found in surface waters. Bacteria can be inhibited or stimulated by sunlight exposure.279 Research focused on UV radiation (UVA and UVB) shows an inhibitory effect on cellular activities due to the mechanisms described in this review article. The effect of UV radiation, however, varies by bacterial group.280,281 Differences in sensitivity to UV radiation between bacterial groups result in changes in bacterial community composition depending on UV dose.280 Researchers have also found that sunlight stimulates bacterial growth by the production of bioavailable organic matter and photorepair of sunlight induced damage.1 Sunlight acts as an energy source, both directly for photosynthetic organisms and indirectly through the photo-transformation of organic matter into a biologically labile form.1,282 The bioavailable organic matter supports growth of certain bacterial groups and repair after sunlight exposure, leading to changes in bacterial community composition.279,280,283 Specific wavelengths of sunlight can stimulate photorepair or photoreactivation.1,284 Sunlight also influences bacterial community composition by controlling bacteriophage populations in surface waters via destruction of viral particles or reduction of viral infectivity during sunlight exposure.106,238,285 Thus, the overall effect of sunlight on bacterial communities in surface waters is the sum of detrimental and beneficial processes. Advances in next-generation sequencing (NGS) technologies are making it cheaper, easier and faster to investigate bacterial communities in surface waters. Understanding the effect of sunlight on bacterial communities will be especially important when analyzing experimental findings238 and building models describing photo-induced chemical and biological processes affecting bacterial communities in aquatic environments.1,28611. Research priorities
Based on our critical review of the current literature on sunlight inactivation of microorganisms, we have identified a number of research areas to prioritize for future work:11.1 UVB measurements and predictions
More accurate approaches for quantifying the UVB portion of natural and simulated sunlight are needed. Improved models for radiative transfer of sunlight into surface waters should be applied to model sunlight inactivation.11.2 Reactivity of microbial building blocks
To enable truly mechanistic descriptions of endogenous and exogenous inactivation, the reactions of biomolecules exposed to solar radiation and PPRI need to be more fully characterized. In addition, an understanding of how reactivity is influenced by the higher-order structure of microorganisms (compared to biomolecules) is needed.11.3 Expansion of microorganisms studied
To date, most of the research on sunlight inactivation has been on a limited number of microorganisms. Our understanding needs to be expanded to a broader suite of health-relevant viruses, bacteria, and protozoa with varied structures and biology, including how the inactivation rates of actual pathogens compare to commonly used indicator organisms for the range of sunlight and water quality conditions. As one example, very little information is available on endogenous and exogenous reaction rates of enveloped viruses.11.4 Endogenous inactivation by solar wavelengths
Additional studies are needed on the mechanisms of endogenous inactivation induced by solar wavelengths. Rate constants for inactivation by sunlight, rather than UVC, should be measured for a wider range of microorganisms. Furthermore, action spectra should be obtained for different microorganisms in the solar spectrum range. The development of methods for wavelength-specific actinometry would be a useful contribution to facilitate comparison of experiments conducted with light sources with different irradiance spectra and waters with different absorption spectra.11.5 PPRI responsible for bacterial inactivation
Whereas the PPRI involved in virus inactivation have been largely characterized, more research on the key exogenous ROS for bacteria is necessary.11.6 Beyond monodispersed microorganisms
Future efforts should seek to understand how sensitizer characteristics and association with microorganisms impacts inactivation rates and mechanisms. Also needed is a better understanding of how aggregation of the microorganisms or sorption on surfaces affects inactivation kinetics in the field.11.7 Particulate organic matter (POM)
To date, most of the focus on the role of organic matter in sunlight inactivation has been on DOM. Wastewater effluent and wetland-derived organic matter are of particular interest as sensitizers, and typically contain high concentrations of particulate organic matter (POM). Association between microorganisms and POM is likely. A further complication is that particulate organic matter scatters radiation, as well as absorbing it, affecting spectral irradiance in waters. Future research could compare photoinactivation in filtered versus unfiltered waters.11.8 Laboratory microorganism effects
More research is needed to identify the factors that alter the susceptibility of laboratory cultures to sunlight, and that underlie differences between laboratory-grown and ‘native’ (environmentally-adapted) bacteria to sunlight. Also, more work is needed to explain why growth phase/physiology affects inactivation of bacterial species. This understanding is important for informing how to conduct experiments with laboratory organisms that can be accurately extrapolated to different field conditions.11.9 Influence of inorganic constituents
Research is needed to characterize the influence of inorganic constituents of waters, notably salinity, halides, iron, dissolved oxygen, and pH, on exogenous inactivation under real-world conditions.11.10 Omics tools
High throughput sequencing, metagenomics, proteomics, transcriptomics, and other omic tools can be further applied to understand pathways of damage that lead to inactivation by sunlight. Also, the ability to characterize changes in the composition of entire microbial communities should be applied to expand the focus beyond readily culturable pathogens and indicator organisms.11.11 Interactions with other inactivation processes
Sunlight can influence other inactivation processes by, for example, increasing the water temperature, enabling photosynthesis, altering the activity of grazers and other predators. These effects need to be incorporated into models to accurately predict overall inactivation.11.12 Validation and improvement of models
More work is needed that compares the results from predictive, mechanistic models with actual measurements of inactivation under a range of conditions in natural and engineered systems. A comparison of mechanistic and statistical modeling approaches would also be insightful. This work will provide insight into the level of detail that is required to accurately predict inactivation, given the variability inherent to real-world systems.11.13 Applications of sunlight inactivation
The improving mechanistic understanding of sunlight inactivation should be exploited to further develop creative and practical disinfection strategies for drinking water, wastewater, and stormwater and better management of natural waters.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
K. L. N., A. B. B., P. A. M., K. M. P., W. A. M., and A. I. S. acknowledge support from the National Science Foundation through the Engineering Research Center for Reinventing the Nation's Urban Water Infrastructure (EEC-1028968). M. C. D. acknowledges support from the National Science Foundation through Grant No. CBET-1236303 and CBET-1254929. This paper has been reviewed in accordance with the U.S. Environmental Protection Agency's (U.S. EPA) peer and administrative review policies and approved for publication. Mention of trade names or commercial products does not constitute an endorsement or recommendation for use by the U.S. EPA.References
- M. A. Moran and R. G. Zepp, in Microbial Ecology of the Oceans, ed. D. Kirchman, Wiley-Liss, Inc., 2000 Search PubMed.
- L. W. Sinton, in Oceans and Health: Pathogens in the Marine Environment, ed. S. Belkin and R. Colwell, Springer, New York, 2005, pp. 69–92 Search PubMed.
- A. Shilton, Pond Treatment Technology, IWA Publishing, London, 2005 Search PubMed.
- J. T. Jasper, M. T. Nguyen, Z. L. Jones, N. S. Ismail, D. L. Sedlak, J. O. Sharp, R. G. Luthy, A. J. Horne and K. L. Nelson, Unit Process Wetlands for Removal of Trace Organic Contaminants and Pathogens from Municipal Wastewater Effluents, Environ. Eng. Sci., 2013, 30, 421–436 CrossRef PubMed.
- K. G. McGuigan, R. M. Conroy, H.-J. Mosler, M. du Preez, E. Ubomba-Jaswa and P. Fernandez-Ibanez, Solar water disinfection (SODIS): A review from bench-top to roof-top, J. Hazard. Mater., 2012, 235, 29–46 CrossRef PubMed.
- R. H. Reed, The inactivation of microbes by sunlight: Solar disinfection as a water treatment process, Adv. Appl. Microbiol., 2004, 54, 333–365 Search PubMed.
- R. Zepp and D. Cline, Rates of Direct Photolysis in Aquatic Environment, Environ. Sci. Technol., 1977, 11, 359–366 CrossRef.
- C. Mobley and L. Sundman, Hydrolight 5. Ecolight 5. Technical Documentation, Sequoia Scientific, Inc., Bellevue, WA, 2008 Search PubMed.
- C. A. Gueymard, Interdisciplinary applications of a versatile spectral solar irradiance model: A review, Energy, 2005, 30, 1551–1576 CrossRef.
- J. Kirk, Light and Photosynthesis in Aquatic Ecosystems, Cambridge University Press, Cambridge, 3rd edn, 2011 Search PubMed.
- R. J. Craggs, R. J. Davies-Colley, C. C. Tanner and J. P. Suklas, Advanced pond system: Performance with high rate ponds of different depths and areas, Water Sci. Technol., 2003, 48, 259–267 CrossRef PubMed.
- C. E. Williamson, S. Madronich, A. Lal, R. G. Zepp, R. M. Lucas, E. P. Overholt, K. C. Rose, S. G. Schladow and J. Lee-Taylor, Climate change-induced increases in precipitation are reducing the potential for solar ultraviolet radiation to inactivate pathogens in surface waters, Sci. Rep., 2017, 7, 13033 CrossRef PubMed.
- G. Miller and R. Zepp, Effects of Suspended Sediments on Photolysis Rates of Dissolved Pollutants, Water Res., 1979, 13, 453–459 CrossRef.
- A. Morel, B. Gentili, H. Claustre, M. Babin, A. Bricaud, J. Ras and F. Tièche, Optical properties of the “clearest” natural waters, Limnol. Oceanogr., 2007, 52, 217–229 CrossRef.
- M. P. Gall, R. J. Davies-Colley and R. A. Merrilees, Exceptional visual clarity and optical purity in a sub-alpine lake, Limnol. Oceanogr., 2013, 58, 443–451 CrossRef.
- C. L. Gallegos, R. J. Davies-Colley and M. Gall, Optical closure in lakes with contrasting extremes of reflectance, Limnol. Oceanogr., 2008, 53, 2021–2034 CrossRef.
- A. I. Silverman, M. T. Nguyen, I. E. Schilling, J. Wenk and K. L. Nelson, Sunlight Inactivation of Viruses in Open-Water Unit Process Treatment Wetlands: Modeling Endogenous and Exogenous Inactivation Rates, Environ. Sci. Technol., 2015, 49, 2757–2766 CrossRef PubMed.
- R. J. Davies-Colley, R. J. Craggs, J. Park, J. P. S. Sukias, J. W. Nagels and R. Stott, Virus removal in a pilot-scale ‘advanced’ pond system as indicated by somatic and F-RNA bacteriophages, Water Sci. Technol., 2005, 51, 107–110 CrossRef PubMed.
- L. I. Grossweiner and K. C. Smith, in The Science of Photobiology, ed. K. C. Smith, Plenum Press, New York and London, 2nd edn, 1989 Search PubMed.
- A. Eisenstark, Mutagenic and lethal effects of near-ultraviolet radiation (290–400 nm) on bacteria and phage, Environ. Mol. Mutagen., 1987, 10, 317–337 CrossRef PubMed.
- D. Vione, M. Minella, V. Maurino and C. Minero, Indirect Photochemistry in Sunlit Surface Waters: Photoinduced Production of Reactive Transient Species, Chem.–Eur. J., 2014, 20, 10590–10606 CrossRef PubMed.
- C. A. Stedmon, S. Markager and H. Kaas, Optical properties and signatures of chromophoric dissolved organic matter (CDOM) in Danish coastal waters, Estuarine, Coastal Shelf Sci., 2000, 51, 267–278 CrossRef.
- K. McNeill and S. Canonica, Triplet state dissolved organic matter in aquatic photochemistry: reaction mechanisms, substrate scope, and photophysical properties, Environ. Sci.: Processes Impacts, 2016, 18, 1381–1399 RSC.
- S. Mostafa and F. L. Rosario-Ortiz, Singlet Oxygen Formation from Wastewater Organic Matter, Environ. Sci. Technol., 2013, 47, 8179–8186 CrossRef PubMed.
- M. M. Dong and F. L. Rosario-Ortiz, Photochemical Formation of Hydroxyl Radical from Effluent Organic Matter, Environ. Sci. Technol., 2012, 46, 3788–3794 CrossRef PubMed.
- L. C. Bodhipaksha, C. M. Sharpless, Y.-P. Chin, M. Sander, W. K. Langston and A. A. MacKay, Triplet Photochemistry of Effluent and Natural Organic Matter in Whole Water and Isolates from Effluent-Receiving Rivers, Environ. Sci. Technol., 2015, 49, 3453–3463 CrossRef PubMed.
- M. Pearson and H. Johns, Suppression of Hydrate and Dimer Formation in Ultraviolet-Irradiated Poly (a + U) Relative to Poly U, J. Mol. Biol., 1966, 20, 215–229 CrossRef PubMed.
- W. J. Schreier, T. E. Schrader, F. O. Koller, P. Gilch, C. E. Crespo-Hernandez, V. N. Swaminathan, T. Carell, W. Zinth and B. Kohler, Thymine dimerization in DNA is an ultrafast photoreaction, Science, 2007, 315, 625–629 CrossRef PubMed.
- P. Klán and J. Wirz, Photochemistry of organic compounds: From concepts to practice, John Wiley & Sons, 2009 Search PubMed.
- J.-L. Ravanat, T. Douki and J. Cadet, Direct and indirect effects of UV radiation on DNA and its components, J. Photochem. Photobiol., B, 2001, 63, 88–102 CrossRef.
- Z. Qiao and K. R. Wigginton, Direct and Indirect Photochemical Reactions in Viral RNA Measured with RT-qPCR and Mass Spectrometry, Environ. Sci. Technol., 2016, 50, 13371–13379 CrossRef PubMed.
- P. Swenson and R. Setlow, Kinetics of Dimer Formation and Photohydration in Ultraviolet-Irradiated Polyuridylic Acid, Photochem. Photobiol., 1963, 2, 419–434 CrossRef.
- D. Mitchell and J. Clarkson, Induction of Photoproducts in Synthetic Polynucleotides by Far and Near Ultraviolet-Radiation, Photochem. Photobiol., 1984, 40, 735–741 CrossRef PubMed.
- Z. Tramer, K. Wierzchowski and D. Shugar, Influence of Polynucleotide Secondary Structure on Thymine Photodimerization, Acta Biochim. Pol., 1969, 16, 83–107 Search PubMed.
- G. J. Fisher and H. E. Johns, in Photochemistry and photobiology of nucleic acids, Academic Press, New York, 1976, vol. 1 Search PubMed.
- J. Burr, A. Chan and E. Park, Nature of Reactive Species in Photohydration of Uracil and Cytosine Derivatives, J. Am. Chem. Soc., 1972, 94, 5866 CrossRef PubMed.
- T. Douki, Low ionic strength reduces cytosine photoreactivity in UVC-irradiated isolated DNA, Photochem. Photobiol. Sci., 2006, 5, 1045 RSC.
- Z. Pan, M. Hariharan, J. D. Arkin, A. S. Jalilov, M. McCullagh, G. C. Schatz and F. D. Lewis, Electron Donor–Acceptor Interactions with Flanking Purines Influence the Efficiency of Thymine Photodimerization, J. Am. Chem. Soc., 2011, 133, 20793–20798 CrossRef PubMed.
- Z. Kuluncsics, D. Perdiz, E. Brulay, B. Muel and E. Sage, Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts, J. Photochem. Photobiol., B, 1999, 49, 71–80 CrossRef.
- O. C. Zafiriou, Sources and reactions of OH and daughter radicals in seawater, J. Geophys. Res., 1974, 79, 4491–4497 CrossRef.
- J. Hoigné, B. C. Faust, W. R. Haag, F. E. Scully and R. G. Zepp, Aquatic Humic Substances as Sources and Sinks of Photochemically Produced Transient Reactants, in Aquatic Humic Substances, ed. P. Suffet and I. H. MacCarthy, American Chemical Society, Washington, D.C., 1988, Advances in Chemistry, vol. 219, pp. 363–381 Search PubMed.
- N. V. Blough and R. G. Zepp, in Active Oxygen in Chemistry, ed. C. S. Foote, J. S. Valentine, A. Greenberg, J. F. Liebman and A. Greenberg, Chapman and Hall, New York, NY, 1995, vol. 2, pp. 280–333 Search PubMed.
- T. Kohn, M. J. Mattle, M. Minella and D. Vione, A modeling approach to estimate the solar disinfection of viral indicator organisms in waste stabilization ponds and surface waters, Water Res., 2016, 88, 912–922 CrossRef PubMed.
- K. M. Parker and W. A. Mitch, Halogen radicals contribute to photooxidation in coastal and estuarine waters, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5868–5873 CrossRef PubMed.
- M. Bodrato and D. Vione, APEX (Aqueous Photochemistry of Environmentally occurring Xenobiotics): a free software tool to predict the kinetics of photochemical processes in surface waters, Environ. Sci.: Processes Impacts, 2014, 16, 732–740 RSC.
- R. P. Schwarzenbach, P. M. Gschwend and D. M. Imboden, Environmental Organic Chemistry, Wiley-Interscience, New York, 2nd edn, 2002 Search PubMed.
- The science of photobiology, ed. K. C. Smith, Springer Science & Business Media, 1989 Search PubMed.
- M. J. Davies, Singlet oxygen-mediated damage to proteins and its consequences, Biochem. Biophys. Res. Commun., 2003, 305, 761–770 CrossRef PubMed.
- A. L. Boreen, B. L. Edhlund, J. B. Cotner and K. McNeill, Indirect photodegradation of dissolved free amino acids: the contribution of singlet oxygen and the differential reactivity of DOM from various sources, Environ. Sci. Technol., 2008, 42, 5492–5498 CrossRef PubMed.
- R. A. Lundeen, E. M.-L. Janssen, C. Chu and K. McNeill, Environmental Photochemistry of Amino Acids, Peptides and Proteins, CHIMIA International Journal for Chemistry, 2014, 68, 812–817 CrossRef PubMed.
- A. Michaeli and J. Feitelson, Reactivity of singlet oxygen toward amino acids and peptides, Photochem. Photobiol., 1994, 59, 284–289 CrossRef PubMed.
- E. Maisonneuve, A. Ducret, P. Khoueiry, S. Lignon, S. Longhi, E. Talla and S. Dukan, Rules Governing Selective Protein Carbonylation, PLoS One, 2009, 4, e7269 CrossRef PubMed.
- J. Jagger, Solar-UV Actions on Living Cells, Praeger Publishers, New York, 1985 Search PubMed.
- C. D. Lytle and J. L. Sagripanti, Predicted inactivation of viruses of relevance to biodefense by solar radiation, J. Virol., 2005, 79, 14244–14252 CrossRef PubMed.
- M. Hessling, B. Spellerberg and K. Hoenes, Photoinactivation of bacteria by endogenous photosensitizers and exposure to visible light of different wavelengths – a review on existing data, FEMS Microbiol. Lett., 2017, 364(2), 1–12 CrossRef PubMed.
- J. J. Cullen and P. J. Neale, in Effects of Ozone Depletion on Aquatic Ecosystems, ed. D.-P. Häder, Academic Press, San Diego, 1997, pp. 91–118 Search PubMed.
- R. Rundel, Action Spectra and Estimation of Biologically Effective UV-Radiation, Physiol. Plant., 1983, 58, 360–366 CrossRef.
- R. G. Zepp, in UV Effects in Aquatic Organisms and Ecosystems, ed. E. W. Helbling and H. Zagarese, Royal Society of Chemistry, 2003, p. 575 Search PubMed.
- R. Sutton and G. Sposito, Molecular Structure in Soil Humic Substances: The New View, Environ. Sci. Technol., 2005, 39, 9009–9015 CrossRef PubMed.
- E. Appiani, R. Ossola, D. E. Latch, P. R. Erickson and K. McNeill, Aqueous singlet oxygen reaction kinetics of furfuryl alcohol: effect of temperature, pH, and salt content, Environ. Sci.: Processes Impacts, 2017, 19, 507–516 RSC.
- D. E. Latch and K. McNeill, Microheterogeneity of singlet oxygen distributions in irradiated humic acid solutions, Science, 2006, 311, 1743–1747 CrossRef PubMed.
- T. Kohn, M. Grandbois, K. McNeill and K. L. Nelson, Association with Natural Organic Matter Enhances the Sunlight-Mediated Inactivation of MS2 Coliphage by Singlet Oxygen, Environ. Sci. Technol., 2007, 41, 4626–4632 CrossRef PubMed.
- T. Kohn and K. L. Nelson, Sunlight-mediated inactivation of MS2 coliphage via exogenous singlet oxygen produced by sensitizers in natural waters, Environ. Sci. Technol., 2007, 41, 192–197 CrossRef PubMed.
- M. T. Nguyen, Ph.D. Dissertation, University of California, Berkeley, 2015.
- S. E. Burns, J. P. Hassett and M. V. Rossi, Binding effects on humic-mediated photoreaction: Intrahumic dechlorination of mirex in water, Environ. Sci. Technol., 1996, 30, 2934–2941 CrossRef.
- S. E. Burns, J. P. Hassett and M. V. Rossi, Mechanistic implications of the intrahumic dechlorination of mirex, Environ. Sci. Technol., 1997, 31, 1365–1371 CrossRef.
- M. Grandbois, D. E. Latch and K. McNeill, Microheterogeneous concentrations of singlet oxygen in natural organic matter isolate solutions, Environ. Sci. Technol., 2008, 42, 9184–9190 CrossRef PubMed.
- C. Chu, R. A. Lundeen, C. K. Remucal, M. Sander and K. McNeill, Enhanced Indirect Photochemical Transformation of Histidine and Histamine through Association with Chromophoric Dissolved Organic Matter, Environ. Sci. Technol., 2015, 49, 5511–5519 CrossRef PubMed.
- T. Zhang and H. Hsu-Kim, Photolytic degradation of methylmercury enhanced by binding to natural organic ligands, Nat. Geosci., 2010, 3, 473–476 CrossRef PubMed.
- P. A. Maurice, M. Manecki, J. B. Fein and J. Schaefer, Fractionation of an aquatic fulvic acid upon adsorption to the bacterium, Bacillus subtilis, Geomicrobiol. J., 2004, 21, 69–78 CrossRef.
- A. Armanious, M. Aeppli, R. Jacak, D. Refardt, T. Sigstam, T. Kohn and M. Sander, Viruses at Solid–Water Interfaces: A Systematic Assessment of Interactions Driving Adsorption, Environ. Sci. Technol., 2016, 50, 732–743 CrossRef PubMed.
- A. F. Carlucci and D. Pramer, An Evaluation of Factors Affecting the Survival of Escherichia coli in Sea Water: I. Experimental Procedures, Appl. Microbiol., 1960, 8, 243 Search PubMed.
- R. S. Fujioka, H. H. Hashimoto, E. B. Siwak and R. H. Young, Effect of sunlight on survival of indicator bacteria in seawater, Appl. Environ. Microbiol., 1981, 41, 690–696 Search PubMed.
- B. J. Dutka, Sensitivity of Legionella pneumophila to sunlight in fresh and marine waters, Appl. Environ. Microbiol., 1984, 48, 970–974 Search PubMed.
- L. M. Evison, Comparative studies on the survival of indicator organisms and pathogens in fresh and sea water, Water Sci. Technol., 1988, 20, 309–315 CrossRef.
- C. M. Davies and L. M. Evison, Sunlight and the survival of enteric bacteria in natural waters, J. Appl. Bacteriol., 1991, 70, 265–274 CrossRef PubMed.
- L. W. Sinton, C. H. Hall, P. A. Lynch and R. J. Davies-colley, Sunlight Inactivation of Fecal Indicator Bacteria and Bacteriophages from Waste Stabilization Pond Effluent in Fresh and Saline Waters, Appl. Environ. Microbiol., 2002, 68, 1122–1131 CrossRef PubMed.
- A. B. Boehm, C. Soetjipto and D. Wang, Solar inactivation of four Salmonella serovars in fresh and marine waters, J. Water Health, 2012, 10, 504–510 CrossRef PubMed.
- R. J. Davies-Colley, R. G. Bell and A. M. Donnison, Sunlight Inactivation of Enterococci and Fecal Coliforms in Sewage Effluent Diluted in Seawater, Appl. Environ. Microbiol., 1994, 60, 2049–2058 Search PubMed.
- L. W. Sinton, R. J. Davies-Colley and R. G. Bell, Inactivation of enterococci and fecal coliforms from sewage and meatworks effluents in seawater chambers., Appl. Environ. Microbiol., 1994, 60, 2040–2048 Search PubMed.
- B. Yuan, M. Pham and T. H. Nguyen, Deposition Kinetics of Bacteriophage MS2 on a Silica Surface Coated with Natural Organic Matter in a Radial Stagnation Point Flow Cell, Environ. Sci. Technol., 2008, 42, 7628–7633 CrossRef PubMed.
- K. M. Parker, J. J. Pignatello and W. A. Mitch, Influence of ionic strength on triplet-state natural organic matter loss by energy transfer and electron transfer pathways, Environ. Sci. Technol., 2013, 47, 10987–10994 CrossRef PubMed.
- F. al Housari, D. Vione, S. Chiron and S. Barbati, Reactive photoinduced species in estuarine waters. Characterization of hydroxyl radical, singlet oxygen and dissolved organic matter triplet state in natural oxidation processes, Photochem. Photobiol. Sci., 2010, 9, 78–86 RSC.
- K. Mopper and X. Zhou, Hydroxyl radical photoproduction in the sea and its potential impact on marine processes, Science, 1990, 250, 661–664 CrossRef PubMed.
- A. Jammoul, S. Dumas, B. D'Anna and C. George, Photoinduced oxidation of sea salt halides by aromatic ketones: a source of halogenated radicals, Atmos. Chem. Phys., 2009, 9, 4229–4237 CrossRef.
- G. E. Adams, J. E. Aldrich, R. H. Bisby, R. B. Cundall, J. L. Redpath and R. L. Willson, Selective free radical reactions with proteins and enzymes: reactions of inorganic radical anions with amino acids, Radiat. Res., 1972, 49, 278–289 CrossRef PubMed.
- J. E. Grebel, J. J. Pignatello and W. A. Mitch, Effect of halide ions and carbonates on organic contaminant degradation by hydroxyl radical-based advanced oxidation processes in saline waters, Environ. Sci. Technol., 2010, 44, 6822–6828 CrossRef PubMed.
- Y. Zhang, R. Del Vecchio and N. V. Blough, Investigating the mechanism of hydrogen peroxide photoproduction by humic substances, Environ. Sci. Technol., 2012, 46, 11836–11843 CrossRef PubMed.
- K. M. Parker, E. S. Reichwaldt, A. Ghadouani and W. A. Mitch, Halogen Radicals Promote the Photodegradation of Microcystins in Estuarine Systems, Environ. Sci. Technol., 2016, 50, 8505–8513 CrossRef PubMed.
- R. J. Davies-Colley, A. M. Donnison, D. J. Speed, C. M. Ross and J. W. Nagels, Inactivation of faecal indicator micro-organisms in waste stabilisation ponds: interactions of environmental factors with sunlight, Water Res., 1999, 33, 1220–1230 CrossRef.
- M. J. Mattle, D. Vione and T. Kohn, Conceptual Model and Experimental Framework to Determine the Contributions of Direct and Indirect Photoreactions to the Solar Disinfection of MS2, phiX174, and Adenovirus, Environ. Sci. Technol., 2015, 49, 334–342 CrossRef PubMed.
- A. I. Silverman, B. M. Peterson, A. B. Boehm, K. McNeill and K. L. Nelson, Sunlight inactivation of human viruses and bacteriophages in coastal waters containing natural photosensitizers, Environ. Sci. Technol., 2013, 47, 1870–1878 CrossRef PubMed.
- D. C. Love, A. Silverman and K. L. Nelson, Human virus and bacteriophage inactivation in clear water by simulated sunlight compared to bacteriophage inactivation at a southern California beach, Environ. Sci. Technol., 2010, 44, 6965–6970 CrossRef PubMed.
- F. Bosshard, F. Armand, R. Hamelin and T. Kohn, Mechanisms of human adenovirus inactivation by sunlight and UVC light as examined by quantitative PCR and quantitative proteomics, Appl. Environ. Microbiol., 2013, 79, 1325–1332 CrossRef PubMed.
- O. C. Romero, A. P. Straub, T. Kohn and T. H. Nguyen, Role of temperature and Suwannee River natural organic matter on inactivation kinetics of rotavirus and bacteriophage MS2 by solar irradiation, Environ. Sci. Technol., 2011, 45, 10385–10393 CrossRef PubMed.
- A. I. Silverman, B. M. Peterson, A. B. Boehm, K. McNeill and K. L. Nelson, Sunlight Inactivation of Human Viruses and Bacteriophages in Coastal Waters Containing Natural Photosensitizers, Environ. Sci. Technol., 2013, 47, 1870–1878 CrossRef PubMed.
- K. R. Wigginton, L. Menin, J. P. Montoya and T. Kohn, Oxidation of virus proteins during UV(254) and singlet oxygen mediated inactivation, Environ. Sci. Technol., 2010, 44, 5437–5443 CrossRef PubMed.
- K. R. Wigginton, B. M. Pecson, T. Sigstam, F. Bosshard and T. Kohn, Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity, Environ. Sci. Technol., 2012, 46, 12069–12078 CrossRef PubMed.
- O. C. Romero-Maraccini, J. L. Shisler and T. H. Nguyen, Solar and Temperature Treatments Affect the Ability of Human Rotavirus Wa To Bind to Host Cells and Synthesize Viral RNA, Appl. Environ. Microbiol., 2015, 81, 4090–4097 CrossRef PubMed.
- T. Sigstam, G. Gannon, M. Cascella, B. M. Pecson, K. R. Wigginton and T. Kohn, Subtle differences in virus composition affect disinfection kinetics and mechanisms, Appl. Environ. Microbiol., 2013, 79, 3455–3467 CrossRef PubMed.
- B. M. Pecson, L. V. Martin and T. Kohn, Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UV-B radiation, and singlet oxygen: advantages and limitations of an enzymatic treatment to reduce false-positive results, Appl. Environ. Microbiol., 2009, 75, 5544–5554 CrossRef PubMed.
- K. R. Wigginton, L. Menin, T. Sigstam, G. Gannon, M. Cascella, H. Ben Hamidane, Y. O. Tsybin, P. Waridel and T. Kohn, UV radiation induces genome-mediated, site-specific cleavage in viral proteins, ChemBioChem, 2012, 13, 837–845 CrossRef PubMed.
- A. C. Eischeid and K. G. Linden, Molecular indications of protein damage in adenoviruses after UV disinfection, Appl. Environ. Microbiol., 2011, 77, 1145–1147 CrossRef PubMed.
- I. P. Boszko and A. J. Rainbow, Removal of UV photoproducts from an adenovirus-encoded reporter gene following infection of unirradiated and UV-irradiated human fibroblasts, Somat. Cell Mol. Genet., 1999, 25, 301–315 CrossRef PubMed.
- R. A. Rodriguez, S. Bounty, S. Beck, C. Chan, C. McGuire and K. G. Linden, Photoreactivation of bacteriophages after UV disinfection: Role of genome structure and impacts of UV source, Water Res., 2014, 55, 143–149 CrossRef PubMed.
- S. W. Wilhelm, M. G. Weinbauer, C. A. Suttle and W. H. Jeffrey, The role of sunlight in the removal and repair of viruses in the sea, Limnol. Oceanogr., 1998, 43, 586–592 CrossRef.
- C. D. Lytle and J.-L. Sagripanti, Predicted inactivation of viruses of relevance to biodefense by solar radiation, J. Virol., 2005, 79, 14244–14252 CrossRef PubMed.
- H. Mamane-Gravetz, K. G. Linden, A. Cabaj and R. Sommer, Spectral sensitivity of Bacillus subtilis spores and MS2 coliphage for validation testing of ultraviolet reactors for water disinfection, Environ. Sci. Technol., 2005, 39, 7845–7852 CrossRef PubMed.
- A. Rauth, The physical state of viral nucleic acid and the sensitivity of viruses to ultraviolet light, Biophys. J., 1965, 5, 257–273 CrossRef PubMed.
- S. E. Beck, R. A. Rodriguez, K. G. Linden, T. M. Hargy, T. C. Larason and H. B. Wright, Wavelength Dependent UV Inactivation and DNA Damage of Adenovirus as Measured by Cell Culture Infectivity and Long Range Quantitative PCR, Environ. Sci. Technol., 2014, 48, 591–598 CrossRef PubMed.
- M. B. Fisher, D. C. Love, R. Schuech and K. L. Nelson, Simulated sunlight action spectra for inactivation of MS2 and PRD1 bacteriophages in clear water, Environ. Sci. Technol., 2011, 45, 9249–9255 CrossRef PubMed.
- L. Sinton, C. Hall, P. A. Lynch and R. J. Davies-Colley, Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters, Appl. Environ. Microbiol., 2002, 68, 1122–1131 CrossRef PubMed.
- O. C. Romero-Maraccini, N. J. Sadik, S. L. Rosado-Lausell, C. R. Pugh, X.-Z. Niu, J.-P. Croué and T. H. Nguyen, Sunlight-induced inactivation of human Wa and porcine OSU rotaviruses in the presence of exogenous photosensitizers, Environ. Sci. Technol., 2013, 47, 11004–11012 CrossRef PubMed.
- T. Kohn and K. L. Nelson, Sunlight-mediated inactivation of MS2 coliphage via exogenous singlet oxygen produced by sensitizers in natural waters, Environ. Sci. Technol., 2007, 41, 192–197 CrossRef PubMed.
- C.-X. Sun, M. Kitajima and K. Y.-H. Gin, Sunlight inactivation of somatic coliphage in the presence of natural organic matter, Sci. Total Environ., 2016, 541, 1–7 CrossRef PubMed.
- S. L. Rosado-Lausell, H. Wang, L. Gutiérrez, O. C. Romero-Maraccini, X.-Z. Niu, K. Y. H. Gin, J.-P. Croué and T. H. Nguyen, Roles of singlet oxygen and triplet excited state of dissolved organic matter formed by different organic matters in bacteriophage MS2 inactivation, Water Res., 2013, 47, 4869–4879 CrossRef PubMed.
- T. Kohn, M. Grandbois, K. McNeill and K. L. Nelson, Association with Natural Organic Matter Enhances the Sunlight-Mediated Inactivation of MS2 Coliphage by Singlet Oxygen, Environ. Sci. Technol., 2007, 41, 4626–4632 CrossRef PubMed.
- S. Bounty, R. A. Rodriguez and K. G. Linden, Inactivation of adenovirus using low-dose UV/H2O2 advanced oxidation, Water Res., 2012, 46, 6273–6278 CrossRef PubMed.
- H. Mamane, H. Shemer and K. G. Linden, Inactivation of E-coli, B-subtilis spores, and MS2, T4, and T7 phage using UV/H2O2 advanced oxidation, J. Hazard. Mater., 2007, 146, 479–486 CrossRef PubMed.
- E. M. Hotze, A. R. Badireddy, S. Chellam and M. R. Wiesner, Mechanisms of bacteriophage inactivation via singlet oxygen generation in UV illuminated fullerol suspensions, Environ. Sci. Technol., 2009, 43, 6639–6645 CrossRef PubMed.
- A. J. Rainbow and S. Mak, DNA damage and biological function of human Adenovirus after UV-irradiation, Int. J. Radiat. Biol., 1973, 24, 59–72 Search PubMed.
- M. Cho, J. Lee, Y. Mackeyev, L. J. Wilson, P. J. J. Alvarez, J. B. Hughes and J.-H. Kim, Visible light sensitized inactivation of MS-2 bacteriophage by a cationic amine-functionalized C60 derivative, Environ. Sci. Technol., 2010, 44, 6685–6691 CrossRef PubMed.
- J. D. Hoerter, A. A. Arnold, D. A. Kuczynska, A. Shibuya, C. S. Ward, M. G. Sauer, A. Gizachew, T. M. Hotchkiss, T. J. Fleming and S. Johnson, Effects of sublethal UVA irradiation on activity levels of oxidative defense enzymes and protein oxidation in Escherichia coli, J. Photochem. Photobiol., B, 2005, 81, 171–180 CrossRef PubMed.
- L. M. Sassoubre, M. M. Ramsey, M. S. Gilmore and A. B. Boehm, Transcriptional response of Enterococcus faecalis to sunlight, J. Photochem. Photobiol., B, 2014, 130, 349–356 CrossRef PubMed.
- R. Webb and M. Brown, Action Spectra for Oxygen-Dependent and Independent Inactivation of Escherichia-Coli-Wp2s from 254-Nm to 460-Nm, Photochem. Photobiol., 1979, 29, 407–409 CrossRef PubMed.
- J. S. McClary, L. M. Sassoubre and A. B. Boehm, Staphylococcus aureus Strain Newman Photoinactivation and Cellular Response to Sunlight Exposure, Appl. Environ. Microbiol., 2017, 83, e01052–17 CrossRef PubMed.
- L. M. Sassoubre, K. L. Nelson and A. B. Boehm, Mechanisms for Photoinactivation of Enterococcus faecalis in Seawater, Appl. Environ. Microbiol., 2012, 78, 7776–7785 CrossRef PubMed.
- J. A. Imlay, Oxidative Stress, EcoSal Plus, 2009 DOI:10.1128/ecosalplus.5.4.4 , Module 5.4.4.
- J. A. Imlay, Pathways of oxidative damage, Annu. Rev. Microbiol., 2003, 57, 395–418 CrossRef PubMed.
- I. Fridovich, Oxygen toxicity: A radical explanation, J. Exp. Biol., 1998, 201, 1203–1209 Search PubMed.
- M. Castro-Alférez, M. I. Polo-López and P. Fernández-Ibáñez, Intracellular mechanisms of solar water disinfection, Sci. Rep., 2016, 6, 38145 CrossRef PubMed.
- J. D. Hoerter, A. A. Arnold, C. S. Ward, M. Sauer, S. Johnson, T. Fleming and A. Eisenstark, Reduced hydroperoxidase (HPI and HPII) activity in the Delta fur mutant contributes to increased sensitivity to UVA radiation in Escherichia coli, J. Photochem. Photobiol., B, 2005, 79, 151–157 CrossRef PubMed.
- M. B. Fisher and K. L. Nelson, Inactivation of Escherichia coli by Polychromatic Simulated Sunlight: Evidence for and Implications of a Fenton Mechanism Involving Iron, Hydrogen Peroxide, and Superoxide, Appl. Environ. Microbiol., 2014, 80, 935–942 CrossRef PubMed.
- K. Kadir and K. L. Nelson, Sunlight mediated inactivation mechanisms of Enterococcus faecalis and Escherichia coli in clear water versus waste stabilization pond water, Water Res., 2014, 50, 307–317 CrossRef PubMed.
- D. MacKay, A. Eisenstark, R. B. Webb and M. S. Brown, Action spectra for lethality in recombination-less strains of Salmonella Typhimurium and Escherichia coli, Photochem. Photobiol., 1976, 24, 337–343 CrossRef PubMed.
- R. Tuveson and L. Sammartano, Sensitivity of Hema Mutant Escherichia-Coli-Cells to Inactivation by Near-Uv Light Depends on the Level of Supplementation with Delta-Aminolevulinic-Acid, Photochem. Photobiol., 1986, 43, 621–626 CrossRef PubMed.
- F. Bosshard, M. Bucheli, Y. Meur and T. Egli, The respiratory chain is the cell's Achilles' heel during UVA inactivation in Escherichia coli, Microbiology, 2010, 156, 2006–2015 CrossRef PubMed.
- G. F. Kramer and B. N. Ames, Oxidative mechanisms of toxicity of low-intensity near-UV light in Salmonella typhimurium, J. Bacteriol., 1987, 169, 2259–2266 CrossRef PubMed.
- M. Castro-Alférez, M. I. Polo-López, J. Marugán and P. Fernández-Ibáñez, Mechanistic model of the Escherichia coli inactivation by solar disinfection based on the photo-generation of internal ROS and the photo-inactivation of enzymes: CAT and SOD, Chem. Eng. J., 2017, 318, 214–223 CrossRef.
- D. Touati, Iron and Oxidative Stress in Bacteria, Arch. Biochem. Biophys., 2000, 373, 1–6 CrossRef PubMed.
- J. Hoerter, A. Pierce, C. Troupe, J. Epperson and A. Eisenstark, Role of enterobactin and intracellular iron in cell lethality during near-UV irradiation in Escherichia coli, Photochem. Photobiol., 1996, 64, 537–541 CrossRef PubMed.
- T. V. Ramabhadran and J. Jagger, Mechanism of growth delay induced in Escherichia coli by near ultraviolet radiation, Proc. Natl. Acad. Sci. U. S. A., 1976, 73, 59–63 CrossRef.
- T. V. Ramabhadran, T. Fossum and J. Jagger, In vivo induction of 4-thiouridine-cytidine adducts in tRNA of E. coli B/r by near-ultraviolet radiation, Photochem. Photobiol., 1976, 23, 315–321 CrossRef PubMed.
- S. Probst-Rued, K. McNeill and M. Ackermann, Thiouridine residues in tRNAs are responsible for a synergistic effect of UVA and UVB light in photoinactivation of Escherichia coli, Environ. Microbiol., 2017, 19, 434–442 CrossRef PubMed.
- L. R. Kelland, S. H. Moss and D. J. G. Davies, Leakage of 86Rb+ after ultraviolet irradiation of Escherichia coli K-12, Photochem. Photobiol., 1984, 39, 329–335 CrossRef PubMed.
- M. J. Peak, J. S. Johnson, R. W. Tuveson and J. G. Peak, Inactivation by monochromatic near-UV radiation of an Escherichia coli hemA8 mutant grown with and without δ-aminolevulinic acid: The role of DNA vs. membrane damage, Photochem. Photobiol., 1987, 45, 473–478 CrossRef PubMed.
- M. Berney, H.-U. Weilenmann and T. Egli, Flow-cytometric study of vital cellular functions in Escherichia coli during solar disinfection (SODIS), Microbiology, 2006, 152, 1719–1729 CrossRef PubMed.
- F. Bosshard, K. Riedel, T. Schneider, C. Geiser, M. Bucheli and T. Egli, Protein oxidation and aggregation in UVA-irradiated Escherichia coli cells as signs of accelerated cellular senescence, Environ. Microbiol., 2010, 12, 2931–2945 CrossRef PubMed.
- J. Calkins, J. Wheeler, C. Keller, E. Colley and J. Hazle, Comparative Ultraviolet Action Spectra (254-320nm) of 5 Wild-Type Eukaryotic Microorganisms and Escherichia-Coli, Radiat. Res., 1988, 114, 307–318 CrossRef PubMed.
- M. B. Fisher, Ph.D. Dissertation, University of California, Berkeley, 2011.
- A. I. Silverman and K. L. Nelson, Modeling the Endogenous Sunlight Inactivation Rates of Laboratory Strain and Wastewater E. coli and Enterococci Using Biological Weighting Functions, Environ. Sci. Technol., 2016, 50, 12292–12301 CrossRef PubMed.
- K. Kadir and K. L. Nelson, Sunlight mediated inactivation mechanisms of Enterococcus faecalis and Escherichia coli in clear water versus waste stabilization pond water, Water Res., 2014, 50, 307–317 CrossRef PubMed.
- G. Y. Lui, D. Roser, R. Corkish, N. J. Ashbolt and R. Stuetz, Point-of-use water disinfection using ultraviolet and visible light-emitting diodes, Sci. Total Environ., 2016, 553, 626–635 CrossRef PubMed.
- M. Pezzoni, R. A. Pizarro and C. S. Costa, Protective role of extracellular catalase (KatA) against UVA radiation in Pseudomonas aeruginosa biofilms, J. Photochem. Photobiol., B, 2014, 131, 53–64 CrossRef PubMed.
- P. A. Maraccini, J. Wenk and A. B. Boehm, Photoinactivation of Eight Health-Relevant Bacterial Species: Determining the Importance of the Exogenous Indirect Mechanism, Environ. Sci. Technol., 2016, 50, 5050–5059 CrossRef PubMed.
- A. L. Santos, C. Moreirinha, D. Lopes, A. C. Esteves, I. Henriques, A. Almeida, M. R. M. Domingues, I. Delgadillo, A. Correia and Â. Cunha, Effects of UV Radiation on the Lipids and Proteins of Bacteria Studied by Mid-Infrared Spectroscopy, Environ. Sci. Technol., 2013, 47, 6306–6315 CrossRef PubMed.
- R. J. Davies-Colley, A. M. Donnison and D. J. Speed, Towards a mechanistic understanding of pond disinfection, Water Sci. Technol., 2000, 42, 149–158 CrossRef.
- R. J. Davies-Colley, A. M. Donnison, D. J. Speed, C. M. Ross and J. W. Nagels, Inactivation of faecal indicator microorganisms in waste stabilisation ponds: Interactions of environmental factors with sunlight, Water Res., 1999, 33, 1220–1230 CrossRef.
- P. a. Maraccini, J. Wenk and A. b. Boehm, Exogenous indirect photoinactivation of bacterial pathogens and indicators in water with natural and synthetic photosensitizers in simulated sunlight with reduced UVB, J. Appl. Microbiol., 2016, 121, 587–597 CrossRef PubMed.
- S. George, M. R. Hamblin and A. Kishen, Uptake pathways of anionic and cationic photosensitizers into bacteria, Photochem. Photobiol. Sci., 2009, 8, 788–795 RSC.
- T. Dahl, W. Midden and P. Hartman, Comparison of Killing of Gram-Negative and Gram-Positive Bacteria by Pure Singlet Oxygen, J. Bacteriol., 1989, 171, 2188–2194 CrossRef PubMed.
- L. C. Seaver and J. A. Imlay, Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli, J. Bacteriol., 2001, 183, 7182–7189 CrossRef PubMed.
- P. A. Maraccini, D. M. Ferguson and A. B. Boehm, Diurnal Variation in Enterococcus Species Composition in Polluted Ocean Water and a Potential Role for the Enterococcal Carotenoid in Protection against Photoinactivation, Appl. Environ. Microbiol., 2012, 78, 305–310 CrossRef PubMed.
- M. T. Nguyen, J. T. Jasper, A. B. Boehm and K. L. Nelson, Sunlight inactivation of fecal indicator bacteria in open-water unit process treatment wetlands: Modeling endogenous and exogenous inactivation rates, Water Res., 2015, 83, 282–292 CrossRef PubMed.
- P. A. Maraccini, D. M. Ferguson and A. B. Boehm, Diurnal Variation in Enterococcus Species Composition in Polluted Ocean Water and a Potential Role for the Enterococcal Carotenoid in Protection against Photoinactivation, Appl. Environ. Microbiol., 2012, 78, 305–310 CrossRef PubMed.
- A. Clauditz, A. Resch, K.-P. Wieland, A. Peschel and F. Götz, Staphyloxanthin Plays a Role in the Fitness of Staphylococcus aureus and Its Ability To Cope with Oxidative Stress, Infect. Immun., 2006, 74, 4950–4953 CrossRef PubMed.
- P. Valero, S. Giannakis, R. Mosteo, M. P. Ormad and C. Pulgarin, Comparative effect of growth media on the monitoring of E. coli inactivation and regrowth after solar and photo-Fenton treatment, Chem. Eng. J., 2017, 313, 109–120 CrossRef.
- F. Bosshard, M. Berney, M. Scheifele, H.-U. Weilenmann and T. Egli, Solar disinfection (SODIS) and subsequent dark storage of Salmonella typhimurium and Shigella flexneri monitored by flow cytometry, Microbiology, 2009, 155, 1310–1317 CrossRef PubMed.
- P. A. Maraccini, M. C. M. Mattioli, L. M. Sassoubre, Y. Cao, J. F. Griffith, J. S. Ervin, L. C. Van De Werfhorst and A. B. Boehm, Solar Inactivation of Enterococci and Escherichia coli in Natural Waters: Effects of Water Absorbance and Depth, Environ. Sci. Technol., 2016, 50, 5068–5076 CrossRef PubMed.
- M. Berney, H. U. Weilenmann, J. Ihssen, C. Bassin and T. Egli, Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection, Appl. Environ. Microbiol., 2006, 72, 2586–2593 CrossRef PubMed.
- P. A. Maraccini, D. Wang, J. S. McClary and A. B. Boehm, Growth-dependent photoinactivation kinetics of Enterococcus faecalis, J. Appl. Microbiol., 2015, 118, 1226–1237 CrossRef PubMed.
- R. Khaengraeng and R. H. Reed, Oxygen and photoinactivation of Escherichia coli in UVA and sunlight, J. Appl. Microbiol., 2005, 99, 39–50 CrossRef PubMed.
- Y. Liu, S. Dong, M. S. Kuhlenschmidt, T. B. Kuhlenschmidt, J. Drnevich and T. H. Nguyen, Inactivation mechanisms of cryptosporidium parvum oocysts by solar ultraviolet irradiation, Environ. Sci.: Water Res. Technol., 2015, 1, 188–198 RSC.
- K. G. McGuigan, F. Méndez-Hermida, J. A. Castro-Hermida, E. Ares-Mazás, S. C. Kehoe, M. Boyle, C. Sichel, P. Fernández-Ibáñez, B. P. Meyer, S. Ramalingham and E. A. Meyer, Batch solar disinfection inactivates oocysts of Cryptosporidium parvum and cysts of Giardia muris in drinking water, J. Appl. Microbiol., 2006, 101, 453–463 CrossRef PubMed.
- B. J. King, D. Hoefel, P. E. Wong and P. T. Monis, Solar radiation induces non-nuclear perturbations and a false start to regulated exocytosis in Cryptosporidium parvum, PLoS One, 2010, 5, e11773 CrossRef PubMed.
- H. Gomez-Couso, M. Fontan-Sainz, K. G. McGuigan and E. Ares-Mazas, Effect of the radiation intensity, water turbidity and exposure time on the survival of Cryptosporidium during simulated solar disinfection of drinking water, Acta Trop., 2009, 112, 43–48 CrossRef PubMed.
- H. Gómez-Couso, M. Fontán-Sainz and E. Ares-Mazás, Thermal contribution to the inactivation of Cryptosporidium in plastic bottles during solar water disinfection procedures, Am. J. Trop. Med. Hyg., 2010, 82, 35 CrossRef PubMed.
- F. Mendez-Hermida, J.-A. Castro-Hermidal, E. Ares-Mazas, N. N. Patel, J. Lonnen, S. Kilvington, S. C. Kehoe and K. G. McGuigan, Solar disinfection (SODIS) of the waterborne protozoan pathogens Cryptosporidium parvum and Acanthamoeba polyphaga in drinking water, Abstr. Gen. Meet. Am. Soc. Microbiol., 2005, 105, 532 Search PubMed.
- M. Fontán-Sainz, H. Gómez-Couso, P. Fernández-Ibáñez and E. Ares-Mazás, Evaluation of the solar water disinfection process (SODIS) against Cryptosporidium parvum using a 25-L static solar reactor fitted with a compound parabolic collector (CPC), Am. J. Trop. Med. Hyg., 2012, 86, 223–228 CrossRef PubMed.
- S. J. Connelly, E. A. Wolyniak, C. E. Williamson and K. L. Jellison, Artificial UV-B and solar radiation reduce in vitro infectivity of the human pathogen Cryptosporidium parvum, Environ. Sci. Technol., 2007, 41, 7101–7106 CrossRef PubMed.
- B. King, D. Hoefel, D. Daminato, S. Fanok and P. Monis, Solar UV reduces Cryptosporidium parvum oocyst infectivity in environmental waters, J. Appl. Microbiol., 2008, 104, 1311–1323 CrossRef PubMed.
- J. Lonnen, S. Kilvington, S. C. Kehoe, F. Al-Touati and K. G. McGuigan, Solar and photocatalytic disinfection of protozoan, fungal and bacterial microbes in drinking water, Water Res., 2005, 39, 877–883 CrossRef PubMed.
- W. Heaselgrave, N. Patel, S. Kilvington, S. C. Kehoe and K. G. McGuigan, Solar disinfection of poliovirus and Acanthamoeba polyphaga cysts in water – a laboratory study using simulated sunlight, Lett. Appl. Microbiol., 2006, 43, 125–130 CrossRef PubMed.
- W. Heaselgrave and S. Kilvington, The efficacy of simulated solar disinfection (SODIS) against Ascaris, Giardia, Acanthamoeba, Naegleria, Entamoeba and Cryptosporidium, Acta Trop., 2011, 119, 138–143 CrossRef PubMed.
- S. Mtapuri-Zinyowera, N. Midzi, C. Muchaneta-Kubara, T. Simbini and T. Mduluza, Impact of solar radiation in disinfecting drinking water contaminated with Giardia duodenalis and Entamoeba histolytica/dispar at a point-of-use water treatment, J. Appl. Microbiol., 2009, 106, 847–852 CrossRef PubMed.
- W. Heaselgrave and S. Kilvington, Antimicrobial activity of simulated solar disinfection against bacterial, fungal, and protozoan pathogens and its enhancement by riboflavin, Appl. Environ. Microbiol., 2010, 76, 6010–6012 CrossRef PubMed.
- W. A. M. Hijnen, E. F. Beerendonk and G. J. Medema, Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: A review, Water Res., 2006, 40, 3–22 CrossRef PubMed.
- B. J. King, A. R. Keegan, P. T. Monis and C. P. Saint, Environmental temperature controls Cryptosporidium oocyst metabolic rate and associated retention of infectivity, Appl. Environ. Microbiol., 2005, 71, 3848–3857 CrossRef PubMed.
- A. M. Johnson, G. D. D. Giovanni and P. A. Rochelle, Comparison of Assays for Sensitive and Reproducible Detection of Cell Culture-Infectious Cryptosporidium parvum and Cryptosporidium hominis in Drinking Water, Appl. Environ. Microbiol., 2012, 78, 156–162 CrossRef PubMed.
- P.-Y. Hong, T. R. Julian, M.-L. Pype, S. C. Jiang, K. L. Nelson, D. Graham, A. Pruden and C. M. Manaia, Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes, Water, 2018, 10, 244 CrossRef.
- A. Pruden, R. T. Pei, H. Storteboom and K. H. Carlson, Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado, Environ. Sci. Technol., 2006, 40, 7445–7450 CrossRef PubMed.
- N. Czekalski, T. Berthold, S. Caucci, A. Egli and H. Buergmann, Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland, Front. Microbiol., 2012, 3, 106 Search PubMed.
- C. W. Xi, Y. L. Zhang, C. F. Marrs, W. Ye, C. Simon, B. Foxman and J. Nriagu, Prevalence of Antibiotic Resistance in Drinking Water Treatment and Distribution Systems, Appl. Environ. Microbiol., 2009, 75, 5714–5718 CrossRef PubMed.
- A. Pruden, D. J. Larsson, A. Amézquita, P. Collignon, K. K. Brandt, D. W. Graham, J. M. Lazorchak, S. Suzuki, P. Silley and J. R. Snape, Management Options for Reducing the Release of Antibiotics and Antibiotic Resistance Genes to the Environment, Environ. Health Perspect., 2013, 121(8), 878–885 CrossRef PubMed.
- M. C. Dodd, Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment, J. Environ. Monit., 2012, 14, 1754–1771 RSC.
- N. Munakata and Y. Ikeda, Inactivation of transforming DNA by ultraviolet irradiation – A study with ultraviolet-sensitive mutants of Bacillus subtilis, Mutat. Res., 1969, 7, 133 CrossRef PubMed.
- E. Cabrera-Juarez, J. K. Setlow, P. A. Swenson and M. J. Peak, Oxygen-independent inactivation of Haemophilus influenzae transforming DNA by monochromatic radiation – Action spectrum, effect of histidine and repair, Photochem. Photobiol., 1976, 23, 309–313 CrossRef PubMed.
- J. K. Setlow, Shape of UV inactivation curve for transforming DNA, Nature, 1977, 268, 169–170 CrossRef PubMed.
- P. H. Chang, B. Juhrend, T. M. Olson, C. F. Marrs and K. R. Wigginton, Degradation of Extracellular Antibiotic Resistance Genes with UV254 Treatment, Environ. Sci. Technol., 2017, 51, 6185–6192 CrossRef PubMed.
- C. W. McKinney and A. Pruden, Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater, Environ. Sci. Technol., 2012, 46, 13393–13400 CrossRef PubMed.
- Y. Yoon, H. J. Chung, D. Y. Wen Di, M. C. Dodd, H.-G. Hur and Y. Lee, Inactivation efficiency of plasmid-encoded antibiotic resistance genes during water treatment with chlorine, UV, and UV/H2O2, Water Res., 2017, 123, 783–793 CrossRef PubMed.
- D. P. Hader and R. P. Sinha, Solar ultraviolet radiation-induced DNA damage in aquatic organisms: potential environmental impact, Mutat. Res. Fund Mol. Mech. Mutagen, 2005, 571, 221–233 CrossRef PubMed.
- R. P. Sinha and D. P. Hader, UV-induced DNA damage and repair: a review, Photochem. Photobiol. Sci., 2002, 1, 225–236 RSC.
- C. A. Engemann, P. L. Keen, C. W. Knapp, K. J. Hall and D. W. Graham, Fate of tetracycline resistance genes in aquatic systems: Migration from the water column to peripheral biofilms, Environ. Sci. Technol., 2008, 42, 5131–5136 CrossRef PubMed.
- C. A. Engemann, L. Adams, C. W. Knapp and D. W. Graham, Disappearance of oxytetracycline resistance genes in aquatic systems, FEMS Microbiol. Lett., 2006, 263, 176–182 CrossRef PubMed.
- C. W. Knapp, W. Zhang, B. S. M. Sturm and D. W. Graham, Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions, Environ. Pollut., 2010, 158, 1506–1512 CrossRef PubMed.
- A. P. Schuch and C. F. Martins Menck, The genotoxic effects of DNA lesions induced by artificial UV-radiation and sunlight, J. Photochem. Photobiol., B, 2010, 99, 111–116 CrossRef PubMed.
- S. P. Walters, K. M. Yamahara and A. B. Boehm, Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: Implications for their use in assessing risk in recreational waters, Water Res., 2009, 43, 4929–4939 CrossRef PubMed.
- N. Al-Jassim, D. Mantilla-Calderon, T. Wang and P.-Y. Hong, Inactivation and Gene Expression of a Virulent Wastewater Escherichia coli Strain and the Nonvirulent Commensal Escherichia coli DSM1103 Strain upon Solar Irradiation, Environ. Sci. Technol., 2017, 51, 3649–3659 CrossRef PubMed.
- National Center for Atmospheric Research, Tropospheric Ultraviolet and Visible (TUV) Radiation Model.
- M. T. Nguyen, A. I. Silverman and K. L. Nelson, Sunlight Inactivation of MS2 Coliphage in the Absence of Photosensitizers: Modeling the Endogenous Inactivation Rate Using a Photoaction Spectrum, Environ. Sci. Technol., 2014, 48, 3891–3898 CrossRef PubMed.
- R. J. Craggs, A. Zwart, J. W. Nagels and R. J. Davies-Colley, Modelling sunlight disinfection in a high rate pond, Ecol. Eng., 2004, 22, 113–122 CrossRef.
- M. Tzortziou, J. R. Herman, C. L. Gallegos, P. J. Neale, A. Subramaniam, L. W. Harding and Z. Ahmad, Bio-optics of the Chesapeake Bay from measurements and radiative transfer closure, Estuar. Coast Shelf Sci., 2006, 68, 348–362 CrossRef.
- M. Castro-Alférez, M. I. Polo-López, J. Marugán and P. Fernández-Ibáñez, Mechanistic modeling of UV and mild-heat synergistic effect on solar water disinfection, Chem. Eng. J., 2017, 316, 111–120 CrossRef.
- M. B. Fisher, D. C. Love, R. Schuech and K. L. Nelson, Simulated Sunlight Action Spectra for Inactivation of MS2 and PRD1 Bacteriophages in Clear Water, Environ. Sci. Technol., 2011, 45, 9249–9255 CrossRef PubMed.
- K. Kadir, Ph.D. Dissertation, University of California, Berkeley, 2010.
- S. Mostafa, M. Rubinato, F. L. Rosario-Ortiz and K. G. Linden, Impact of Light Screening and Photosensitization by Surface Water Organic Matter on Enterococcus Faecalis Inactivation, Environ. Eng. Sci., 2016, 33, 365–373 CrossRef.
- O. US EPA, GCSOLAR, http://www2.epa.gov/exposure-assessment-models/gcsolar, accessed 1 October 2015.
- C. Pellieux, A. Dewilde, C. Pierlot and J.-M. Aubry, in Methods in Enzymology, ed. H. S. Lester Packer, Academic Press, 2000, vol. 319, pp. 197–207 Search PubMed.
- E. G. Mbonimpa, B. Vadheim and E. R. Blatchley III, Continuous-flow solar UVB disinfection reactor for drinking water, Water Res., 2012, 46, 2344–2354 CrossRef PubMed.
- W. Haag and J. Hoigne, Singlet Oxygen in Surface Waters. 3. Photochemical Formation and Steady-State Concentrations in Various Types of Waters, Environ. Sci. Technol., 1986, 20, 341–348 CrossRef PubMed.
- M. B. Fisher, M. Iriarte and K. L. Nelson, Solar water disinfection (SODIS) of Escherichia coli, Enterococcus spp., and MS2 coliphage: Effects of additives and alternative container materials, Water Res., 2012, 46, 1745–1754 CrossRef PubMed.
- D. Dulin and T. Mill, Development and Evaluation of Sunlight Actinometers, Environ. Sci. Technol., 1982, 16, 815–820 CrossRef PubMed.
- A. Leifer, The kinetics of environmental aquatic photochemistry: theory and practice, American Chemical Society, Washington, DC, 1988 Search PubMed.
- A. H. Geeraerd, V. P. Valdramidis and J. F. Van Impe, GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves, Int. J. Food Microbiol., 2005, 102, 95–105 CrossRef PubMed.
- J. R. Bolton and K. G. Linden, Standardization of Methods for Fluence (UV Dose) Determination in Bench-Scale UV Experiments, J. Environ. Eng., 2003, 129, 209–215 CrossRef.
- M. Berney, H.-U. Weilenmann, A. Simonetti and T. Egli, Efficacy of solar disinfection of Escherichia coli, Shigella flexneri, Salmonella Typhimurium and Vibrio cholerae, J. Appl. Microbiol., 2006, 101, 828–836 CrossRef PubMed.
- Monitoring Bathing Waters, A Practical Guide to the Design and Implementation of Assessments and Monitoring Programs, World Health Organization, 2000 Search PubMed.
- A. B. Boehm, K. M. Yamahara, D. C. Love, B. M. Peterson, K. McNeill and K. L. Nelson, Covariation and Photoinactivation of Traditional and Novel Indicator Organisms and Human Viruses at a Sewage-Impacted Marine Beach, Environ. Sci. Technol., 2009, 43, 8046–8052 CrossRef PubMed.
- A. B. Boehm, S. B. Grant, J. H. Kim, S. L. Mowbray, C. D. McGee, C. D. Clark, D. M. Foley and D. E. Wellman, Decadal and Shorter Period Variability of Surf Zone Water Quality at Huntington Beach, California, Environ. Sci. Technol., 2002, 36, 3885–3892 CrossRef PubMed.
- Z. Ge, R. L. Whitman, M. B. Nevers, M. S. Phanikumar and M. N. Byappanahalli, Nearshore hydrodynamics as loading and forcing factors for Escherichia coli contamination at an embayed beach, Limnol. Oceanogr., 2012, 57, 362–381 CrossRef.
- R. L. Whitman, M. B. Nevers, G. C. Korinek and M. N. Byappanahalli, Solar and Temporal Effects on Escherichia coli Concentration at a Lake Michigan Swimming Beach, Appl. Environ. Microbiol., 2004, 70, 4276–4285 CrossRef PubMed.
- A. de Brauwere, N. K. Ouattara and P. Servais, Modeling Fecal Indicator Bacteria Concentrations in Natural Surface Waters: A Review, Crit. Rev. Environ. Sci. Technol., 2014, 44, 2380–2453 CrossRef.
- C. J. A. Campos, S. R. Kershaw and R. J. Lee, Environmental Influences on Faecal Indicator Organisms in Coastal Waters and Their Accumulation in Bivalve Shellfish, Estuar. Coast, 2013, 36, 834–853 CrossRef.
- J. Flannery, P. Rajko-Nenow, S. Keaveney, V. O'Flaherty and W. Doré, Simulated sunlight inactivation of norovirus and FRNA bacteriophage in seawater, J. Appl. Microbiol., 2013, 115, 915–922 CrossRef PubMed.
- R. J. Newton, M. J. Bootsma, H. G. Morrison, M. L. Sogin and S. L. McLellan, A Microbial Signature Approach to Identify Fecal Pollution in the Waters Off an Urbanized Coast of Lake Michigan, Microb. Ecol., 2013, 65, 1011–1023 CrossRef PubMed.
- T. Unno, J. Jang, D. Han, J. H. Kim, M. J. Sadowsky, O.-S. Kim, J. Chun and H.-G. Hur, Use of Barcoded Pyrosequencing and Shared OTUs To Determine Sources of Fecal Bacteria in Watersheds, Environ. Sci. Technol., 2010, 44, 7777–7782 CrossRef PubMed.
- L. M. Sassoubre, K. M. Yamahara and A. B. Boehm, Temporal Stability of the Microbial Community in Sewage-Polluted Seawater Exposed to Natural Sunlight Cycles and Marine Microbiota, Appl. Environ. Microbiol., 2015, 81, 2107–2116 CrossRef PubMed.
- A. B. Boehm, N. J. Ashbolt, J. M. Colford, L. E. Dunbar, L. E. Fleming, M. A. Gold, J. A. Hansel, P. R. Hunter, A. M. Ichida, C. D. Mcgee, J. A. Soller and S. B. Weisberg, A sea change ahead for recreational water quality criteria, J. Water Health, 2009, 7, 9–20 CrossRef PubMed.
- V. M. Yau, K. C. Schiff, B. F. Arnold, J. F. Griffith, J. S. Gruber, C. C. Wright, T. J. Wade, S. Burns, J. M. Hayes, C. McGee, M. Gold, Y. Cao, A. B. Boehm, S. B. Weisberg and J. M. Colford, Effect of submarine groundwater discharge on bacterial indicators and swimmer health at Avalon Beach, CA, USA, Water Res., 2014, 59, 23–36 CrossRef PubMed.
- M. E. Verbyla and J. R. Mihelcic, A review of virus removal in wastewater treatment pond systems, Water Res., 2015, 71, 107–124 CrossRef PubMed.
- A. I. Silverman, M. O. Akrong, P. Drechsel and K. L. Nelson, On-farm treatment of wastewater used for vegetable irrigation: bacteria and virus removal in small ponds in Accra, Ghana, J. Water Reuse Desalin., 2014, 4, 276–286 CrossRef.
- T. Curtis, D. Mara and S. Silva, Influence of pH, Oxygen, and Humic Substances on Ability of Sunlight to Damage Fecal-Coliforms in Waste Stabilization Pond Water, Appl. Environ. Microbiol., 1992, 58, 1335–1343 Search PubMed.
- H. E. Maynard, S. K. Ouki and S. C. Williams, Tertiary lagoons: a review of removal mechanisms and performance, Water Res., 1999, 33, 1–13 CrossRef.
- A. Carratalà, A. D. Calado, M. J. Mattle, R. Meierhofer, S. Luzi and T. Kohn, Solar Disinfection of Viruses in Polyethylene Terephthalate Bottles, Appl. Environ. Microbiol., 2016, 82, 279–288 CrossRef PubMed.
- M. Castro-Alférez, M. Inmaculada Polo-López, J. Marugán and P. Fernández-Ibáñez, Validation of a solar-thermal water disinfection model for Escherichia coli inactivation in pilot scale solar reactors and real conditions, Chem. Eng. J., 2018, 331, 831–840 CrossRef.
- M. Agullo-Barcelo, M. I. Polo-Lopez, F. Lucena, J. Jofre and P. Fernandez-Ibanez, Solar Advanced Oxidation Processes as disinfection tertiary treatments for real wastewater: Implications for water reclamation, Appl. Catal., B, 2013, 136, 341–350 CrossRef.
- M. B. Fisher, C. R. Keenan, K. L. Nelson and B. M. Voelker, Speeding up solar disinfection (SODIS): effects of hydrogen peroxide, temperature, pH, and copper plus ascorbate on the photoinactivation of E-coli, J. Water Health, 2008, 6, 35–51 CrossRef PubMed.
- P. S. Hartman and A. Eisenstark, Killing of Escherichia coli K-12 by near-ultraviolet radiation in the presence of hydrogen peroxide - Role of double-strand DNA breaks in absence of recombinational repair, Mutat. Res., 1980, 72, 31–42 CrossRef PubMed.
- A. Eisenstark, R. L. Buzard and P. S. Hartman, Inactivation of phage by near-ultraviolet radiation and hydrogen peroxide, Photochem. Photobiol., 1986, 44, 603–606 CrossRef PubMed.
- C. Sichel, P. Fernandez-Ibanez, M. de Cara and J. Tello, Lethal synergy of solar radiation and H2O2 on wild Fusarium solani spores in distilled and natural well water, Water Res., 2009, 43, 1841–1850 CrossRef PubMed.
- F. Sciacca, J. A. Rengifo-Herrera, J. Wethe and C. Pulgarin, Dramatic enhancement of solar disinfection (SODIS) of wild Salmonella sp in PET bottles by H2O2 addition on natural water of Burkina Faso containing dissolved iron, Chemosphere, 2010, 78, 1186–1191 CrossRef PubMed.
- J. Ndounla, D. Spuhler, S. Kenfack, J. Wéthé and C. Pulgarin, Inactivation by solar photo-Fenton in pet bottles of wild enteric bacteria of natural well water: Absence of re-growth after one week of subsequent storage, Appl. Catal., B, 2013, 129, 309–317 CrossRef.
- S. Giannakis, M. I. Polo López, D. Spuhler, J. A. Sánchez Pérez, P. Fernández Ibáñez and C. Pulgarin, Solar disinfection is an augmentable, in situ-generated photo-Fenton reaction—Part 1: A review of the mechanisms and the fundamental aspects of the process, Appl. Catal., B, 2016, 199, 199–223 CrossRef.
- A. W. Vermilyea and B. M. Voelker, Photo-Fenton Reaction at Near Neutral pH, Environ. Sci. Technol., 2009, 43, 6927–6933 CrossRef PubMed.
- J. J. Pignatello, E. Oliveros and A. MacKay, Advanced oxidation processes for organic contaminant destruction based on the Fenton reaction and related chemistry, Crit. Rev. Environ. Sci. Technol., 2006, 36, 1–84 CrossRef.
- J. I. Nieto-Juarez, K. Pierzchla, A. Sienkiewicz and T. Kohn, Inactivation of MS2 coliphage in Fenton and Fenton-like systems: role of transition metals, hydrogen peroxide and sunlight, Environ. Sci. Technol., 2010, 44, 3351–3356 CrossRef PubMed.
- A.-G. Rincón and C. Pulgarin, Comparative evaluation of Fe3+ and TiO2 photoassisted processes in solar photocatalytic disinfection of water, Appl. Catal., B, 2006, 63, 222–231 CrossRef.
- A.-G. Rincon and C. Pulgarin, Fe3+ and TiO2 solar-light-assisted inactivation of E-coli at field scale - Implications in solar disinfection at low temperature of large quantities of water, Catal. Today, 2007, 122, 128–136 CrossRef.
- W. Koster, ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B-12, Res. Microbiol., 2001, 152, 291–301 CrossRef PubMed.
- J. E. Forsyth, P. Zhou, Q. Mao, S. S. Asato, J. S. Meschke and M. C. Dodd, Enhanced Inactivation of Bacillus subtilis Spores during Solar Photolysis of Free Available Chlorine, Environ. Sci. Technol., 2013, 47, 12976–12984 CrossRef PubMed.
- P. Zhou, G. D. Di Giovanni, J. S. Meschke and M. C. Dodd, Enhanced Inactivation of Cryptosporidium parvum Oocysts during Solar Photolysis of Free Available Chlorine, Environ. Sci. Technol. Lett., 2014, 1, 453–458 CrossRef.
- A. N. Pisarenko, B. D. Stanford, S. A. Snyder, S. B. Rivera and A. K. Boal, Investigation of the use of Chlorine Based Advanced Oxidation in Surface Water: Oxidation of Natural Organic Matter and Formation of Disinfection Byproducts, J. Adv. Oxid. Technol., 2013, 16, 137–150 Search PubMed.
- S. Loeb, R. Hofmann and J.-H. Kim, Beyond the Pipeline: Assessing the Efficiency Limits of Advanced Technologies for Solar Water Disinfection, Environ. Sci. Technol. Lett., 2016, 3, 73–80 CrossRef.
- M. A. Alotaibi and W. Heaselgrave, Solar Disinfection of Water for Inactivation of Enteric Viruses and its Enhancement by Riboflavin, Food Environ. Virol., 2011, 3, 70–73 CrossRef.
- C. K. Remucal and K. McNeill, Photosensitized Amino Acid Degradation in the Presence of Riboflavin and Its Derivatives, Environ. Sci. Technol., 2011, 45, 5230–5237 CrossRef PubMed.
- P. D. Wood and L. J. Johnston, Photoionization and photosensitized electron-transfer reactions of psoralens and coumarins, J. Phys. Chem. A, 1998, 102, 5585–5591 CrossRef.
- A. S. Harding and K. J. Schwab, Using Limes and Synthetic Psoralens to Enhance Solar Disinfection of Water (SODIS): A Laboratory Evaluation with Norovirus, Escherichia coli, and MS2, Am. J. Trop. Med. Hyg., 2012, 86, 566–572 CrossRef PubMed.
- C. V. Hanson, Photochemical inactivation of viruses with psoralens: an overview, Blood Cells, 1992, 18, 7–25 Search PubMed.
- C. Pulgarin, Fe vs. TiO2 Photo-assisted Processes for Enhancing the Solar Inactivation of Bacteria in Water, Chimia, 2015, 69, 7–9 CrossRef PubMed.
- O. Legrini, E. Oliveros and A. M. Braun, Photochemical processes for water treatment, Chem. Rev., 1993, 93, 671–698 CrossRef.
- J. A. Byrne, P. A. Fernandez-Ibanez, P. S. M. Dunlop, D. M. A. Alrousan and J. W. J. Hamilton, Photocatalytic Enhancement for Solar Disinfection of Water: A Review, Int. J. Photoenergy, 2011, 1–12 CrossRef.
- S. Helali, M. I. Polo-López, P. Fernández-Ibáñez, B. Ohtani, F. Amano, S. Malato and C. Guillard, Solar photocatalysis: A green technology for E. coli contaminated water disinfection. Effect of concentration and different types of suspended catalyst, J. Photochem. Photobiol., A, 2014, 276, 31–40 CrossRef.
- G. P. Tegos, T. N. Demidova, D. Arcila-Lopez, H. Lee, T. Wharton, H. Gali and M. R. Hamblin, Cationic fullerenes are effective and selective antimicrobial photosensitizers, Chem. Biol., 2005, 12, 1127–1135 CrossRef PubMed.
- M. Cho, J. D. Fortner, J. B. Hughes and J. H. Kim, Escherichia coli Inactivation by Water-Soluble, Ozonated C-60 Derivative: Kinetics and Mechanisms, Environ. Sci. Technol., 2009, 43, 7410–7415 CrossRef PubMed.
- A. R. Badireddy, J. F. Budarz, S. Chellam and M. R. Wiesner, Bacteriophage Inactivation by UV-A Illuminated Fullerenes: Role of Nanoparticle-Virus Association and Biological Targets, Environ. Sci. Technol., 2012, 46, 5963–5970 CrossRef PubMed.
- C. Liu, D. Kong, P.-C. Hsu, H. Yuan, H.-W. Lee, Y. Liu, H. Wang, S. Wang, K. Yan, D. Lin, P. A. Maraccini, K. M. Parker, A. B. Boehm and Y. Cui, Rapid water disinfection using vertically aligned MoS2 nanofilms and visible light, Nat. Nanotechnol., 2016, 11, 1098 CrossRef PubMed.
- E. L. Cates, Photocatalytic Water Treatment: So Where Are We Going with This?, Environ. Sci. Technol., 2017, 51, 757–758 CrossRef PubMed.
- M. J. Lindell, H. W. Graneli and L. J. Tranvik, Effects of sunlight on bacterial growth in lakes of different humic content, Aquat. Microb. Ecol., 1996, 11, 135–141 CrossRef.
- L. Alonso-Saez, J. M. Gasol, T. Lefort, J. Hofer and R. Sommaruga, Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in northwestern Mediterranean coastal waters, Appl. Environ. Microbiol., 2006, 72, 5806–5813 CrossRef PubMed.
- F. Joux, W. H. Jeffrey, P. Lebaron and D. L. Mitchell, Marine bacterial isolates display diverse responses to UV-B radiation, Appl. Environ. Microbiol., 1999, 65, 3820–3827 Search PubMed.
- M. Lindell, W. Graneli and L. Tranvik, Enhanced Bacterial-Growth in Response to Photochemical Transformation of Dissolved Organic-Matter, Limnol. Oceanogr., 1995, 40, 195–199 CrossRef.
- R. Benner and B. Biddanda, Photochemical transformations of surface and deep marine dissolved organic matter: Effects on bacterial growth, Limnol. Oceanogr., 1998, 43, 1373–1378 CrossRef.
- W. H. Jeffrey, J. P. Kase and S. W. Wilhelm, in The effects of UV radiation in the marine environment, ed. S. de Mora, S. Demers and M. Vernet, 2000 Search PubMed.
- K. E. Wommack, R. T. Hill, T. A. Muller and R. R. Colwell, Effects of sunlight on bacteriophage viability and structure, Appl. Environ. Microbiol., 1996, 62, 1336–1341 Search PubMed.
- Y. Huot, W. H. Jeffrey, R. F. Davis and J. J. Cullen, Damage to DNA in bacterioplankton: A model of damage by ultraviolet radiation and its repair as influenced by vertical mixing,, Photochem. Photobiol., 2000, 72, 62–74 CrossRef PubMed.
Footnotes |
| † Research Center for Reinventing the Nation's Urban Water Infrastructure, ReNUWIt, Stanford University, Stanford, CA, USA. |
| ‡ Hydrolight5 was used for generating this figure, because the RADTRANX routine can account for overcast skies; however, a limitation of this model is that the lowest wavelength is 300 nm. |
| This journal is © The Royal Society of Chemistry 2018 |