Characterizing single chain nanoparticles (SCNPs): a critical survey†
Eva
Blasco‡
 *ab,
Bryan T.
Tuten‡
ac,
Hendrik
Frisch‡
c,
Albena
Lederer
*de and
Christopher
Barner-Kowollik
*ab,
Bryan T.
Tuten‡
ac,
Hendrik
Frisch‡
c,
Albena
Lederer
*de and
Christopher
Barner-Kowollik
 *abc
*abc
aMacromolecular Architectures, Institut für Technische Chemie und Polymerchemie, Karlsruhe Institute of Technology (KIT), Engesserstr. 18, 76131 Karlsruhe, Germany. E-mail: eva.blasco@kit.edu
bInstitut für Biologische Grenzflächen, Karlsruhe Institute of Technology (KIT), Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany
cSchool of Chemistry, Physics and Mechanical Engineering, Queensland University of Technology (QUT), 2 George Street, QLD 4000, Brisbane, Australia. E-mail: christopher.barnerkowollik@qut.edu.au
dLeibniz Institut für Polymerforschung Dresden, Hohe Str. 6, D-01069 Dresden, Germany
eTechnische Universität Dresden, D-01062 Dresden, Germany. E-mail: lederer@ipfdd.de
First published on 28th August 2017
Abstract
We provide the results of a critical literature survey on the reported sizes of single chain polymer nanoparticles (SCNPs), an emerging class of functional nanomaterials with sub-30 nm diameters. Comparing different size evaluation techniques (DLS, 2D DOSY NMR, viscometry as well as microscopic techniques) by plotting the SCNPs’ estimated diameters, D, versus their measured (apparent) number average molecular weight, Mn, we demonstrate the vast data scatter that besets their analysis. We show that while relative reductions in measured diameter certainly indicate chain collapse, accurately describing the absolute size of SCNPs in solution remains a challenging task. Critically, conformation-size relationships emerge depending on the method used for size determination. We submit that the vast majority of reported sizes are only indicative of the relative size reduction during chain collapse and that absolute size determination approaches currently in use need to be further refined.
The field of single chain polymer nanoparticles (SCNPs)1–6 (occasionally also referred to as nanogels) has seen substantial growth over the last 15 years based on a plethora of experimental techniques becoming available for the synthesis of well-defined functional precursor macromolecules, most prominently reversible deactivation radical polymerization (RDRP)7,8 in combination with versatile modular ligation processes. SCNPs are intramolecular, cross-linked single polymer chains whose properties are distinctly different from their linear parent polymers. In contrast to cyclic polymers containing exactly one connection,9,10 the properties of SCNPs are not exclusively dominated by the absence of free chain ends, but by both the nature and quantity of intramolecular crosslinks.
The ultimate aim of synthetic SCNP design is to achieve full molecular control over the morphology and folding behavior of the precursor chains, ideally mimicking the functionality and precision of naturally occurring biomolecules.11 Clearly, although impressive progress has been made over the last decade, this aim remains largely elusive.
One of the major challenges in the field is the characterization of the obtained nanoparticles, which can be beset with problems reaching from their molecular characterization, their mass determination and a reliable morphological assessment. Chemically, nuclear magnetic resonance (NMR) spectroscopy as well as – most recently – high resolution mass spectrometry12 are employed, while size – or rather the changes in size – is typically assessed via size exclusion chromatography (SEC) coupled to viscometry and MALS detectors, dynamic light scattering (DLS) and, less frequently, pulse field gradient NMR methods such as 2D DOSY.3 In certain cases, atomic force microscopy (AFM) as well as transmission electron microscopy (TEM) has been employed, too. Critically, a set of the obtained size values (most from DLS and SEC measurements) has been used in a pioneering assessment by Pomposo and colleagues to derive information on SCNP shape and the expected size reduction upon intrachain collapse in both reversible and irreversible collapse scenarios.13,14 By invoking Flory-theory arguments, a functioning relation was suggested that – within experimental error limits – predicts the experimentally observed relative collapses well. Interestingly, and perhaps not surprisingly, most studies focus on discussing the relative changes that occur in coil dimensions when going from the non-crosslinked precursor to the compact SCNP state. However, the absolute values of the obtained sizes are typically not discussed. The reason for this lack of a discussion of the absolute sizes is connected to the lack of information that is to be expected in terms of the final size of the SCNPs, which varies as a function of the employed solvent as well as the number of cross-linking points – as quantified by Pomposo and colleagues – yet also depends on the prepolymer, type of cross-linking chemistry and the specifically employed reaction conditions. In the current contribution, we provide a concise overview of the thus far reported sizes of SCNPs as obtained via DLS,15–49 DOSY,18,19,21,50 SEC coupled to a viscometry detector51–58 as well as microscopy44,45,47,50 and comment on the observed numbers. It is hoped that the summary of sizes and their visualization provided herein will aid the community in further understanding SCNP folding and lead to improved protocols for size determination.
In our survey of the literature available data on the size of the obtained SCNPs (denoted as the diameter, D, assessed via viscometry, AFM, TEM and DOSY as well as DLS based on the Stokes–Einstein relation), we have made the following observations, which will be discussed and evaluated in detail in the following: (i) The molecular weights of the precursor polymers ranges from as small as 2000 Da to over 200 kDa, constituting a very wide spread of number average molecular weights. However, caution is advised when discussing these molecular weights, as only in cases where the precursor chain exclusively consists of (functional) polystyrenes or poly(methyl methacrylates) and the analysis is based on polystyrene or poly(methyl methacrylate) SEC calibration, these numbers are beset with a small error. Due to many SCNPs having a non-polystyrene or non-PMMA based backbone, relative changes in size are almost exclusively discussed as the primary methodology to demonstrate the intra-molecular folding of linear polymers into SCNPs. In some cases absolute molecular weight methods are employed to determine the precursor size, yet most studies report relative molecular weights. (ii) For (apparent) identical molecular weights, there exists a wide spread of observed sizes for the SCNPs. For example, for a molecular weight of close to 25 kDa, the literature indicates D values that range from 7.4 nm (in a polyether system with a functionalization degree of 18% using thiol–yne cross-linking chemistry)56 to 17.8 nm (in a polycyclooctadiene system cross-linked with 1 mol% rhodium chloride complexation)30 or, even more pronounced for 50 kDa from 6.8 nm (in a poly(methyl methacrylate) system consisting of nominally 26% eneamine cross-links)29 to 19.8 nm (in a poly(acrylate) system with 9% complementary hydrogen bonding via UPy moieties).24 Similar spreads can be found for lower molecular weights, too. For example, for an apparent molecular weight of close to 15 kDa, values ranging from 3.8 to 8.0 nm have been reported (in a poly(azobenzene) ADMET polymer with nominally 50% NITEC cross-linking19vs. a polystyrene system with exactly two complementary hydrogen bonding cross-links),22 and even for 10 kDa spreads from above 1 to 4.4 nm are literature known.21,23 While some variation is expected in terms of the different folding chemistries employed, the coil dynamics and the number of established cross-links, this variation is indeed remarkable. Although estimations of the SCNPs density based on assumptions should be treated with strong caution as it requires knowledge of the particles shape and a highly reliable value for both Dh and the number average molecular weight, high Dh values generally indicate a very loosely packed particle. For example, based on spherical particles, densities of approx. 0.01 g cm−3 are estimated in some cases (e.g., Dh 17.8, Mn 25 kDa),30 which is 14 times less dense than a well-solvated poly(styrene) chain of similar Mn in cyclohexane, which features a Dh of close to 8.2 nm (density approximately 0.14 g cm−3).60 While such a distinct difference in SCNP density will be affected by inaccuracies of the apparent Mn as previously discussed in (i), inaccuracies of the Dh have a more drastic effect on the density as r is raised to the power of 3. (iii) When going to lower molecular weight systems, below 20 kDa, the reported apparent Dh values in some cases appear too small, leading to densities that exceed those of the bulk material. Clearly, measuring reliable D values in such size regimes is extremely challenging, yet some of these values have been confirmed with DOSY measurements19,21,50 and even TEM47,50 in some systems suggesting highly compact particles, as also noted by Pomposo and colleagues.35,59 In any case, apparently too strong reductions in size do not appear to be uncommon.
To visualize these observations, we have constructed a graph which combines a large number of literature reported DLS (black), SEC viscometry (red), DOSY (blue) and microscopy (green) data by plotting the obtained D values vs. the (apparent) molecular weights (Mn). D refers to the diameters measured from the different techniques, i.e., Dh (DLS and DOSY), equivalent sphere diameter (viscosity) or the visually relevant geometrical diameter (microscopy). Due to the low polydispersity of most of the reported functional polymers employed for the preparation of SCNPs, we consider that the difference between Mn and Mw is negligible. Fig. 1 depicts the above noted spread very well, while – not surprisingly – a general trend towards smaller D values with decreasing molecular weight is evident.
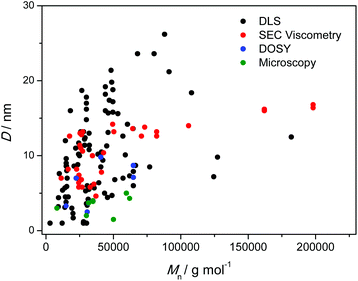 | ||
| Fig. 1 D vs. the (apparent) Mn of reported SCNPs (for the full collation of all data points within the figure, refer to Table S1 in the ESI†). | ||
It is an interesting exercise to plot constant density lines into Fig. 1 to arrive at Fig. 2a. Here, we plotted the theoretical Dh as a function of Mn for constant density values using a simplistic solid sphere model. As an upper limit, we selected a density of 1.4 g cm−3 as some proteins have been reported to reach such densities.61,62 To further compare the SCNPs with their natural analogs, we calculated the Dh for a general data set spanning more than 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different proteins from their experimentally determined Rg values and plotted against Mn (Fig. 2b and c).63 This set of protein data displays a relatively well-defined border of Dh values towards an Mn dependent maximum density threshold. Especially in the Mn regime >100 kDa, this maximum density aligns very well with the solid sphere model of a constant density of 1.4 g cm−3, while protein densities far exceeding even 1.4 g cm−3 are observed for low molecular weights. Some studies have indeed argued that ideal SCNPs should be as closely packed as the corresponding bulk material (approx. 1 g cm−3 or even slightly above) and provide experimental evidence for this notion.35 However, densities exceeding its corresponding bulk material and the extensive protein envelope appear to be physically inaccessible. To address the question of where the low density limit for an SCNP lies, which can still be termed as SCNP, we take reference from one of our earlier studies, where we estimate the Dh of open and closed configurations of a simple polystyrene folding system merely tethered at its chain ends via all atomistic molecular dynamics simulations (violet dot, Fig. 2b and c).64 On the basis of a folded circular 5 kDa polystyrene chain, a density of 0.3 g cm−3 is approximated, providing a hint that higher order cross-linked true SCNP structures should feature densities well in excess of 0.3 g cm−3. Nevertheless, we also include constant density functions for 0.1 as well as 0.5 g cm−3, which we suggest as minimum SCNP density in Fig. 2a–c. Inspection of these plots makes it immediately evident that – based on densities between 0.5 and 1 g cm−3 – a very high number of SCNP systems are below the density threshold of 0.5 g cm−3 and even below the density of linear polystyrene in a good solvent65 (pink dots, Fig. 2b and c). In contrast, a few systems show apparent densities that exceed the 1.4 g cm−3 upper limit and some of the results lie outside the possible conformational space exceeding the density of proteins of comparable molar masses (Fig. 2b and c). Since no apparent patterns seemed to emerge with regard to an ‘allowed’ SCNP density envelope, we explored individual plots based on the employed characterization method (Fig. 3). Inspection of DLS (Fig. 3a) and viscometric based data (Fig. 3b) yields surprising results. Perhaps the most startling observation is the lack of trends observed in the expected densities of the SCNPs when measured with DLS. In fact, more measurements lie outside the expected density realm than lie within it (14 too dense, 42 too loose and 32 within the envelope), even if we allow for densities down to 0.1 g cm−3 to be counted as SCNPs, which is an unlikely assertion. It should be noted that similar to calculating Mnvia comparison to a given standard, DLS also invokes critical assumptions, i.e. that all SCNPs are hard spheres in solution and it does not take into account the conformation of the measured objects. This assumption only allows for the calculation of Rh (and in turn Dh) based on the Stokes–Einstein relation and neglects the radius of gyration (Rg) of a polymer in solution thus leaving the true nature of the SCNPs’ size incomplete (refer also to the ESI†). Furthermore, strong scattering of large particles can suppress the detection of smaller particles, which is a major problem when number distributions are to be derived. In order to obtain reliable data for a wide range of sizes, angle dependent dynamic light scattering should be a viable option. Thus, for very small macromolecules, the data should be taken cautiously. The Pomposo team have made remarkable strides addressing this issue by incorporating SAXS and SANS techniques in their characterization repertoire.66–69
000 different proteins from their experimentally determined Rg values and plotted against Mn (Fig. 2b and c).63 This set of protein data displays a relatively well-defined border of Dh values towards an Mn dependent maximum density threshold. Especially in the Mn regime >100 kDa, this maximum density aligns very well with the solid sphere model of a constant density of 1.4 g cm−3, while protein densities far exceeding even 1.4 g cm−3 are observed for low molecular weights. Some studies have indeed argued that ideal SCNPs should be as closely packed as the corresponding bulk material (approx. 1 g cm−3 or even slightly above) and provide experimental evidence for this notion.35 However, densities exceeding its corresponding bulk material and the extensive protein envelope appear to be physically inaccessible. To address the question of where the low density limit for an SCNP lies, which can still be termed as SCNP, we take reference from one of our earlier studies, where we estimate the Dh of open and closed configurations of a simple polystyrene folding system merely tethered at its chain ends via all atomistic molecular dynamics simulations (violet dot, Fig. 2b and c).64 On the basis of a folded circular 5 kDa polystyrene chain, a density of 0.3 g cm−3 is approximated, providing a hint that higher order cross-linked true SCNP structures should feature densities well in excess of 0.3 g cm−3. Nevertheless, we also include constant density functions for 0.1 as well as 0.5 g cm−3, which we suggest as minimum SCNP density in Fig. 2a–c. Inspection of these plots makes it immediately evident that – based on densities between 0.5 and 1 g cm−3 – a very high number of SCNP systems are below the density threshold of 0.5 g cm−3 and even below the density of linear polystyrene in a good solvent65 (pink dots, Fig. 2b and c). In contrast, a few systems show apparent densities that exceed the 1.4 g cm−3 upper limit and some of the results lie outside the possible conformational space exceeding the density of proteins of comparable molar masses (Fig. 2b and c). Since no apparent patterns seemed to emerge with regard to an ‘allowed’ SCNP density envelope, we explored individual plots based on the employed characterization method (Fig. 3). Inspection of DLS (Fig. 3a) and viscometric based data (Fig. 3b) yields surprising results. Perhaps the most startling observation is the lack of trends observed in the expected densities of the SCNPs when measured with DLS. In fact, more measurements lie outside the expected density realm than lie within it (14 too dense, 42 too loose and 32 within the envelope), even if we allow for densities down to 0.1 g cm−3 to be counted as SCNPs, which is an unlikely assertion. It should be noted that similar to calculating Mnvia comparison to a given standard, DLS also invokes critical assumptions, i.e. that all SCNPs are hard spheres in solution and it does not take into account the conformation of the measured objects. This assumption only allows for the calculation of Rh (and in turn Dh) based on the Stokes–Einstein relation and neglects the radius of gyration (Rg) of a polymer in solution thus leaving the true nature of the SCNPs’ size incomplete (refer also to the ESI†). Furthermore, strong scattering of large particles can suppress the detection of smaller particles, which is a major problem when number distributions are to be derived. In order to obtain reliable data for a wide range of sizes, angle dependent dynamic light scattering should be a viable option. Thus, for very small macromolecules, the data should be taken cautiously. The Pomposo team have made remarkable strides addressing this issue by incorporating SAXS and SANS techniques in their characterization repertoire.66–69
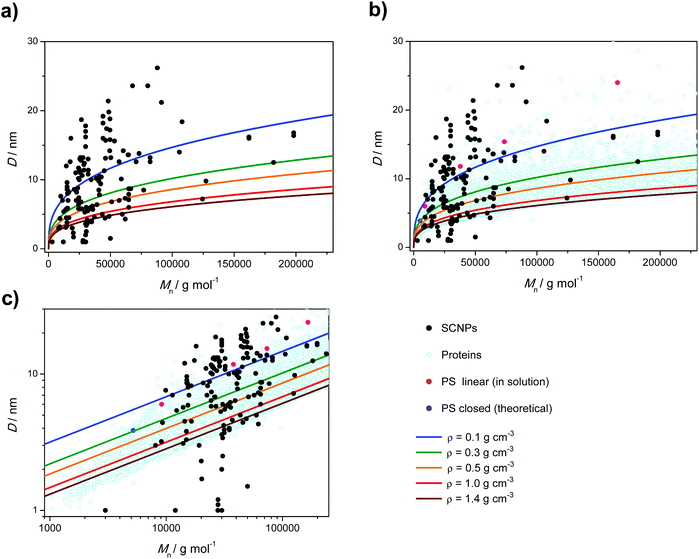 | ||
Fig. 2 (a) D (from all surveyed methods) vs. (apparent) Mn of the reported SCNPs including density functions; (b) D vs. (apparent) Mn of the reported SCNPs including Dh values of 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different proteins (light blue circles),63 linear polystyrene in a good solvent (pink dots)65 and theoretical Dh for 5 kDa polystyrene in a closed conformation (violet dot);64 (c) D vs. (apparent) Mn of the reported SCNPs (log–log plot). 000 different proteins (light blue circles),63 linear polystyrene in a good solvent (pink dots)65 and theoretical Dh for 5 kDa polystyrene in a closed conformation (violet dot);64 (c) D vs. (apparent) Mn of the reported SCNPs (log–log plot). | ||
 | ||
| Fig. 3 Diameter (D) vs. (apparent) Mn of SCNPs measured by (a) DLS; (b) viscometry; (c) 2D DOSY-NMR and (d) microscopy. The blue line indicates a density of 0.1 g cm−3 and the red line 1 g cm−3. | ||
Interestingly and outside the SCNP field, a recent study on cyclic polyenes reports a Dh of 4.4 nm for an Mn of 45.6 kDa, corresponding to a nominal density of 1.67 g cm−3.70 For the linear counterpart, a Dh of 5.2 nm (Mn = 47.3 kDa) was reported, which still suggests a nominal density of 1.09 g cm−3. Thus, here and in the SCNP field, it remains to be established what the exact cause of these apparently too compact – and as noted above too loose – structures is, ranging from DLS measurements beset with a large error, molecular weight determinations with considerable uncertainties or – in the case of very low densities – a possible ineffectiveness of the crosslinking process. However, it is important to point out that in all systems where the collapse is covalently driven, size exclusion chromatography measurements unambiguously confirm the collapse of the precursor chain.
Viscometric measurements are an interesting alternative and do not rely on the assumptions made in DLS. Interestingly, size data derived from viscometry – mainly reported by the Berda team – seem to follow the allowable density realm more closely up to 200 kDa. However, calculating a D via viscometry requires knowledge of a particular polymer's behavior within a specific solvent in order to yield its intrinsic viscosity, which in essence is the inverse density function of the polymeric material ([mL g−1]) in solution. D can be calculated via the Einstein–Simha relation via Vη = M[η]/(2.5NA) and Dη = 2 (3Vh/4π), where [η] is intrinsic viscosity, M the molar mass, Vη the equivalent sphere radius71 (in some reports equally treated as Vh) volume and NA Avogadro's constant. Note that the prerequisite of knowing the materials density in order to deduce D could be the reason that the viscometric size measurements for the SCNPs follow the expected density trends to a much closer degree. The contrast between these two characterization methods is indeed remarkable, however, one must be cautious when using the viscometric examples as the number of SCNPs fully characterized with viscometry are much fewer than those characterized via DLS. Further examples will need to be characterized via viscometry in order to assess if the true D of SCNPs is best measured via viscometry.
The limited number of examples measured with DOSY and microscopy makes it difficult to draw final conclusions from their stand-alone plots (Fig. 3c and d). Although examples exist of D values measured via microscopy techniques in the expected size regime, these numbers cannot be directly compared with Dh as Dh, by definition, is a measurement of a polymer in solution. Due to the fact that AFM and TEM are generally devoid of solvent and drop casted onto surfaces (cryo-TEM being one exception), the observed sizes can be expected to deviating by some degree. Nevertheless, the small sizes reported by microscopic methods are remarkable, all falling below the high density limit.
In summary, a careful survey and analysis of the SCNP literature and the size data contained therein has revealed interesting and important aspects on how SCNPs are currently characterized. Outside of the methods that provide chemical characterization, the approaches available for characterizing SCNPs rely on various techniques assessing their absolute size, yet most studies are concerned with changes in size only. Here, we provide an analysis of the absolute sizes as they relate to their apparent Mn and estimate the potential implications for the apparent density of these particles. We demonstrate that if only DLS and relative Mn are reviewed, a complex picture with a wide spread of absolute sizes emerges. However, we have also evaluated the literature findings on SCNP sizes based on viscometry, DOSY and microscopic methods. This analysis appears to suggest that techniques evaluating the SCNP's intrinsic viscosity may provide the most reliable results. While we can certainly not offer a conclusive answer for the observed absolute size behavior, we submit that the careful analysis provided herein is critical for moving the field towards not only relative size change observations and their rationalization, but also absolute radii discussions, which are critical for the design of functional biomimetic entities.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
C. B.-K. acknowledges support from the Queensland University of Technology (QUT), the Australian Research Council (ARC) in the context of a Laureate Fellowship as well as continued support from the Karlsruhe Institute of Technology (KIT) via the STN and BIFTM programs of the Helmholtz association. In addition, support from the SFB 1176 funded by the German Research Council (DFG) in the context of projects A1 and A2 is gratefully acknowledged. H. F. acknowledges generous funding from the German National Academy of Sciences Leopoldina. Prof. Dr Manfred Schmidt (University of Mainz) is thanked for interesting discussions regarding particle densities. Kilian Wüst (KIT/UNSW) is acknowledged for carefully commenting on the final version of the manuscript. In addition, the authors thank all the PhD and postdoctoral researchers in the QUT/KIT SCNP sub-group.References
- O. Altintas and C. Barner-Kowollik, Macromol. Rapid Commun., 2012, 33, 958 CrossRef CAS PubMed.
- O. Altintas and C. Barner-Kowollik, Macromol. Rapid Commun., 2016, 37, 29 CrossRef CAS PubMed.
- C. K. Lyon, A. Prasher, A. M. Hanlon, B. T. Tuten, C. A. Tooley, P. G. Frank and E. B. Berda, Polym. Chem., 2015, 6, 181 RSC.
- A. M. Hanlon, C. K. Lyon and E. B. Berda, Macromolecules, 2016, 49, 2 CrossRef CAS.
- M. Gonzalez-Burgos, A. Latorre-Sanchez and J. A. Pomposo, Chem. Soc. Rev., 2015, 44, 6122 RSC.
- S. Mavila, O. Eivgi, I. Berkovich and N. G. Lemcoff, Chem. Rev., 2016, 116, 878 CrossRef CAS PubMed.
- W. A. Braunecker and K. Matyjaszewski, Prog. Polym. Sci., 2007, 32, 93 CrossRef CAS.
- R. K. Roy and J.-F. Lutz, J. Am. Chem. Soc., 2014, 136, 12888 CrossRef CAS PubMed.
- Z. Jia and M. J. Monteiro, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 2085 CrossRef CAS.
- X.-Y. Tu, M.-Z. Liu and H. Wei, J. Polym. Sci., Part A: Polym. Chem., 2016, 54, 1447 CrossRef CAS.
- M. Ouchi, N. Badi, J.-F. Lutz and M. Sawamoto, Nat. Chem., 2011, 3, 917 CrossRef CAS PubMed.
- J. Steinkoenig, H. Rothfuss, A. Lauer, B. T. Tuten and C. Barner-Kowollik, J. Am. Chem. Soc., 2017, 139, 51 CrossRef CAS PubMed.
- J. A. Pomposo, J. Rubio-Cervilla, A. J. Moreno, F. Lo Verso, P. Bacova, A. Arbe and J. Colmenero, Macromolecules, 2017, 50, 1732 CrossRef CAS.
- J. A. Pomposo, I. Perez-Baena, F. Lo Verso, A. J. Moreno, A. Arbe and J. Colmenero, ACS Macro Lett., 2014, 3, 767 CrossRef CAS.
- J. Willenbacher, K. N. R. Wuest, J. O. Mueller, M. Kaupp, H.-A. Wagenknecht and C. Barner-Kowollik, ACS Macro Lett., 2014, 3, 574 CrossRef CAS.
- J. Willenbacher, O. Altintas, V. Trouillet, N. Knofel, M. J. Monteiro, P. W. Roesky and C. Barner-Kowollik, Polym. Chem., 2015, 6, 4358 RSC.
- O. Altintas, J. Willenbacher, K. N. R. Wuest, K. K. Oehlenschlaeger, P. Krolla-Sidenstein, H. Gliemann and C. Barner-Kowollik, Macromolecules, 2013, 46, 8092 CrossRef CAS.
- T. S. Fischer, D. Schulze-Sünninghausen, B. Luy, O. Altintas and C. Barner-Kowollik, Angew. Chem., Int. Ed., 2016, 55, 11276 CrossRef CAS PubMed.
- J. T. Offenloch, J. Willenbacher, P. Tzvetkova, C. Heiler, H. Mutlu and C. Barner-Kowollik, Chem. Commun., 2017, 53, 775 RSC.
- O. Altintas, P. Krolla-Sidenstein, H. Gliemann and C. Barner-Kowollik, Macromolecules, 2014, 47, 5877 CrossRef CAS.
- C. Heiler, J. T. Offenloch, E. Blasco and C. Barner-Kowollik, ACS Macro Lett., 2017, 6, 56 CrossRef CAS.
- O. Altintas, E. Lejeune, P. Gerstel and C. Barner-Kowollik, Polym. Chem., 2012, 3, 640 RSC.
- E. Blasco, B. Yameen, A. Q. Quick, P. Krolla-Sidenstein, A. Welle, M. Wegener and C. Barner-Kowollik, Macromolecules, 2015, 48, 8718 CrossRef CAS.
- P. J. M. Stals, M. A. J. Gillissen, R. Nicolay, A. R. A. Palmans and E. W. Meijer, Polym. Chem., 2013, 4, 2584 RSC.
- F. Wang, H. Pu, M. Jin, H. Pan, Z. Chang, D. Wan and J. Du, J. Polym. Sci., Part A: Polym. Chem., 2015, 53, 1832 CrossRef CAS.
- E. A. Appel, J. Dyson, J. del Barrio, Z. Walsh and O. A. Scherman, Angew. Chem., Int. Ed., 2012, 51, 4185 CrossRef CAS PubMed.
- F. Wang, H. Pu and X. Che, Chem. Commun., 2016, 52, 3516 RSC.
- C. Song, L. Li, L. Dai and S. Thayumanavan, Polym. Chem., 2015, 6, 4828 RSC.
- A. Sanchez-Sanchez, D. A. Fulton and J. A. Pomposo, Chem. Commun., 2014, 50, 1871 RSC.
- S. Mavila, C. E. Diesendruck, S. Linde, L. Amir, R. Shikler and N. G. Lemcoff, Angew. Chem., Int. Ed., 2013, 52, 5767 CrossRef CAS PubMed.
- J. Jeong, Y.-J. Lee, B. Kim, K. S. Jung and H.-J. Paik, Polym. Chem., 2015, 6, 3392 RSC.
- S. Mavila, I. Rozenberg and N. G. Lemcoff, Chem. Sci., 2014, 5, 4196 RSC.
- F. Wang, H. Pu, M. Jin and D. Wan, Macromol. Rapid Commun., 2016, 37, 330 CrossRef CAS PubMed.
- J. Lu, N. ten Brummelhuis and M. Weck, Chem. Commun., 2014, 50, 6225 RSC.
- L. Oria, R. Aguado, J. A. Pomposo and J. A. Colmenero, Adv. Mater., 2010, 22, 3038 CrossRef CAS PubMed.
- D. Chao, X. Jia, B. T. Tuten, C. Wang and E. B. Berda, Chem. Commun., 2013, 49, 4178 RSC.
- A. Sanchez-Sanchez and J. A. Pomposo, J. Nanomater., 2015, 2015, 7 Search PubMed.
- I. Perez-Baena, I. Asenjo-Sanz, A. Arbe, A. J. Moreno, F. Lo Verso, J. Colmenero and J. A. Pomposo, Macromolecules, 2014, 47, 8270 CrossRef CAS.
- A. R. De Luzuriaga, N. Ormategui, H. J. Grande, I. Odriozola, J. A. Pomposo and I. Loinaz, Macromol. Rapid Commun., 2008, 29, 1156 CrossRef.
- X. Jiang, H. Pu and P. Wang, Polymer, 2011, 52, 3597 CrossRef CAS.
- P. Wang, H. Pu and M. S. Jin, J. Polym. Sci., Part A: Polym. Chem., 2011, 49, 5133 CrossRef CAS.
- D. Mecerreyes, V. Lee, C. J. Hawker, J. L. Hedrick, A. Wursch, W. Volksen, T. Magbitang, E. Huang and R. D. Miller, Adv. Mater., 2001, 13, 204 CrossRef CAS.
- T.-K. Nguyen, S. J. Lam, K. K. K. Ho, N. Kumar, G. G. Qiao, S. Egan, C. Boyer and E. H. H. Wong, ACS Infect. Dis., 2017, 3, 237 CrossRef CAS PubMed.
- N. Ormategui, I. Garcia, D. Padro, G. Cabanero, H. J. Grande and I. Loinaz, Soft Matter, 2012, 8, 734 RSC.
- C. T. Adkins, H. Muchalski and E. Harth, Macromolecules, 2009, 42, 5786 CrossRef CAS.
- N. D. Knöfel, H. Rothfuss, J. Willenbacher, C. Barner-Kowollik and P. W. Roesky, Angew. Chem., Int. Ed., 2017, 56, 4950 CrossRef PubMed.
- J. N. Dobish, S. K. Hamilton and E. Harth, Polym. Chem., 2012, 3, 857 RSC.
- L. Buruaga and J. A. Pomposo, Polymers, 2011, 3, 1673 CrossRef CAS.
- J. Rumulus and M. Weck, Macromol. Rapid Commun., 2013, 34, 1518 CrossRef PubMed.
- A. M. Hanlon, R. Chen, K. J. Rodriguez, C. Willis, J. G. Dickinson, M. Cashman and E. B. Berda, Macromolecules, 2017, 50, 2996 CrossRef CAS.
- C. K. Lyon, E. O. Hill and E. B. Berda, Macromol. Chem. Phys., 2016, 217, 501 CrossRef CAS.
- A. Prasher, C. M. Loynd, B. T. Tuten, P. G. Frank, D. Chao and E. B. Berda, J. Polym. Sci., Part A: Polym. Chem., 2016, 54, 209 CrossRef CAS.
- C. A. Tooley, S. Pazicni and E. B. Berda, Polym. Chem., 2015, 6, 7646 RSC.
- P. G. Frank, B. T. Tuten, A. Prasher, D. Chao and E. B. Berda, Macromol. Rapid Commun., 2014, 35, 249 CrossRef CAS PubMed.
- B. T. Tuten, D. Chao, C. K. Lyon and E. B. Berda, Polym. Chem., 2012, 3, 3068 RSC.
- S. Basasoro, M. Gonzalez-Burgos, A. J. Moreno, F. L. Verso, A. Arbe, J. Colmenero and J. A. Pomposo, Macromol. Rapid Commun., 2016, 37, 1060 CrossRef CAS PubMed.
- J. Rubio-Cervilla, F. Barroso-Bujans and J. A. Pomposo, Macromolecules, 2016, 49, 90 CrossRef CAS.
- A. M. Hanlon, I. Martin, E. B. Bright, J. Chouinard, K. J. Rodriguez, G. E. Patenotte and E. B. Berda, Polym. Chem., 2017, 8, 5120 RSC.
- I. Perez-Baena, I. Loinaz, D. Padro, I. Garcia, H. J. Grande and I. Odriozola, J. Mater. Chem., 2010, 20, 6916 RSC.
- G. Hadziioannou, P. M. Cotts, G. ten Brinke, C. C. Han, P. Lutz, C. Strazielle, P. Rempp and A. J. Kovacs, Macromolecules, 1987, 20, 493 CrossRef CAS.
- P. G. Squire and M. E. Himmel, Arch. Biochem. Biophys., 1979, 196, 165 CrossRef CAS PubMed.
- K. Gekko and H. Noguchi, J. Phys. Chem., 1979, 83, 2706 CrossRef CAS.
- L. Hong and J. Lei, J. Polym Sci., Part B: Polym. Phys., 2009, 47, 207 CrossRef CAS.
- D. Danilov, C. Barner-Kowollik and W. Wenzel, Chem. Commun., 2015, 51, 6002 RSC.
- M. Duval, P. Lutz and C. Strazielle, Makromol. Chem., Rapid Commun., 1985, 6, 71 CrossRef CAS.
- I. Perez-Baena, F. Barroso-Bujans, U. Gasser, A. Arbe, A. J. Moreno, J. Colmenero and J. A. Pomposo, ACS Macro Lett., 2013, 2, 775 CrossRef CAS.
- A. Sanchez-Sanchez, S. Akbari, A. Etxeberria, A. Arbe, U. Gasser, A. J. Moreno, J. Colmenero and J. A. Pomposo, ACS Macro Lett., 2013, 2, 491 CrossRef CAS.
- A. Sanchez-Sanchez, S. Akbari, A. J. Moreno, F. L. Verso, A. Arbe, J. Colmenero and J. A. Pomposo, Macromol. Rapid Commun., 2013, 34, 1665 CrossRef CAS.
- A. J. Moreno, F. Lo Verso, A. Arbe, J. A. Pomposo and J. Colmenero, J. Phys. Chem. Lett., 2016, 7, 838 CrossRef CAS PubMed.
- C. D. Roland, H. Li, K. A. Abboud, K. B. Wagener and A. S. Veige, Nat. Chem., 2016, 8, 791 CrossRef CAS PubMed.
- W. Burchard, Adv. Polym. Sci., 1999, 143, 113 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: A detailed table containing number average molecular weights, Mn, sizes, as well as the techniques employed for the characterization of the SCNPs and the details for the calculation of protein Dh values can be found in this section. See DOI: 10.1039/c7py01278k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |
