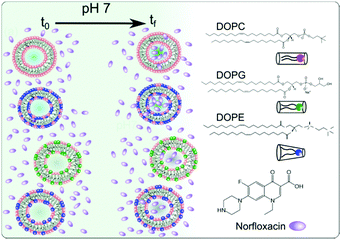
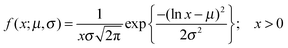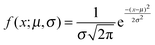Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dependence of norfloxacin diffusion across bilayers on lipid composition†
Sowmya
Purushothaman
,
Jehangir
Cama
and
Ulrich F.
Keyser
*
Biological and Soft Systems, Department of Physics, University of Cambridge, Cavendish Laboratory, JJ Thomson Avenue, Cambridge, CB3 0HE, UK. E-mail: ufk20@cam.ac.uk
First published on 15th December 2015
Abstract
Antibiotic resistance is a growing concern in medicine and raises the need to develop and design new drug molecules that can efficiently inhibit bacterial replication. Spurring the passive uptake of the drug molecules is an obvious solution. However our limited understanding of drug–membrane interactions due to the presence of an overwhelming variety of lipids constituting cellular membranes and the lack of facile tools to probe the bio-physical interactions between drugs and lipids imposes a major challenge towards developing new drug molecules that can enter the cell via passive diffusion. Here, we used a label-free micro-fluidic platform combined with giant unilamellar lipid vesicles to investigate the permeability of membranes containing mixtures of DOPE and DOPG in DOPC, leading to a label-free measurement of passive membrane-permeability of autofluorescent antibiotics. A fluoroquinolone drug, norfloxacin was used as a case study. Our results indicate that the diffusion of norfloxacin is strongly dependent on the lipid composition which is not expected from the traditional octanol–lipid partition co-efficient assay. The anionic lipid, DOPG, slows the diffusion process whereas the diffusion across liposomes containing DOPE increases with higher DOPE concentration. Our findings emphasise the need to investigate drug–membrane interactions with focus on the specificity of drugs to lipids for efficient drug delivery, drug encapsulation and targeted drug-delivery.
Introduction
The development of antibiotics has been instrumental in combating a large number of infectious diseases over the past century.1 However, with the excessive and at times unnecessary use of antibiotics, antibacterial resistance is on the rise. The rapid spread of resistance and simultaneous decline in the discovery of novel antimicrobials pose a challenge to drug development and treatment of bacterial infections.A large number of antibiotics function by targeting critical intracellular processes after overcoming the cellular membrane barrier.2 Small molecules and drugs can overcome this barrier by (i) binding to active transporters to cross the membrane, (ii) targeting bacteria-specific membrane components involved in cell-wall synthesis,3 (iii) passive diffusion across lipid membranes or (iv) via liposomal carriers that act as cargos to transport them to the target cells.4,5 One of the origins of antibiotic resistance is the reduction of bacterial membrane permeability to drugs. Reduced uptake in turn reduces drug accumulation within the cell leading to decreased cell susceptibility. The permeability is reduced when there are alterations in the target membrane-composition,6,7 mutations in porins present in the outer membrane or when there is an over-expression of efflux pumps that are mainly present in the inner membrane.4,8
The factors that cause these mutations and adaptations leading to drug resistance are poorly understood.9 Efficient translocation of drugs across the membrane is therefore one of the major challenges in drug development. Designing new drugs with increased rates of diffusion and accumulation faces a further setback because of the multitude of macromolecules present in the membrane, many of which are not even identified.10–12 While designing drugs requires an understanding of the drug–membrane interaction mechanisms, drugs themselves can potentially influence the activity of various membrane proteins or enzymes.13,14 Understanding lipid–drug interactions is also important for developing efficient liposomal drug-delivery systems. Therapeutically, liposomal drug delivery is one of the most successful delivery systems owing to its bio-compatibility and low toxicity.15–18 Despite their wide applicability, there are limited laboratory-based techniques to probe liposome–drug bio-compatibility for efficient drug-encapsulation.19–21
There is extensive literature where a variety of elegant techniques such as NMR,22 X-ray diffraction,23 FTIR,22 CD,6,24 fluorescence anisotropy, FRET25 and FPE26 were used to show that the interaction of drugs with membranes is lipid-composition specific.27,28 Most of the techniques give valuable information about drug–lipid interactions at a molecular level. The studies have revealed that the charged state, hydrophobicity and the headgroup structure dictate the passage of drugs via membranes.29 However, these techniques can often only elucidate interactions of drugs with single component lipid systems in contrast to the multi-component membrane architecture found in nature. The analysis and interpretation of the data obtained from these techniques is non-trivial and often cannot be directly used to extract permeability. The bio-chemical diversity combined with the lack of technologies to probe into the bio-physical characteristics of lipid–drug complexes make the understanding of the mechanisms underpinning lipid–drug interactions challenging. Unlocking the drug–lipid interactions by developing techniques that can facilitate screening of lipids based on properties such as the bio-compatibility of lipids to a specific drug molecule, its permeability, stability and toxicity is therefore crucial for drug development as well as for the design of optimised drug-delivery systems.
In addition, while most of these studies involve labelling the molecules, other more popular techniques such as the partition co-efficient measurement30 or HPLC31,32 investigate only the bulk properties based on the octanol–drug interaction33,34 as opposed to a more relevant lipid–drug interaction. Amphiphilicity is the key feature for treating octanol as a simple model to membrane lipids. However, this crude chemical similarity cannot encompass the complex structural variations found in lipids. Octanol itself possesses an internal structure leading to the formation of inverse micellar aggregates (of 18.5–21.5 Å diameters)35 which could lead to the underestimation of partition co-efficients.36 Therefore, the prediction of molecular interactions of drug molecules with respect to octanol cannot fully reflect the drug–membrane interactions occurring in biological systems as shown in this article.
Microfluidic devices are emerging as powerful tools in bio-sensing and medical diagnostics.37,38 Major advantages of microfluidic devices are the ease of sample handling, modularity, portability, potential high-throughput capability and cost effectiveness, all of which are desirable for bio-sensing and analysis.39,40–42 In recent years the application of the miniature microfluidic devices has also been extended to drug screening43 and testing targeted delivery systems for anti-cancer drugs.44 Combining liposomal membrane systems with microfluidics can therefore provide an ideal tool to probe the structure–function relationship between lipid membranes and drugs.
In this work, we built upon the previously described in-house microfluidic setup45 combined with giant unilamellar vesicles as model membranes to probe the diffusion of norfloxacin across vesicular lipid membranes with specific compositions inspired by the E. coli inner membrane composition. Norfloxacin is widely used in the treatment of infectious conditions such as urinary tract infections,46 prostate, skin, pulmonary and digestive infections.47–49 It belongs to the fluoroquinolone family of drugs that target bacterial enzymes involved in DNA replication thereby inhibiting cell division.50 The exact mechanism of its action, particularly from its uptake by the membrane through drug–lipid interactions to drug–DNA complex formation however is not well established.51 The use of a microfluidic platform for the above-mentioned advantages combined with the liposomes52 of various compositions provides a facile tool to measure the diffusion across a variety of lipid compositions.
We investigated the effect of the bilayer forming lipid DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), non-bilayer forming lipid DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) and charged (anionic at pH 7) lipid DOPG (1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt)) on the permeability of norfloxacin across bilayers (see Fig. 1 for molecular structures). The compositions were chosen close to their concentrations found in the inner membrane of E. coli. Furthermore, to keep the hydrophobic area constant, DOPE, DOPG and DOPC lipids were used since all of them contain 18 carbon chains with a cis unsaturation at the 9th carbon position. It should be noted that the real membrane, however, contains a mixture of PE or PG lipids with varying chain lengths or degrees of unsaturation. PE constitutes about 70% of the total lipids found in the E. coli membrane whereas PG forms about 23% of the total lipids. Our approach exploits the auto-fluorescence properties of norfloxacin thereby enabling a label-free measurement of drug permeation across the lipid bilayer.45,53 The permeation of norfloxacin is investigated at two pH conditions, at pH 5 at which most of the norfloxacin molecules are positively charged and at pH 7 at which norfloxacin is uncharged/zwitterionic.54
We show that the lipid composition does in fact play a crucial role in the passive diffusion of small molecules such as norfloxacin (see Fig. 1). In particular, we found that the charged lipid slows the diffusion at pH 7 whereas zwitterionic lipids at the same concentrations increase the permeability. This is in contrast to the observations made for ciprofloxacin, another fluoroquinolone drug, where the drug was found to interact more strongly with anionic lipids at a similar pH.29,55 Furthermore, we found that the diffusion also depends on the amount of PE present in the membrane with lower concentrations retarding the diffusion. It is thus clear that changing the lipid composition of a membrane has a considerable effect on its permeability to norfloxacin. These results reinforce the importance of studying drug transport in a lipid based model system in order to better understand transport properties across cellular membranes.
Materials
DOPG, DOPE, and ITO glass slides were purchased from Sigma. DOPC was purchased from Avanti Polar Lipids. We used 100 mM sucrose in 5 mM phosphate buffer for pH 7 or 100 mM sucrose in 5 mM acetic acid buffer for pH 5.Preparation of GUVs
GUVs were prepared using the standard electro-formation technique. Briefly, lipids were mixed in the desired ratios, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 DOPC
30 DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE, 30
DOPE, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 DOPC
70 DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE, 70
DOPE, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 DOPC
30 DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPG and pure DOPC in chloroform to obtain a final concentration of 0.5 mg ml−1 lipid mixture. A thin layer of the lipid film was deposited on ITO glass slides and left to dry for two hours in a vacuum desiccator. The dried film was rehydrated in the buffer and an AC electric field was applied at 2.6 V peak-to-peak at 10 Hz for 3 hours followed by 4.4 V at 4 Hz for 30 minutes. The vesicles were stored at 4 °C and were used within 48 hours.
DOPG and pure DOPC in chloroform to obtain a final concentration of 0.5 mg ml−1 lipid mixture. A thin layer of the lipid film was deposited on ITO glass slides and left to dry for two hours in a vacuum desiccator. The dried film was rehydrated in the buffer and an AC electric field was applied at 2.6 V peak-to-peak at 10 Hz for 3 hours followed by 4.4 V at 4 Hz for 30 minutes. The vesicles were stored at 4 °C and were used within 48 hours.
Optical setup
A custom-built UV epi-fluorescence microscope was used to perform all the experiments.45 The optical setup consists of a broadband white light source (EQ99FC, Energetiq), whose output is routed through a monochromator (Monoscan 2000, OceanOptics) to select the desired excitation wavelength (340 nm). The excitation beam is directed onto the microfluidic device via a dichroic mirror (DLHS UV 351–355, Qi-Optiq, Germany) and a 60× water immersion UPLSAPO Olympus objective (NA 1.2). The emitted light is recorded using an Evolve 512 EMCCD camera (2 ms exposure, bin 2, 65 fps) using Micromanager 1.4,56 an open source microscopy platform. UV fused silica lenses were used for light navigation to optimise transmission in the UV region.Microfluidic chip design
The microfluidic chip (Fig. 2) was constructed using standard photo- and soft-lithography techniques.57 Two designs were used for the experiments (Fig. 2(a) and (b)). For experiments carried out at pH 7, a small chip containing a ∼28 mm long channel folded 4 times was used whereas for experiments carried out at pH 5, where the diffusion was much slower, a chip with an ∼380 mm long channel folded 15 times (see Fig. 2(b)) was used. In both the chips, a series of pillars in the inlet reservoirs were designed to filter out vesicles of size greater than 40 μm diameter from entering the channel. The microfluidic flows were controlled by applying suction at the outlet reservoir using a neMESYS syringe pump system with a 250 μl Duran borosilicate glass syringe (ILS, Germany). At the inlets, pipette tips with 40 μl of vesicle stock solution and 40 μl of 2 mM norfloxacin solution, respectively, were fed into the two reservoirs. Once the fluorescence intensities were uniform, data collection was initiated. The flows were reduced from 30 μl h−1 down to 3 μl h−1 for visualisation and collection of videos.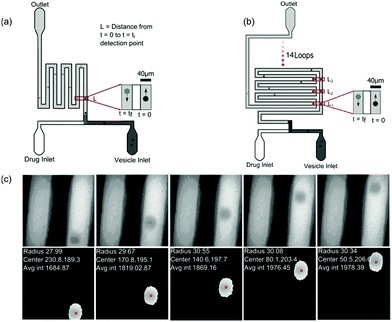 | ||
| Fig. 2 Experimental setup. (a) Design of the small microfluidic chip used in the experiments carried out at pH 7. The schematic also depicts the intensity variations measured across two observation points along the channel. (b) Design of the chip used for measurements carried out at pH 5. The chip has a longer channel and hence vesicles could be observed for longer periods of time of about 60 s or more. (c) Typical vesicles observed along the channel. Our automated MATLAB routine identifies vesicles (events) and tracks the vesicles across frames. The routine finds the centre of the vesicles (pixels), radius (μm) and velocity (mm s−1) from the frame rate and traced centre position information from all frames where the event was detected (for more details, see ESI†). The data are then further processed using an automated LABVIEW routine (ESI†) that processes the data by removing the outliers, fitting and finally calculating the permeability for each data set. | ||
Data analysis
In order to calculate the permeability coefficient, we determined the normalised fluorescence intensity differences between the exterior and the interior of the vesicles at t = 0 (ΔI1) and later at tf (ΔI2) given by the following equations (see Fig. 2(c)):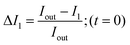 | (1) |
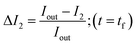 | (2) |
A MATLAB routine extracts the list of files and frame numbers containing vesicles and calculates the radius, velocity, centre co-ordinates, circularity and the total average intensity after background subtraction of the detected vesicles. We developed further post-processing routines to remove the outliers, fit data and finally calculate the permeability co-efficient thereby fully automating the routines. A detailed description of the process can be found in the ESI.†
After solving the diffusion model, we obtained the following equation that was used for analysis:45
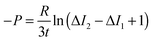 | (3) |
Results
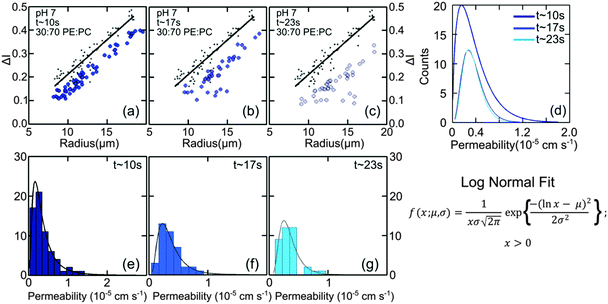
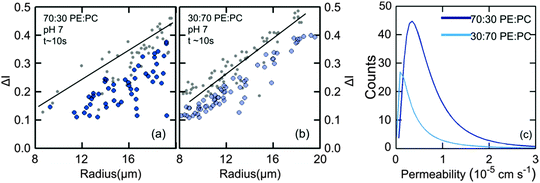
Vesicles containing different compositions of DOPE and DOPG lipids were made as detailed before. The permeability values calculated using the procedure described in the previous section were based on the calculation of the increase in intensity inside the vesicle as the auto-fluorescent norfloxacin permeates the vesicle from the external solution. At the initial viewpoint, defined as time t = 0, the vesicles have just been exposed to norfloxacin and appear dark when viewed at 340 nm excitation (shown in grey in Fig. 3(a–c)). As the vesicle travels along the micro-fluidic channel, norfloxacin from the surroundings permeates through the membrane making the vesicle appear brighter.The normalised intensity difference between the outer region (Iout) and the intensity calculated at the center (I1 at t = 0 and I2 at t = tf respectively) of the vesicles containing 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 DOPE
70 DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC lipids is shown in Fig. 3(a–c). Here, each data point represents one individual vesicle measurement. The initial relative intensity ΔI1 at t = 0 of the vesicles is shown as grey points, with a linear fit shown as a black line to facilitate the visual comparison of the data points at t = 0 (ΔI1) and tf (ΔI2). As the vesicles travel longer distances (Fig. 3(b) and (c)) the difference between Iout and I2 decreases in comparison to the difference between Iout and I1. The difference in the intensities between the two observation points is a direct measurement of the amount of norfloxacin permeating the membrane. A histogram of the permeability values obtained for all the tracked vesicles is plotted as shown in Fig. 3(d) and in the subsequent graphs presented in the article. The bin size is chosen based on the Rice rule where the number of bins is 2n1/3, n being the number of data points. For pH 7 measurements, a density distribution function is fit to the histogram using MATLAB's ‘histfit’ function by selecting the lognormal distribution which is given by:
DOPC lipids is shown in Fig. 3(a–c). Here, each data point represents one individual vesicle measurement. The initial relative intensity ΔI1 at t = 0 of the vesicles is shown as grey points, with a linear fit shown as a black line to facilitate the visual comparison of the data points at t = 0 (ΔI1) and tf (ΔI2). As the vesicles travel longer distances (Fig. 3(b) and (c)) the difference between Iout and I2 decreases in comparison to the difference between Iout and I1. The difference in the intensities between the two observation points is a direct measurement of the amount of norfloxacin permeating the membrane. A histogram of the permeability values obtained for all the tracked vesicles is plotted as shown in Fig. 3(d) and in the subsequent graphs presented in the article. The bin size is chosen based on the Rice rule where the number of bins is 2n1/3, n being the number of data points. For pH 7 measurements, a density distribution function is fit to the histogram using MATLAB's ‘histfit’ function by selecting the lognormal distribution which is given by:
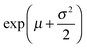 and the variance by exp(2μ + σ2)(exp(σ2) − 1). The histograms with the respective fits are shown in Fig. 3(e–g). Fig. 3 clearly demonstrates that the vesicles remain intact for several minutes in the channel and the permeability of norfloxacin measured at various observation points gives a consistent value of 0.44 ± 0.03 × 10−5 cm s−1, validating the technique.
and the variance by exp(2μ + σ2)(exp(σ2) − 1). The histograms with the respective fits are shown in Fig. 3(e–g). Fig. 3 clearly demonstrates that the vesicles remain intact for several minutes in the channel and the permeability of norfloxacin measured at various observation points gives a consistent value of 0.44 ± 0.03 × 10−5 cm s−1, validating the technique.
Fig. 4(d–f) presents the data obtained at pH 7 at which norfloxacin is predominantly uncharged or zwitterionic,54 and can permeate through the lipid membrane more easily. As a control, we also performed experiments at pH 5 at which norfloxacin is in a charged state and hence cannot permeate through lipid membranes easily. The results are presented in Fig. 4(a–c) for all the lipid compositions investigated in this work. The three control measurements show an almost perfect overlap between the normalised intensity ΔI1 at t = 0 and the normalised intensity ΔI2 calculated after a minute's exposure to the drug (Fig. 4(a–c)). This proves that the vesicles are intact and the transport is negligible as expected for the charged form of norfloxacin at pH 5. The corresponding histogram fits are shown in Fig. 4(g). For pH 5 experiments the histogram was fitted using a ‘Gaussian’ distribution:
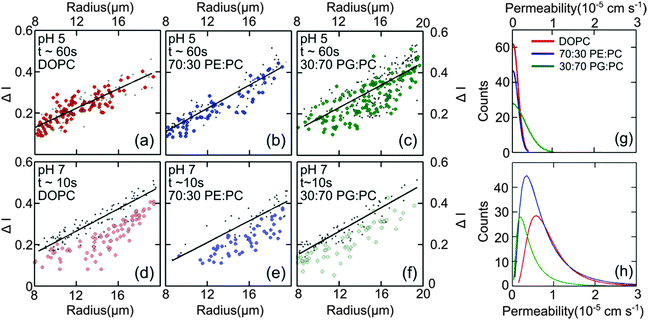 | ||
Fig. 4 Diffusion of norfloxacin in buffers at pH 5 (a–c) and 7 (d–f) for a distance of 45.5 mm (pH 5) and 7.4 mm and at different lipid compositions (a and d) 100% DOPC, (b and e) 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 DOPE 30 DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC and (c and f) 30 DOPC and (c and f) 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 DOPG 70 DOPG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC. (a–c) Control measurements for all lipid mixtures showing that the vesicles are impermeable for charged norfloxacin at pH 5 at these time scales. For all lipid mixtures ΔI measurements coincide perfectly with the fit (black line) representing ΔI(t0) proving negligible diffusion of norfloxacin for up to 60 s. (d and e) In stark contrast, ΔI shifts to lower values for all three mixtures at pH 7 when the uncharged norfloxacin is present. Obviously the change in ΔI is more pronounced in pure DOPC membranes indicating higher permeability for norfloxacin (d). The data shown here are the results of a single experiment. Additional independent runs were performed for each composition (see ESI†). (g) Permeability histograms as fitted to the distribution for pH 5. The most probable values are 0 as expected for the control measurement. (h) Histograms for the data at pH 7 indicating that permeability depends on the lipid mixture. DOPC. (a–c) Control measurements for all lipid mixtures showing that the vesicles are impermeable for charged norfloxacin at pH 5 at these time scales. For all lipid mixtures ΔI measurements coincide perfectly with the fit (black line) representing ΔI(t0) proving negligible diffusion of norfloxacin for up to 60 s. (d and e) In stark contrast, ΔI shifts to lower values for all three mixtures at pH 7 when the uncharged norfloxacin is present. Obviously the change in ΔI is more pronounced in pure DOPC membranes indicating higher permeability for norfloxacin (d). The data shown here are the results of a single experiment. Additional independent runs were performed for each composition (see ESI†). (g) Permeability histograms as fitted to the distribution for pH 5. The most probable values are 0 as expected for the control measurement. (h) Histograms for the data at pH 7 indicating that permeability depends on the lipid mixture. | ||
The most probable permeability for all three lipids is 0, as expected for the control measurements.
In contrast, our results at pH 7, shown in Fig. 4(d–f) for pure DOPC (Fig. 4(d)), 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 DOPE
30 DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC (Fig. 4(e)), and 30
DOPC (Fig. 4(e)), and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 DOPG
70 DOPG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC (Fig. 4(f)) show significant changes in ΔI indicating the permeation of norfloxacin into vesicles. It is also important to note here that the final observation point for our pH 7 measurements was only after 10 s. The diffusion time is thus six times shorter than the timescale in all control experiments presented in Fig. 4(a–c), indicating a relatively faster permeation of the drug. As before, we use log-normal fits to the permeability histograms (Fig. 4(h)).
DOPC (Fig. 4(f)) show significant changes in ΔI indicating the permeation of norfloxacin into vesicles. It is also important to note here that the final observation point for our pH 7 measurements was only after 10 s. The diffusion time is thus six times shorter than the timescale in all control experiments presented in Fig. 4(a–c), indicating a relatively faster permeation of the drug. As before, we use log-normal fits to the permeability histograms (Fig. 4(h)).
We observed that the DOPC membrane was the most permeable at pH 7 with a value of 0.81 ± 0.01 × 10−5 cm s−1 followed by the vesicles containing a mixture of DOPC and DOPE, with high amounts of DOPE (70%) having a value of 0.68 ± 0.07 × 10−5 cm s−1. However at lower concentrations of DOPE (30%), the permeability dropped to 0.44 ± 0.03 × 10−5 cm s−1 (see Fig. 6). At lower amounts of the charged lipid, DOPG (30%), the permeability was found to be 0.43 ± 0.02 × 10−5 cm s−1. At pH 5, the permeability was not affected by lipid composition at these timescales (see Fig. 4(a–c and g)). These data are remarkable as they clearly show that there is a significant difference in permeability values for different lipid mixtures. These results cannot be predicted from the measurements of the partition coefficient in octanol.
In order to widen the range of lipid mixtures we also compare results of mixtures of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 DOPE
30 DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC lipids (Fig. 5(a)) and 30
DOPC lipids (Fig. 5(a)) and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 DOPE
70 DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC. At the lower DOPE concentration, the permeability of the vesicles was lower by a factor of two (0.44 ± 0.03 × 10−5 cm s−1) than the pure DOPC counterparts (0.81 ± 0.01 × 10−5 cm s−1). Interestingly, when the concentration of the non-lamellar lipid was increased, the vesicles became more permeable (0.68 ± 0.07 × 10−5 cm s−1) compared to the vesicles with lower amounts of DOPE (Fig. 5). Therefore, we find that the diffusion of norfloxacin is faster by roughly 30% for vesicles made from mixtures containing less DOPC in comparison to vesicles containing more DOPC in DOPE. This is interesting, as our original data suggest that diffusion through DOPC is faster. However, one clearly cannot conclude that the DOPC content determines permeability.
DOPC. At the lower DOPE concentration, the permeability of the vesicles was lower by a factor of two (0.44 ± 0.03 × 10−5 cm s−1) than the pure DOPC counterparts (0.81 ± 0.01 × 10−5 cm s−1). Interestingly, when the concentration of the non-lamellar lipid was increased, the vesicles became more permeable (0.68 ± 0.07 × 10−5 cm s−1) compared to the vesicles with lower amounts of DOPE (Fig. 5). Therefore, we find that the diffusion of norfloxacin is faster by roughly 30% for vesicles made from mixtures containing less DOPC in comparison to vesicles containing more DOPC in DOPE. This is interesting, as our original data suggest that diffusion through DOPC is faster. However, one clearly cannot conclude that the DOPC content determines permeability.
It is worth mentioning that since DOPC vesicles showed the fastest increase in intensity inside the vesicle attributed to the permeation of norfloxacin, all the measurements for DOPE and DOPG compositions were therefore taken for the same time scales of approximately 10 s (7.4 mm along the channel).
The overall results regarding the extracted permeability values obtained for all lipid compositions in this study are summarised in Fig. 6.
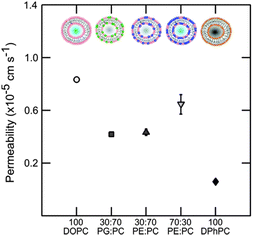 | ||
| Fig. 6 Permeability of vesicles containing DOPC, DOPE and DOPG lipids at various concentrations at pH 7. Pure DOPC is the most permeable. The effects of small amounts of the non-lamellar lipid DOPE or DOPG are similar indicating no preference of norfloxacin to a charged moiety. At higher concentrations of DOPE the permeability goes up significantly indicating the possible effect of curvature frustration. The errorbars are within the legend size. The permeability value of DPhPC obtained from the literature45 is shown for comparison. The permeability of norfloxacin is clearly dependent on the type of lipid and the relative concentration. | ||
Discussion
Passive transport of drug molecules across lipid membranes is a complex process as it depends on the changes in the intermolecular forces acting in lipid membranes in response to the physiological conditions (temperature, pH and pressure) as well as the chemical properties of drug molecules (solutes). The complexity increases due to the lack of techniques to probe intermolecular forces and the bio-chemical mechanisms of drug–membrane interactions. While a thorough investigation of these bio-chemical and bio-physical factors is desirable for drug development and design, our technique nevertheless provides an alternative for screening drug molecules for various lipid compositions. Our experiments also validate the label-free and quasi high-throughput micro-fluidic technique to determine the permeability co-efficient for norfloxacin across vesicles with a wide range of lipid compositions. The experiment proves that lipid composition significantly affects the passive transport of norfloxacin across the membrane.The experiments were performed at pH 5 and 7. At pH 5 norfloxacin is positively charged and hence has a very low affinity for the hydrophobic and non-polar core of the lipid membrane. Consequently the permeability of norfloxacin at pH 5 across lipid vesicles is very low and not measureable in the time scales observed here. We will therefore examine the structural differences between lipids that contribute to the variations in permeability of vesicles for norfloxacin at pH 7. The results of the permeability measurements with the standard errors from independent experiments (see ESI† for additional data sets) are combined in Fig. 6. To emphasize the strong dependence of permeability of norfloxacin on the lipid composition, we also presented the permeability of norfloxacin into DPhPC lipid vesicles obtained from our previous measurement.45
The discussion is divided into four parts: (1) the large disparity between the permeability of DOPC and the previously used DPhPC lipid vesicles for norfloxacin. (2) The effect of the presence of low amounts of the non-lamellar lipid DOPE in DOPC vesicles. This is found to reduce the permeability of the membrane to norfloxacin. (3) A comparison of assays containing small amounts of DOPG, an anionic lipid at pH 7 in DOPC and low amounts of the non-lamellar lipid DOPE in DOPC and finally (4) higher permeability at high DOPE concentrations.
For the first part, our investigation shows that DOPC is more permeable to norfloxacin in comparison to the previously measured value for DPhPC (see Fig. 6). The difference in the permeability of the two lipids is more than one order of magnitude which is surprisingly high for PC membranes with the same head groups. If we consider the structural differences between DOPC and DPhPC, both contain the same head group but differ in their hydrophobic chain structure (Fig. 1). DPhPC contains 16 carbon chains with branched methyl groups at each node in contrast to an 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 carbon chain with a cis-double bond at the 9th carbon position in the case of DOPC.
1 carbon chain with a cis-double bond at the 9th carbon position in the case of DOPC.
We are able to rule out photo-oxidation of the double bonds in DOPC as a possible cause since the vesicles were intact at pH 5 for more than 60 s. Molecular dynamics simulations58,59 and X-ray diffraction experiments indicate that the branched hydrocarbon chain in the case of DPhPC does not alter the partition coefficient significantly in comparison to their straight-chain counterparts.58 Furthermore, the thickness of the bilayer of DPhPC is similar to that of DOPC (∼36 Å).60 These factors evidently indicate that the most widely used solubility diffusion model which relates the permeability to the thickness of the membrane cannot be used to explain the process.
A possible explanation could be drawn based on an improved three-slab model which incorporates area per lipid molecule to describe the permeability of small molecules across lipid membranes.61 The branched chain region of DPhPC has bends at the tetra carbon positions leading to interposed chains. This reduces the trans–gauche isomerisation and provides structural stability. Furthermore, molecular dynamics simulations showed that there were fewer cavities present in the hydrocarbon region which restricts the mobility of small neutral molecules across the bilayer.58,59 The area per lipid molecule for DPhPC is much larger (80 Å2)60 than DOPC (72 Å2)60 considering that DPhPC has only 16 carbon chains. The three-slab model predicts that the resistance to the mobility of the solute predominantly comes from the interfacial region. Since the effective free volume (owing to its large area) is much smaller for DPhPC, the permeability values for small neutral molecules such as norfloxacin are smaller compared to its straight chain counterparts. The experiments and the simulation conclude that the differences in the permeability of DPhPC and its straight chain counterparts are predominantly due to the restricted diffusion arising from the presence of methyl groups in the hydrocarbon chain region. The methyl groups therefore increase the diffusion time for molecules across DPhPC bilayers in comparison to the DOPC bilayers where there are no branched structures in the chain region. Our experiments are in line with the simulation58,59 and X-ray diffraction61 results describing the permeability of small molecules. We observe that the diffusion of norfloxacin across DPhPC is much slower than across DOPC bilayers.
Next, we inspect the permeability of vesicles containing small amounts of DOPE (30%). DOPE has the same chain configuration as DOPC but has a much smaller headgroup. The characteristic features of DOPE are its ability to form non-lamellar structures and the propensity of forming hydrogen bonds between the amine group present in the headgroup and water resulting in a small headgroup.62 This acts like a glue and keeps the lipid molecules closely packed near the headgroup region. When a small amount of DOPE is added, the diffusion is therefore reduced since norfloxacin cannot access the hydrophobic region as easily as it can in the case of DOPC. This could explain the small permeability coefficient in the case of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 mixtures of DOPE
70 mixtures of DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC vesicles. This also indicates that not only the type of lipid but also the amount of lipids present in a membrane influences the permeability of norfloxacin or small molecules across membranes. This re-affirms the complex non-linear behaviour of drug molecules with lipid membranes.
DOPC vesicles. This also indicates that not only the type of lipid but also the amount of lipids present in a membrane influences the permeability of norfloxacin or small molecules across membranes. This re-affirms the complex non-linear behaviour of drug molecules with lipid membranes.
When we compare the mixtures of DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC (3
DOPC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) and DOPG
7) and DOPG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPC (3
DOPC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) vesicles, the former containing a non-lamellar lipid and the latter containing an anionic lipid, the effect of both kinds of lipids on the permeability of bilayers to norfloxacin seems to be very similar. In the case of the anionic vesicle (DOPC
7) vesicles, the former containing a non-lamellar lipid and the latter containing an anionic lipid, the effect of both kinds of lipids on the permeability of bilayers to norfloxacin seems to be very similar. In the case of the anionic vesicle (DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPG), the electrostatic interaction due to the presence of anionic headgroups restricts the passage of neutral norfloxacin across the bilayer. Previous studies have shown that PG lipids have high propensity of forming interlipid hydrogen bonds. Molecular dynamics simulations show that about 30% of PG headgroups are involved in intramolecular hydrogen bonding where the hydroxyl headgroup donates a hydrogen to a phosphate (and to some extent the glycero-ester) oxygen atom. Another 45% of PG molecules are involved in hydrogen bonding with the PC lipid. The overall effect of this is increasing order or having a more regular arrangement of lipids thereby making it less permeable to norfloxacin.63
DOPG), the electrostatic interaction due to the presence of anionic headgroups restricts the passage of neutral norfloxacin across the bilayer. Previous studies have shown that PG lipids have high propensity of forming interlipid hydrogen bonds. Molecular dynamics simulations show that about 30% of PG headgroups are involved in intramolecular hydrogen bonding where the hydroxyl headgroup donates a hydrogen to a phosphate (and to some extent the glycero-ester) oxygen atom. Another 45% of PG molecules are involved in hydrogen bonding with the PC lipid. The overall effect of this is increasing order or having a more regular arrangement of lipids thereby making it less permeable to norfloxacin.63
Interestingly, the experiment indicates that at the same concentration of either type of lipid, the effect on norfloxacin permeability is not affected by the composition of the anionic or non-bilayer lipid. At much higher concentrations of DOPE (70%) however (Fig. 5 and 6), a higher permeability was observed (Fig. 6). At 70% DOPE, the lipid membrane is in a highly stressed state.64 Higher concentration of non-bilayer lipids increases the drive to form non-lamellar phases. Since the lipids are bound in a lamellar conformation, the membrane experiences high curvature elastic stress.65 The interaction of vesicles with another neutral molecule therefore paves the way for the molecule into the vesicle relatively quickly compared to vesicles with low amounts of DOPE.66 However, it is also likely that the vesicles may contain a range of DOPE concentrations. The spread in our data is broad indicating that the effect could be due to both curvature frustration and varied composition. While this remains a qualitative description, a thorough analysis of drug–membrane interactions will provide more insight into their behaviour.
To summarize, the passive permeability of norfloxacin across a bilayer depends extensively on the type of lipid and relative abundance in a non-trivial way. Our experiments also indicate that not only the head group but also the hydrophobic chain structure affects the permeation of drugs across membranes in a complex way. The presence of charged DOPG lipid could be one of the key factors to slow down the transport of norfloxacin across the bilayer. On the other hand, from a therapeutic viewpoint, these results indicate that for more efficient drug transport antibiotics should be tested on different lipid mixtures to obtain accurate results. In future, a comparison of the permeability of the individual lipid components presented here can be compared to the permeability values obtained for bilayers formed using membrane extracts (e.g. E. coli membrane extract) to understand the cumulative role of lipid composition in regulating the permeation of antibiotics through membranes.
Conclusion
We have shown that microfluidics is a powerful tool to measure the passive membrane permeability of drugs, in particular, norfloxacin across lipid membranes. The technique was successfully used to measure permeability coefficients for a statistically significant number of vesicles for a wide range of lipid compositions without the need to investigate the structural nuances of lipids and drugs. The modularity of the technique also demonstrates the potential to be further extended into multiplexed devices for a full lipodome analysis for a variety of drugs making the selection of drug–lipid pairs less time consuming. The information obtained from the method can aid in the development of more efficient encapsulation methodologies in liposomes and designing drugs based on their preference to interact with specific lipid molecules. The technique is a viable alternative to the established octanol–drug partition co-efficient technique which cannot account for the influence of lipid structure. It also has the advantage of being able to be developed into high throughput screening assay which may not be provided by other recent methods.67 Our results demonstrate that lipid composition as well as constitution greatly affects the permeation of molecules. In contrast to ciprofloxacin, which is another fluroquinolone drug, the diffusion of norfloxacin across membranes is slower in the presence of an anionic lipid emphasising the fact that drug–membrane interactions are complex and their understanding is crucial for the development of drugs. Our technique can be extended to understand the permeability and kinetics of uptake of a range of fluorescent small molecules67 or nanoparticles with excitation ranging from the visible to near ultra-violet region.Acknowledgements
SP and UFK acknowledge funding from an ERC starting grant, Passmembrane 261101 and an EPSRC grant GRAPHTED, EP/K016636/1, and JC acknowledges the support from an Internal Graduate Studentship, Trinity College, Cambridge and a Research Studentship from the Cambridge Philosophical Society. The authors would like to thank John Seddon, Department of Chemistry, Imperial College London, London, for critical reading of the manuscript.References
- M. L. Cohen, Changing patterns of infectious disease, Nature, 2000, 406, 762–767 CrossRef CAS PubMed.
- C. T. Walsh and T. A. Wencewicz, Prospects for new antibiotics: a molecule-centered perspective, J. Antibiot., 2014, 67, 7–22 CrossRef CAS PubMed.
- E. Breukink and B. de Kruijff, Lipid II as a target for antibiotics, Nat. Rev. Drug Discovery, 2006, 5, 321–332 CrossRef CAS PubMed.
- J.-M. Bolla, et al., Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria, FEBS Lett., 2011, 585, 1682–1690 CrossRef CAS PubMed.
- H. Nikaido, Molecular Basis of Bacterial Outer Membrane Permeability Revisited, Microbiol. Mol. Biol. Rev., 2003, 67, 593–656 CrossRef CAS PubMed.
- A. H. Delcour, Outer membrane permeability and antibiotic resistance, Biochim. Biophys. Acta, 2009, 1794, 808–816 CrossRef CAS PubMed.
- P. M. Romsicki, Y. Romsicki and F. J. Sharom, The Membrane Lipid Environment Modulates Drug Interactions with the P-Glycoprotein Multidrug Transporter, Biochemistry, 1999, 38, 6887–6896 CrossRef PubMed.
- D. C. Hooper, Emerging Mechanisms of Fluoroquinolone Resistance, Emerging Infect. Dis., 2001, 7, 337–341 CrossRef CAS PubMed.
- R. Schmieder and R. Edwards, Insights into antibiotic resistance through metagenomic approaches, Future Microbiol., 2012, 7, 73–89 CrossRef CAS PubMed.
- D. M. Engelman, Membranes are more mosaic than fluid, Nature, 2005, 438, 578–580 CrossRef CAS PubMed.
- D. Oursel, et al., Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization, Rapid Commun. Mass Spectrom., 2007, 21, 1721–1728 CrossRef CAS PubMed.
- C. Alexander and E. T. Rietschel, Bacterial lipopolysaccharides and innate immunity, J. Endotoxin Res., 2001, 7, 167–202 CAS.
- S. Urban and S. M. Moin, A subset of membrane-altering agents and γ-secretase modulators provoke nonsubstrate cleavage by rhomboid proteases, Cell Rep., 2014, 8, 1241–1247 CrossRef CAS PubMed.
- C. Bourgaux and P. Couvreur, Interactions of anticancer drugs with biomembranes: what can we learn from model membranes?, J. Controlled Release, 2014, 190, 127–138 CrossRef CAS PubMed.
- S. Isabel and G. Paula, Encapsulation of Fluoroquinolones in Phosphatidylcholine: Cholesterol Liposomes, J. Pharm. Drug. Delivery Res., 2013, 2–5 Search PubMed.
- T. M. Allen and P. R. Cullis, Liposomal drug delivery systems: from concept to clinical applications, Adv. Drug Delivery Rev., 2013, 65, 36–48 CrossRef CAS PubMed.
- Y. Kaneda, Virosomes: evolution of the liposome as a targeted drug delivery system, Adv. Drug Delivery Rev., 2000, 43, 197–205 CrossRef CAS PubMed.
- V. P. Torchilin, Recent advances with liposomes as pharmaceutical carriers, Nat. Rev. Drug Discovery, 2005, 4, 145–160 CrossRef CAS PubMed.
- K. Eyer, et al., A liposomal fluorescence assay to study permeation kinetics of drug-like weak bases across the lipid bilayer, J. Controlled Release, 2014, 173, 102–109 CrossRef CAS PubMed.
- M. Kansy, F. Senner and K. Gubernator, Physicochemical High Throughput Screening: Parallel Artificial Membrane Permeation Assay in the Description of Passive Absorption Processes, J. Med. Chem., 1998, 41, 1007–1010 CrossRef CAS PubMed.
- A. Cern, Y. Barenholz, A. Tropsha and A. Goldblum, Computer-aided design of liposomal drugs: in silico prediction and experimental validation of drug candidates for liposomal remote loading, J. Controlled Release, 2014, 173, 125–131 CrossRef CAS PubMed.
- H. Bensikaddour, et al., Interactions of ciprofloxacin with DPPC and DPPG: fluorescence anisotropy, ATR-FTIR and 31P NMR spectroscopies and conformational analysis, Biochim. Biophys. Acta, 2008, 1778, 2535–2543 CrossRef CAS PubMed.
- T. P. Norden and S. Engstrom, Cubic phases for studies of drug partition into lipid bilayers, Eur. J. Pharm. Sci., 1999, 8, 243–254 CrossRef.
- J. A. Custódio, L. M. Almeida and V. M. C. Madeira, A reliable and rapid procedure to estimate drug partitioning in biomembranes, Biochem. Biophys. Res. Commun., 1991, 176, 1079–1085 CrossRef.
- M. Manrique-Moreno, et al., The membrane-activity of Ibuprofen, Diclofenac and Naproxen: a physico-chemical study with lecithin phospholipids, Biochim. Biophys. Acta, 2009, 1788, 1296–1303 CrossRef PubMed.
- A. M. Seddon, et al., Drug interactions with lipid membranes, Chem. Soc. Rev., 2009, 38, 2509–2519 RSC.
- M. Deleu, J.-M. Crowet, M. N. Nasir and L. Lins, Complementary biophysical tools to investigate lipid specificity in the interaction between bioactive molecules and the plasma membrane: a review, Biochim. Biophys. Acta, 2014, 1838, 3171–3190 CrossRef CAS PubMed.
- C. Peetla, A. Stine and V. Labhasetwar, Biophysical Interactions with Model Lipid Membranes: Applications in Drug Discovery and Drug Delivery, Mol. Pharm., 2009, 6, 8053–8058 Search PubMed.
- J. L. Vazquez, M. T. Montero, J. Trias and J. Hernandez-Borrell, 6-Fluoroquinolone – liposome interactions: fluorescence quenching study using iodide, Int. J. Pharm., 1998, 171, 75–86 CrossRef CAS.
- A. Leo, C. Hansch and D. Elkins, Partition coefficients and their uses, Chem. Rev., 1971, 71, 525–616 CrossRef CAS.
- C. Y. Yang, S. J. Cai, H. Liu and C. Pidgeon, Immobilized artificial membranes – screens for drug membrane interactions, Adv. Drug Delivery Rev., 1996, 23, 229–256 CrossRef.
- F. Beigi, I. Gottschalk and C. L. Ha, Immobilized liposome and biomembrane partitioning chromatography of drugs for prediction of drug transport, Int. J. Pharm., 1998, 164, 129–137 CrossRef CAS.
- M. La Rotonda, et al., Relationships between Octanol–Water Partition Data, Chromatographic Indices and Their Dependence on pH in a Set of Nonsteroidal Anti-Inflammatory Drugs, Quant. Struct.-Act. Relat., 1983, 2, 168–173 CrossRef CAS.
- N. F. Ii and V. D. Montesano, Interactions of Nonsteroidal Antiinflammatory Drugs with Phospholipids: Comparison between Octanol/Buffer Partition Coefficients and Chromatographic Indexes on Immobilized Artificial Membranes, J. Pharma Res., 1997, 86, 225–229 Search PubMed.
- S. E. Debolt and P. A. Kollman, Investigation of Structure, Dynamics and Solvation in 1-Octanol and Its Water-Saturated Solution: Molecular Dynamics and Free-Energy Perturbation Studies, J. Am. Chem. Soc., 1995, 117, 5316–5340 CrossRef CAS.
- J. L. M. Hermens, J. H. M. de Bruijn and D. N. Brooke, The octanol–water partition coefficient: strengths and limitations, Environ. Toxicol. Chem., 2013, 32, 732–733 CrossRef CAS PubMed.
- C. Rivet, H. Lee, A. Hirsch, S. Hamilton and H. Lu, Microfluidics for medical diagnostics and biosensors, Chem. Eng. Sci., 2011, 66, 1490–1507 CrossRef CAS.
- C. D. Chin, et al., Microfluidics-based diagnostics of infectious diseases in the developing world, Nat. Med., 2011, 17, 1015–1019 CrossRef CAS PubMed.
- M. Hashimoto, et al., Rapid PCR in a continuous flow device, Lab Chip, 2004, 4, 638–645 RSC.
- N. Goedecke, et al., A high-performance multilane microdevice system designed for the DNA forensics laboratory, Electrophoresis, 2004, 25, 1678–1686 CrossRef CAS PubMed.
- C. Chou, et al., Sorting by diffusion: an asymmetric obstacle course for continuous molecular separation, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 31–34 Search PubMed.
- E. T. Lagally, C. A. Emrich and R. A. Mathies, Fully integrated PCR-capillary electrophoresis microsystem for DNA analysis, Lab Chip, 2001, 1, 102–107 RSC.
- J. H. Tsui, W. Lee, S. H. Pun, J. Kim and D.-H. Kim, Microfluidics-assisted in vitro drug screening and carrier production, Adv. Drug Delivery Rev., 2013, 65, 1575–1588 CrossRef CAS PubMed.
- I. U. Khan, C. A. Serra, N. Anton and T. Vandamme, Microfluidics: a focus on improved cancer targeted drug delivery systems, J. Controlled Release, 2013, 172, 1065–1074 CrossRef CAS PubMed.
- J. Cama, C. Chimerel, S. Pagliara, A. Javer and U. F. Keyser, A label-free microfluidic assay to quantitatively study antibiotic diffusion through lipid membranes, Lab Chip, 2014, 14, 2303–2308 RSC.
- K. E. N. B. Waites, K. A. Y. C. Canupp and M. J. Devivo, Efficacy and tolerance of Norfloxacin in treatment of complicated urinary tract infection in outpatients with neurogenic bladder secondary to spinal cord injury, Uropharamacology, 1991, XXXVIII, 589–596 Search PubMed.
- S. Sirinavin, et al., Norfloxacin and Azithromycin for Treatment of Nontyphoidal Salmonella Carriers, Clin. Infect. Dis., 2003, 35, 685–691 CrossRef PubMed.
- J. Fernández, et al., Norfloxacin vs. ceftriaxone in the prophylaxis of infections in patients with advanced cirrhosis and hemorrhage, Gastroenterology, 2006, 131, 1049–1056 CrossRef PubMed.
- H. H. Gadebusch and D. L. Shungu, Norfloxacin, the first of a new class of fluoroquinolone antimicrobials, revisited, Int. J. Antimicrob. Agents, 1991, 1, 3–28 CrossRef CAS PubMed.
- K. Drlica, X. Zhao, P. Health and N. York, DNA Gyrase, Topoisomerase IV and the 4-Quinolones, Microbiol. Mol. Biol. Rev., 1997, 61, 377–392 CAS.
- G. S. V. Muniz, L. R. Teixeira and S. R. W. Louro, Interaction of the antibiotic norfloxacin with ionic micelles: pH-dependent binding, Eur. Biophys. J., 2014, 43, 477–483 CrossRef CAS PubMed.
- M. Z. Islam, J. M. Alam, Y. Tamba, M. A. S. Karal and M. Yamazaki, The single GUV method for revealing the functions of antimicrobial, pore-forming toxin and cell-penetrating peptides or proteins, Phys. Chem. Chem. Phys., 2014, 16, 15752–15767 RSC.
- M. Björnmalm, Y. Yan and F. Caruso, Engineering and evaluating drug delivery particles in microfluidic devices, J. Controlled Release, 2014, 190, 139–149 CrossRef PubMed.
- H. Nikaido, Prevention of Drug Access to Bacterial Targets: Permeability Barriers and Active Efflux, Science, 1994, 264, 383–388 Search PubMed.
- J. Hernández-Borrell and M. T. Montero, Does ciprofloxacin interact with neutral bilayers? An aspect related to its antimicrobial activity, Int. J. Pharm., 2003, 252, 149–157 CrossRef.
- N. Stuurman, N. Amdodaj and R. D. Vale, MicroManager: open source software forlight microscope imaging, Microsc. Today, 2007, 15, 42–43 Search PubMed.
- D. Qin, Y. Xia and G. M. Whitesides, Soft lithography for micro- and nanoscale patterning, Nat. Protoc., 2010, 5, 491–502 CrossRef CAS PubMed.
- W. Shinoda, M. Mikami, T. Baba and M. Hato, Molecular Dynamics Study on the Effects of Chain Branching on the Physical Properties of Lipid Bilayers: 2. Permeability, J. Phys. Chem. B, 2004, 108, 9346–9356 CrossRef CAS.
- W. Shinoda, M. Mikami, T. Baba and M. Hato, Molecular Dynamics Study on the Effect of Chain Branching on the Physical Properties of Lipid Bilayers: Structural Stability, J. Phys. Chem. B, 2003, 107, 14030–14035 CrossRef CAS.
- S. Tristram-Nagle, et al., Structure and Permeability of Fully Hydrated DiphytanoylPC, Chem. Phys. Lipids, 2010, 163, 630–637 CrossRef CAS PubMed.
- J. C. Mathai, S. Tristram-Nagle, J. F. Nagle and M. L. Zeidel, Structural determinants of water permeability through the lipid membrane, J. Gen. Physiol., 2008, 131, 69–76 CrossRef CAS PubMed.
- H. I. Petrache, et al., Structure and Fluctuations of Charged Phosphatidylserine Bilayers in the Absence of Salt, Biophys. J., 2004, 86, 1574–1586 CrossRef CAS PubMed.
- L. Janosi and A. A. Gorfe, Simulating POPC and POPC/POPG Bilayers: Conserved Packing and Altered Surface Reactivity, J. Chem. Theory Comput., 2010, 6, 3267–3273 CrossRef CAS PubMed.
- R. M. Epand, Functional Roles of Non-Lamellar Forming Lipids, Chem. Phys. Lipids, 1996, 81, 101–104 CrossRef CAS.
- R. S. Cantor, Lipid Composition and the Lateral Pressure Profile in Bilayers, Biophys. J., 1999, 76, 2625–2639 CrossRef CAS PubMed.
- M. Orsi and J. W. Essex, Physical properties of mixed bilayers containing lamellar and nonlamellar lipids: insights from coarse-grain molecular dynamics simulations, Faraday Discuss., 2013, 161, 249 RSC.
- P. Kuhn, K. Eyer, S. Allner, D. Lombardi and P. S. Dittrich, A microfluidic vesicle screening platform: monitoring the lipid membrane permeability of tetracyclines, Anal. Chem., 2011, 83, 8877–8885 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sm02371h |
| This journal is © The Royal Society of Chemistry 2016 |