Open Access Article
Open Access ArticleThe discovery of 9/8-ribbons, β/γ-peptides with curved shapes governed by a combined configuration-conformation code†
Claire M.
Grison
a,
Sylvie
Robin
ab and
David J.
Aitken
*a
aCP3A Organic Synthesis Group, ICMMO-UMR 8182, Université Paris-Sud, Université Paris-Saclay, 15 Rue Georges Clemenceau, 91405 Orsay cedex, France. E-mail: david.aitken@u-psud.fr; Fax: +33-(0)169156278; Tel: +33-(0)169153238
bUniversité Paris Descartes, UFR Sciences Pharmaceutiques et Biologiques, 4 Avenue de l'Observatoire, 75270 Paris cedex 06, France
First published on 15th September 2015
Abstract
The de novo design of a β/γ-peptidic foldamer motif has led to the discovery of an unprecedented 9/8-ribbon featuring an uninterrupted alternating C9/C8 hydrogen-bonding network. The ribbons adopt partially curved topologies determined synchronistically by the β-residue configuration and the γ-residue conformation sets.
The emergence of peptide-based foldamers has had a major impact on the design of molecular architectures shaped by intramolecular non-covalent interactions, principally hydrogen bonds (H-bonds).1,2 The studies of different types of homo-, hetero- and hybrid oligopeptides containing custom-build β- or γ-amino acid building blocks have furnished a considerable collection of folded conformations. Most of these periodic structures are helical; indeed, helices have been prominent in non-peptide foldamers areas too.2,3 Other secondary structural patterns such as strands or turns have been established, but there are very few descriptions of peptide-based foldamer ribbons, which can be defined as flattened structures featuring a succession of regular, well-defined short-range H-bond patterns. Only one 8-ribbon β-peptide structure has been described,4 while two 9-ribbons have been discovered for γ-peptides.5 A bent γ-peptide 7-ribbon, featuring intra-residue H-bonds, was observed for a very short homooligomer of a highly constrained γ-amino acid.6 Very recently, a mixed 7/8-ribbon was described for a short α/β-hybrid peptide.7
The search for new types of foldamers remains a key objective in order to expand the array of organized secondary structures, particularly with unusual architectures. A bottom-up design of “foldable” oligomers should exploit predictable built-in conformational restraints which lead to local folding in a predictable manner.
The β/γ-hybrid peptide manifold provides an interesting case which lends itself to further exploration. Confirming theoretical predictions,8 the 11/13-helix9 and the 13-helix10 have been demonstrated as stable structures. The latter is of particular interest due to its analogy with Nature's α-helix and successful isosteric replacement of short α-peptide segments by β/γ-peptide motifs in α-helical peptides has been carried out.11 Conspicuously, however, no other folding manifolds based on shorter range interactions have been demonstrated nor anticipated for β/γ-peptides.
We speculated that it should be possible to rationally design a regular β/γ-peptide folding pattern based on short-range H-bonds. Applying a bottom-up design principle, we reasoned that the β-amino acid component should impart a dominant backbone constraint inducing strong (i − 1 → i + 1) 8-membered ring H-bonds (C8), while the γ-amino acid component should be essentially unconstrained in order to best adapt its backbone torsion angles to accommodate (i − 1 → i + 1) 9-membered ring (C9) folding pattern. Appropriate building blocks for such a study emerged as the highly constrained β-amino acid (1R,2R)-2-aminocyclobutane carboxylic acid (tACBC),12 which has been shown to promote strong C8 structural features in peptides,13,14 and γ-aminobutyric acid (GABA); although oligomers of GABA do not appear to adopt a regular folded structure and are adopted as flexible linkers in medicinal chemistry,15 a GABA residue is able to support a C9 secondary features in γ-peptides.16
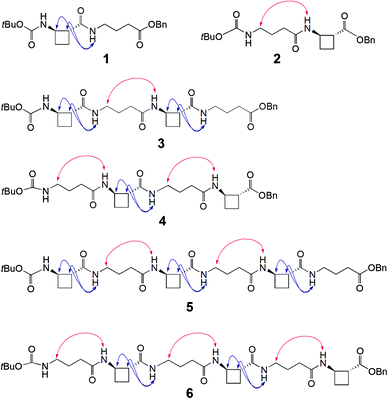
To test this hypothesis two series of peptides were proposed (Fig. 1): Boc-(tACBC-GABA)n-OBn (n = 1, 2, 3; peptides 1, 3, 5, respectively) and Boc-(GABA-tACBC)n-OBn (n = 1, 2, 3; peptides 2, 4, 6, respectively). These peptides were synthesised using standard solution-state methods (see ESI†) and their conformational analysis was conducted using solution-state spectroscopic techniques and molecular modelling.‡
1H NMR spectra of all peptides in CDCl3 were sufficiently well defined and signals were conveniently dispersed, allowing confirmation of the structures and unambiguous attribution of all signals pertinent for conformational analysis, using standard 1D and 2D NMR sequences. ROESY experiments were then performed and the observed correlations are illustrated in Fig. 1. Solution-state IR absorption spectra of all peptides were recorded in the same solvent and are illustrated in Fig. 2.§
As anticipated, in dipeptide 1 the tACBC-1 residue induced a strong 8-membered ring H-bonded (C8) feature, indicated by diagnostic ROESY interactions and corroborated by a low DMSO-d6 amide NH titration coefficient in 1H NMR studies in CDCl3 (see ESI† for details). The IR absorption spectrum of 1 showed a low frequency (H-bonded) amide NH band (3297 cm−1) in addition to a free carbamate NH absorption (3447 cm−1). In the ROESY analysis of tetrapeptide 3 two tACBC-induced C8 interactions were evident, gratifyingly accompanied by a correlation between H(γ) of GABA-2 and the NH of tACBC-3 which indicated a 9-membered ring H-bond (C9) and thus a C8/C9/C8 conformer. This was corroborated by low 1H NMR DMSO-d6 titration coefficients for the three amide NHs but not for the carbamate NH of tACBC-1. In the H-bonded region of the IR spectrum, the C8 absorption band at 3280 cm−1 now had a shoulder at 3350 cm−1, attributed to the NH of tACBC-3 implicated in the C9 interaction. ROESY analysis of hexapeptide 5 showed the appropriate sequence of correlations for an uninterrupted C8/C9/C8/C9/C8 network of H-bonded interactions while DMSO-d6 titrations confirmed that only the tACBC-1 carbamate NH was not H-bonded. The IR absorption spectrum clearly showed the C8 band at 3286 cm−1 with a pronounced C9 shoulder at 3340 cm−1.
In dipeptide 2, the tACBC-2 NH showed a ROESY correlation with H(γ) of GABA-1 and a lower 1H NMR DMSO-d6 titration coefficient than that of the GABA NH, while an H-bonded amide NH absorption (3312 cm−1) was observed in the IR spectrum. These data suggest a contribution from a C9 conformer implicating the GABA residue, which was again gratifying given its flexibility. Tetrapeptide 4 showed appropriate ROESY correlations to implicate a C9/C8/C9 conformer, in which only the N-terminal GABA-1 NH was entirely free on the basis of DMSO-d6 titration coefficients. In the IR spectrum, the broad absorption in the range 3370–3240 cm−1 comprises two C9 and one C8 H-bonded NH functions. ROESY analysis of hexapeptide 6 displayed an uninterrupted C9/C8/C9/C8/C9 H-bonded interaction series, substantiated by DMSO-d6 titrations and complemented by the broad NH absorption band in the range 3370–3240 cm−1 in the IR spectrum.
Molecular modelling of peptides 3–6 fully supported the strong propensity for the formation of a continuous network of alternating C9/C8 interactions in tACBC/GABA peptides and revealed some engrossing facets of the GABA residues' behaviour and the peptides' topologies. A hybrid Monte Carlo Multiple Minima (MCMM) molecular mechanics conformational search was carried out in chloroform medium using Macromodel and the MMFF force field without restraints. From 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 generated structures the lowest energy conformers (up to 10 kJ mol−1) were retained and sorted according to their conformer family type; in all cases the conformational landscape was dominated by C9/C8-conformer families. It was notable that contributions from helical conformers were non-existent; occasionally C13 features were detected but were part of conformers with significantly higher energies (see ESI†). The C9/C8-conformers of each peptide were subjected to ab initio geometrical optimization by DFT using GAUSSIAN 09 and the B3LYP/6-311G(d,p) basis set in a chloroform medium.
000 generated structures the lowest energy conformers (up to 10 kJ mol−1) were retained and sorted according to their conformer family type; in all cases the conformational landscape was dominated by C9/C8-conformer families. It was notable that contributions from helical conformers were non-existent; occasionally C13 features were detected but were part of conformers with significantly higher energies (see ESI†). The C9/C8-conformers of each peptide were subjected to ab initio geometrical optimization by DFT using GAUSSIAN 09 and the B3LYP/6-311G(d,p) basis set in a chloroform medium.
Peptide 3 gave a single low-energy conformer (Fig. 3). The rigid tACBC units displayed highly uniform (φ, θ, ψ) torsion angle values (88°, −101°, 32°) very close to those which characterize this residue in an optimized 8-helix foldamer.13a GABA-2 adopted a favourable g+, g+ local conformation for the (θ, ζ) torsion angles, facilitating formation of the (i − 1 → i + 1) 9-membered ring H-bond and concomitant orientation of its NH towards an (i − 2) carbonyl and of its C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O towards an (i + 2) amide NH. Thus a fully structured C8/C9/C8 conformer was in evidence, with C
O towards an (i + 2) amide NH. Thus a fully structured C8/C9/C8 conformer was in evidence, with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N distances in the range 1.87–1.91 Å. The structure does not have a helical topology, being flattened and resembling a ribbon. Although the ester C-terminal of GABA-4 was not involved in H-bonding, the (θ, ζ) torsion angles also adopted a g+, g+ local conformation.
O⋯H–N distances in the range 1.87–1.91 Å. The structure does not have a helical topology, being flattened and resembling a ribbon. Although the ester C-terminal of GABA-4 was not involved in H-bonding, the (θ, ζ) torsion angles also adopted a g+, g+ local conformation.
 | ||
| Fig. 3 Top-view of the low energy conformers of β/γ-peptides 3 (left) and 5 (right), showing the alternating C9/C8 H-bonding network. | ||
Peptide 5 also gave a single low-energy conformer (Fig. 3). The tACBC units behaved essentially as above, dictating local C8 structures. GABA-2 behaved as its eponym in peptide 3, whereas GABA-4 adopted a g−, g− local conformation for the (θ, ζ) torsion angles in order to accommodate its central C9 feature and both of the adjacent C8 structures. The uninterrupted C8/C9/C8/C9/C8 network, suggested by the spectroscopic studies, was clearly in evidence with all five C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N distances in the range 1.85–1.91 Å. The unsymmetrical 9/8-ribbon architecture neatly disposed the tACBC and GABA residues alternately on either side of the propagation axis.
O⋯H–N distances in the range 1.85–1.91 Å. The unsymmetrical 9/8-ribbon architecture neatly disposed the tACBC and GABA residues alternately on either side of the propagation axis.
The conformational analysis of peptides 4 and 6 was illuminating. Peptide 4 showed four C9/C8/C9 conformers (Fig. 4), differing by no more than 0.4 kJ mol−1 in energy. The tACBC-2 residue imposed a C8 local structure as before while each GABA residue adopted a conformer which allowed formation of a C9 feature: this was achieved when the (θ, ζ) torsion angles corresponded to g+, g+ (hereafter G+) or g−, g− (G−) local conformations, leading to all four possible combinations (G+G+, G+G−, G−G+, G−G−) on the low energy conformer landscape of 4. Each of the three H-bonds were near-planar in all four conformers, with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N distances in the range 1.83–1.93 Å.
O⋯H–N distances in the range 1.83–1.93 Å.
 | ||
| Fig. 4 Side-view of the four low energy conformers of β/γ-peptide 4 showing the C9/C8/C9 H-bonding networks. Atoms not relevant to H-bonding have been removed for clarity. | ||
Peptide 6 showed eight C9/C8/C9/C8/C9 conformers, separated by less than 0.6 kJ mol−1 in energy, a family within which all eight possible G+ and G− combinations for the three GABA residues were present (Fig. 5). Once again, near-planar H-bonds were present in all conformers, with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N distances in the narrow range 1.85–1.93 Å.
O⋯H–N distances in the narrow range 1.85–1.93 Å.
Inspection and comparison of the 9/8-ribbon conformer families of 4 and 6 revealed a fascinating feature: the ribbons showed a certain degree of curvature in the plane perpendicular to that of the propagation of the 9/8-ribbon axis. The extent of curvature for any given conformer could be characterized in terms of the relative orientations of consecutive H-bonded rings, qualified as “straight” (–) or “bent” (∩);¶ the latter relationship induces an incremental curvature of the conformer structure. This phenomenon does not correlate with any single GABA conformation (G+ or G−) as such, but instead with the combined local conformation types of the GABA pair on either side of the intervening tACBC residue in a given C9/C8/C9 segment. An unambiguous code exists between GABA-(i)/GABA-(i + 2) conformation pairs and the relative orientations of the pairs of H-bonded rings around which they are folded (Table 1). Thus, for example, in conformer 4C the (G+G+) GABA conformer pair translates to a “straight” GABA-1-tACBC-2 fragment followed by a “bent” tACBC-2-GABA-3 fragment. In conformer 6B, with a (G+G+G−) GABA conformer set, the first pair (G+G+) correlates to a “straight-bent” GABA-1-tACBC-2-GABA-3 segment while the second pair (G+G−) translates to a “straight-straight” GABA-3-tACBC-4-GABA-5 segment.
| Peptide conformer | GABA conformer set (G+ or G−) | Relative orientations of successive H-ring |
|---|---|---|
| 4A | + − | – – |
| 4B | − − | ∩ – |
| 4C | + + | – ∩ |
| 4D | − + | ∩ ∩ |
| 6A | + − − | – – ∩ – |
| 6B | + + − | – ∩ – – |
| 6C | + − + | – – ∩ ∩ |
| 6D | − − − | ∩ – ∩ – |
| 6E | ++ + | – ∩ – ∩ |
| 6F | − + − | ∩ ∩ – – |
| 6G | − + + | ∩ ∩ – ∩ |
| 6H | − − + | ∩ – ∩ ∩ |
Intriguingly, the mathematical nature of this code makes it impossible to generate a 9/8-ribbon conformer having three or more consecutive “straight” or “bent” fragments. This suggests that these 9/8-ribbons cannot lie flat in a plane nor adopt highly-rounded structures; by benefiting from the continuous H-bonding network, they are intrinsically destined to adopt partly curved architectures. Retrospectively, the code allows the assignment of the central C9/C8/C9 fragment of the low energy conformer of peptide 5: with a (G+G−) pair, a “straight-straight” topology is adopted.
In summary, these results illustrate the successful application of a bottom-up foldamer design leading to the unprecedented 9/8-ribbon. The unsymmetrical core structure disposes its β- and γ-residues on opposite sides of the ribbon propagation axis, which is partially curved in the perpendicular plane. The facile coexistence of several low energy 9/8-ribbon conformers is rendered possible due to the local flexibility of the GABA components, while the global topology portfolio remains within the limits of a coded curvature, which is governed by a combination of the stereochemical factors imparted by both the rigid tACBC components (configuration) and the GABA components (conformation).17 These observations underline the interest of ribbons in the foldamer field and further the understanding of the subtle factors which control molecular shape.
We are grateful to Mr J.-P. Baltaze (ICMMO) for help with NMR experiments. The award of a French MESR doctoral research scholarship (to C.M.G.) is acknowledged.
Notes and references
- (a) T. A. Martinek and F. Fülöp, Chem. Soc. Rev., 2012, 41, 687 RSC; (b) L. K. A. Pilsl and O. Reiser, Amino Acids, 2011, 41, 709 CrossRef CAS PubMed; (c) P. G. Vasudev, S. Chatterjee, N. Shamala and P. Balaram, Chem. Rev., 2011, 111, 657 CrossRef CAS PubMed; (d) F. Bouillère, F. S. Thétiot-Laurent, C. Kouklovsky and V. Alezra, Amino Acids, 2011, 41, 687 CrossRef PubMed; (e) D. Seebach and J. Gardiner, Acc. Chem. Res., 2008, 41, 1366 CrossRef CAS PubMed; (f) W. S. Horne and S. H. Gellman, Acc. Chem. Res., 2008, 41, 1399 CrossRef CAS PubMed; (g) D. Seebach, D. F. Hook and A. Glättli, Biopolymers, 2006, 84, 23 CrossRef CAS PubMed; (h) R. P. Cheng, S. H. Gellman and W. F. DeGrado, Chem. Rev., 2001, 101, 3219 CrossRef CAS PubMed.
- (a) Foldamers: Structure, Properties, and Applications, ed. S. Hecht and I. Huc, Wiley-VCH Verlag GmbH & Co., Weinheim, Germany, 2007 Search PubMed; (b) G. Guichard and I. Huc, Chem. Commun., 2011, 47, 5933 RSC; (c) D. J. Hill, M. J. Mio, R. B. Prince, T. S. Hughes and J. S. Moore, Chem. Rev., 2001, 101, 3893 CrossRef CAS PubMed.
- (a) C. M. Goodman, S. Choi, S. Shandler and W. F. DeGrado, Nat. Chem. Biol., 2007, 3, 252 CrossRef CAS PubMed; (b) A. Roy, P. Prabhakaran, P. K. Baruah and G. J. Sanjayan, Chem. Commun., 2011, 47, 11593 RSC; (c) H. Juwarker, J.-m. Suk and K.-S. Jeong, Chem. Soc. Rev., 2009, 38, 3316 RSC; (d) J. S. Laursen, J. Engel-Andreasen and C. A. Olsen, Acc. Chem. Res., 2015, 48 DOI:10.1021/acs.accounts.5b00257.
- S. Abele, P. Seiler and D. Seebach, Helv. Chim. Acta, 1999, 82, 1559 CrossRef CAS.
- (a) P. G. Vasudev, K. Ananda, N. Shamala and P. Balaram, Angew. Chem., Int. Ed., 2005, 44, 4972 CrossRef CAS PubMed; (b) J. Farrera-Sinfreu, L. Zaccaro, D. Vidal, X. Salvatella, E. Giralt, M. Pons, F. Albericio and M. Royo, J. Am. Chem. Soc., 2004, 126, 6048 CrossRef CAS PubMed.
- A. Kothari, M. K. N. Qureshi, E. M. Beck and M. D. Smith, Chem. Commun., 2007, 2814 RSC.
- S. Chandrasekhar, K. V. M. Rao, M. Seenaiah, P. Naresh, A. S. Devi and B. Jagadeesh, Chem. – Asian J., 2014, 9, 457 CrossRef CAS PubMed.
- C. Baldauf, R. Günther and H.-J. Hofmann, J. Org. Chem., 2006, 71, 1200 CrossRef CAS PubMed.
- G. V. M. Sharma, V. B. Jadhav, K. V. S. Ramakrishna, P. Jayaprakash, K. Narsimulu, V. Subash and A. C. Kunwar, J. Am. Chem. Soc., 2006, 128, 14657 CrossRef CAS PubMed.
- (a) L. Guo, A. M. Almeida, W. Zhang, A. G. Reidenbach, S. H. Choi, I. A. Guzei and S. H. Gellman, J. Am. Chem. Soc., 2010, 132, 7868 CrossRef CAS PubMed; (b) P. G. Vasudev, K. Ananda, S. Chatterjee, S. Aravinda, N. Shamala and P. Balaram, J. Am. Chem. Soc., 2007, 129, 4039 CrossRef CAS PubMed.
- (a) R. R. Araghi, C. Jäckel, H. Cölfen, M. Salwiczek, A. Völkel, S. C. Wagner, S. Wieczorek, C. Baldauf and B. Koksch, ChemBioChem, 2010, 11, 335 CrossRef PubMed; (b) I. L. Karle, A. Pramanik, A. Bannerjee, S. Bhattacharjya and P. Balaram, J. Am. Chem. Soc., 1997, 119, 9087 CrossRef CAS.
- (a) V. Declerck and D. J. Aitken, Amino Acids, 2011, 41, 587 CrossRef CAS PubMed; (b) C. Fernandes, E. Pereira, S. Faure and D. J. Aitken, J. Org. Chem., 2009, 74, 3217 CrossRef CAS PubMed; (c) C. Fernandes, C. Gauzy, Y. Yang, O. Roy, E. Pereira, S. Faure and D. J. Aitken, Synthesis, 2007, 2222 CAS.
- (a) A. Altmayer-Henzien, V. Declerck, J. Farjon, D. Merlet, R. Guillot and D. J. Aitken, Angew. Chem., Int. Ed., 2015, 54, 10807 CrossRef CAS PubMed; (b) M. Alaudddin, E. Gloaguen, V. Brenner, B. Tardivel, M. Mons, A. Zehnacker-Rentien, V. Declerck and D. J. Aitken, Chem. – Eur. J., 2015, 21 DOI:10.1002/chem.201501794.
- (a) E. Gorrea, G. Pohl, P. Nolis, S. Celis, K. K. Burusco, V. Branchadell, A. Perczel and R. M. Ortuño, J. Org. Chem., 2012, 77, 9795 CrossRef CAS PubMed; (b) E. Torres, E. Gorrea, E. Da Silva, P. Nolis, V. Branchadell and R. M. Ortuño, Org. Lett., 2009, 11, 2301 CrossRef CAS PubMed.
- (a) W. Nomura, T. Koseki, N. Ohashi, T. Mizuguchi and H. Tamamura, Org. Biomol. Chem., 2015, 13, 8734 RSC; (b) S. R. Barkow, I. Levchenko, T. A. Baker and R. T. Sauer, Chem. Biol., 2009, 16, 605 CrossRef CAS PubMed; (c) D. Y. Maeda, S. S. Mahajan, W. M. Atkins and J. A. Zebala, Bioorg. Med. Chem. Lett., 2006, 16, 3780 CrossRef CAS PubMed.
- (a) G. V. M. Sharma, P. Jayaprakash, K. Narsimulu, A. R. Sankar, K. R. Reddy, K. R. Krishna and A. C. Kunwar, Angew. Chem., Int. Ed., 2006, 45, 2944 CrossRef CAS PubMed; (b) G. Dado and S. H. Gellman, J. Am. Chem. Soc., 1994, 116, 1054 CrossRef CAS.
- For an inspired configuration-based coded approach for the design of helical peptide foldamers, see: I. M. Mándity, E. Wéber, T. A. Martinek, G. Olajos, G. K. Tóth, E. Vass and F. Fülöp, Angew. Chem., Int. Ed., 2009, 48, 2171 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthetic procedures, IR and NMR analyses, molecular modelling data. See DOI: 10.1039/c5cc07136d |
| ‡ Amino acid residues are numbered in this paper using the conventional manner for peptides. All peptides were adequately soluble in chloroform but very poorly soluble in more polar solvents and not at all in water. |
| § No change in the IR spectral profiles were observed over the range 1–10 mM, suggesting the absence of intermolecular interactions. |
| ¶ For peptides 4 and 6, these qualitative appreciations correspond to dihedral angles between successive O⋯H–N planes in the ranges 164° to 178° (–) and 122° to 136° (∩) for C9 → C8, and in the ranges 160° to 168° (–) and −136° to −144° (∩) for C8 → C9. See ESI† for more details. |
| This journal is © The Royal Society of Chemistry 2015 |