Open Access Article
Open Access ArticleHepatocyte-like cells and liver organoids: the application of iPSCs and their derivants for treating liver diseases
Ruobing
Ju†
a,
Siyuan
Tian†
a,
Yulong
Shang
a,
Shuoyi
Ma
a,
Miao
Zhang
a,
Jingyi
Liu
b,
Keshuai
Sun
c,
Lina
Cui
*a,
Xia
Zhou
*a and
Ying
Han
 *a
*a
aState Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, Xijing Hospital of Digestive Diseases, Air Force Military Medical University, Xi’an, 710032, Shaanxi, China. E-mail: cuilina-2007@163.com; zhouxialyg@163.com; hanying1@fmmu.edu.cn; Fax: +86 29 82539041; Tel: +86 29 84771506
bDepartment of Radiation Oncology, Xijing Hospital, Air Force Military Medical University, Xi’an, 710032, Shaanxi, China
cDepartment of Gastroenterology, The Air Force Hospital From Eastern Theater of PLA, Nanjing, 210002, Jiangsu, China
First published on 10th September 2024
Abstract
Liver diseases have become a great burden to human health because of their high morbidity and mortality rates. Orthotopic liver transplantation, which has always been considered the primary treatment for end-stage liver disease, has limitations in clinical practice. The development of cell therapy, especially induced pluripotent stem cells (iPSCs), holds promise in treating liver diseases. It has been reported that hepatocyte-like cells and liver organoids derived from iPSCs can be applied to establish disease models, test drug hepatotoxicity or directly perform specific functions as grafts. In this article, we systematically reviewed two differentiated derivants of iPSCs and show the prospective application of differentiated products in order to provide an experimental and theoretical basis for clinical treatment.
1. Introduction
Over the past decades, liver diseases have become a major cause of high morbidity and mortality around the world. About 350 million people worldwide suffer from liver diseases currently. It is estimated that more than one-fifth of the Chinese population experiences liver diseases, such as hepatitis B virus infection, metabolic dysfunction-associated steatotic liver disease (MASLD), alcohol-related liver disease, drug-induced liver injury (DILI) and liver cancer.1 Orthotopic liver transplantation is the only curative treatment for end-stage liver diseases.2 However, the application of liver transplantation is limited because of donor shortage, immunological rejection, and the complexity of operation. Therefore, the emergence of cell therapy may weaken these restrictions.3 Several preclinical and clinical studies have demonstrated that cell therapy will be a promising treatment for liver diseases.Cell therapy can provide cells with regenerative capacity to restore hepatic functions. Cell sources include autologous primary hepatocytes, xenogeneic hepatocytes or different types of stem cells. In fact, primary hepatocytes are not only difficult to isolate and expand in vitro but are also restricted by the need for cryogenic storage and long-term immunosuppressive therapy.4 Meanwhile, xenografts pose ethical and immune rejection issues, which are difficult to solve. In terms of stem cells, induced pluripotent stem cells (iPSCs) have the potential for multidirectional differentiation and low immunogenicity, thus becoming a type of relatively innovative stem cell source with high application prospect.5
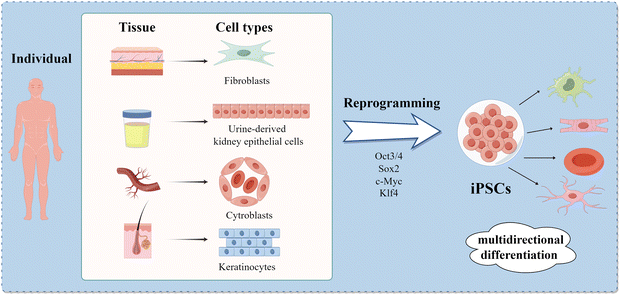
IPSCs are a type of pluripotent stem cells with the potential of multiple differentiation and possess infinite self-renewal capacity. In 2006, Takahashi first cultured iPSCs by introducing four specific transcription factors (Oct3/4, Sox2, c-Myc, and Klf4) into mouse embryonic cells through a retroviral vector.6 In 2007, iPSCs were first created by reprogramming human somatic cells.7,8 This technology won the 2012 Nobel Prize in Physiology or Medicine, and then, a wide array of association studies on iPSCs have been performed worldwide. Almost all cells of somatic tissues can be programmed into iPSCs, including skin tissues, hair, blood and even urine.9–12 As iPSCs are widely derived from individuals, the old issues that immune exclusion or ethical crisis can be improved based on this feature of iPSCs.13
It has been demonstrated that iPSCs can differentiate into neural cells,14,15 cardiac cells,16 vascular cells,17 functional hepatocytes18 and various other cellular types (Fig. 1).
The multidirectional differentiation ability of iPSCs enables their potential applications in neurodegenerative diseases, cardiac diseases, liver diseases and some inherited bleeding disorders.19–22 Besides, the emergence of new organoids has deepened research in the iPSCs field. Organoids are three-dimensional (3D) structures that highly replicate cell–cell and cell-extracellular matrix interactions and simulate the pathophysiology and disease process in vitro. Organoids derived from iPSCs and adult stem cells are studied for disease modeling, drug screening and toxicity testing.23
In recent years, iPSCs have shown differentiation ability in terms of liver diseases. The derivants, namely hepatocyte-like cells (HLCs) and liver organoids (LOs), are prospective tools for research on hepatitis B, MASLD, metabolic dysfunction-associated steatohepatitis (MASH) and liver tumors. In this review, we have summarized the methods used for the generation of HLCs and LOs from iPSCs. We also highlight their applications in treating various liver diseases, aiming to provide a basis for clinical treatment.
2. Generation of HLCs derived from iPSCs
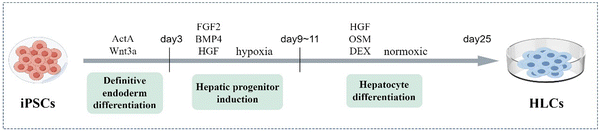
The liver has numerous cellular types, of which hepatocytes are the most dominant cell type, accounting for more than 80% of cells and covering most of the hepatic functions. HLCs are generated from stem cells to function like fetal hepatocytes. Although HLCs at low maturity currently, they are conducive to long-term culture and research due to their ability to stably maintain the differentiation degree quite stably and expand indefinitely.24In 2009, Song et al. and Si-Tayeb's team demonstrated that human iPSCs can be directly induced into HLCs by stepwise differentiation.18,25 The initial scheme consisted of four stages: endoderm induction, hepatic specification, hepatoblastic expansion and hepatic maturation. The differentiation protocol stimulated some relevant pathways that imitate key stages in liver development by the addition of different cytokines or growth factors, including activin A, fibroblast growth factor 2 (FGF2), bone morphogenetic protein 4 (BMP4), hepatocyte growth factor (HGF), oncostatin M (OSM), and glucocorticoid dexamethasone (DEX).25 Identifying ways to induce HLCs with the primary hepatocyte phenotype and function is key to the culture process, as well as research progress. The common hepatic differentiation protocol of iPSCs lasts for about twenty five days and can be divided into three stages: definitive endoderm differentiation, hepatic progenitor induction, and hepatocyte differentiation26,27(Fig. 2). Firstly, the definitive endodermal formation is induced by activin A, which can regulate the expression of the iPSC-specific gene NANOG, thereby contributing to the pluripotency maintenance of iPSCs.28 Sometimes, Wnt3a is added at this stage to promote the differentiation of iPSCs into endodermal cells. After about 3 days, they are exposed to FGF2 and BMP4 under hypoxic conditions to determine the differentiation direction, and HGF is added to encourage hepatic progenitor cell proliferation.29 Then, under normoxic conditions, HLC maturation is promoted in the hepatocyte culture medium with OSM and DEX.27 This approach produces more mature HLCs from human iPSCs efficiently, uniformly, and repeatably.
To confirm the effectiveness of differentiation in each stage, it is crucial to measure the expression levels of some specific hepatic markers. Similar to primary human hepatocytes, iPSC-induced HLCs successfully express several key markers, such as alpha-1 antitrypsin (AAT), hepatocyte nuclear factor 4 alpha, and alpha fetoprotein.30,31 However, HLCs stay in a relatively immature state and retain some characteristics of fetal hepatocytes.26 There are some defects in the functional characteristics of HLCs. A notable issue is that the activity of cytochrome P450 (CYP450) isozyme in HLCs is significantly lower than that in primary human hepatocytes.32 CYP450 takes part in the process of drug metabolism in the liver, and hence, its absence is a serious disadvantage while culturing hepatocytes in vitro. Additionally, Laemmle et al. found the lack of aquaporin 9 expression in HLCs. Aquaporin 9 is a membrane channel protein that mediates the urea cycle. Its deficiency is the reason for less urea secretion by HLCs than primary human hepatocytes.33 Alternatively, low levels of α-fetoprotein secretion and alcohol dehydrogenase inactivity are also characteristics of low maturity of HLCs. Meanwhile, the expression of CYP3A and CYP2D6 are also significant markers.34
Although the production protocol of iPSCs-HLCs needs improvements to enhance maturity, much progress has been made toward their clinical application. Human iPSCs provide a powerful platform for disease modeling, personalized medicine and cell-based therapy. Recently, various gene engineering tools, especially clustered regularly interspaced short palindromic repeats (CRISPR), have been widely applied.35 Further, considerable progress has been made in rectifying the genetic shortcomings. The CRISPR/Cas9 system combined with a guide RNA can target a specific genetic site and effectively repair genetic defects.36 Taking advantage of this technology, patient-derived iPSCs can be differentiated into particular cell types, such as modified HLCs, and then directly infused into patients, or specific mutations can be introduced to establish disease models.37 However, great challenges exist in the clinical translation of these processes. In the future, more clinical trials must be conducted to drive iPSC-HLCs development.
3. Generation of liver organoids (LOs) from iPSCs
Organoid is a 3D structure derived from (pluripotent) stem cells; the progenitor and differentiated cells self-organize through cell–cell and cell-matrix interactions to recapitulate aspects of the native tissue architecture and function in vitro.38 As 3D cultivation provides high cell–cell and cell-matrix interactions that involve the dynamic regulation of signaling pathways and paracrine signals, the structure of LOs is more similar to the hepatic physiological microenvironment state.39,40 Thus, the 3D culture can mimic the complex liver architecture and function better than single two-dimensional (2D) monocultures, which makes LOs promising tools for preclinical applications and regenerative medicine (Table 1). In the past, organoids have been derived from two types of stem cells: pluripotent stem cells (either embryonic stem cells or iPSCs) and organ-specific adult stem cells.23 At present, the cell sources of organoids include pluripotent stem cells, progenitor cells, primary cells, and tumor specimens from patients.41 Among these, iPSC-HLCs are ideal cell sources for LOs due to their features, such as patient specificity and unlimited supply.42 They can be used to study the occurrence, development and treatment of liver diseases, especially in the establishment of disease models and drug models.| Parameters | 2D cultivation methods | 3D cultivation methods |
|---|---|---|
| Abbreviations: 2D, two-dimensional; 3D, three-dimensional. | ||
| Derivation of cells | • Cell lines from a single cell | • Pluripotent stem cells |
| • Multiple cell types | ||
| Components | • Only cell lines | • Cells |
| • Medium with growth factors | ||
| • Matrix or scaffold material | ||
| Model effect | • Lack the capability of simulating the architectural features of organoids | • Replicate high cell–cell and cell-extracellular matrix interactions |
| • Low maturity | • Involve the dynamic regulation of signaling pathways and paracrine signals | |
| • Have significant inherited, epigenetic, and functional changes | • Have organ structure and genetic stability | |
| Preparation cost | • Low | • High |
| Expansion | • Easy but not precise | • Long-term and stable |
| Application characteristics | • Widespread but high failure rate | • In the experimental phase |
| • Bright prospect | ||
The first organoid was that of intestinal organs generated by researchers using mouse intestinal stem cells.43 In the following years, the protocol for culturing organoids was successfully applied to other organs, including the stomach, liver, prostate, pancreas and lungs.44 LOs were first reported in 2013. Huch's team used Wnt signaling to drive the long-term expansion of LOs that retained many features of the original epithelial structure.45 In the same year, Takebe et al. obtained vascularized and functional LOs from iPSCs for the first time, which could connect with the host blood vessels within 48 hours after transplantation.46 There have been important research breakthroughs towards generating LOs based on iPSCs technology in the past decade, which have been beneficial to clinical practice. Herein, we describe the approach to LO development from iPSCs and show how they can be applied to study liver diseases.
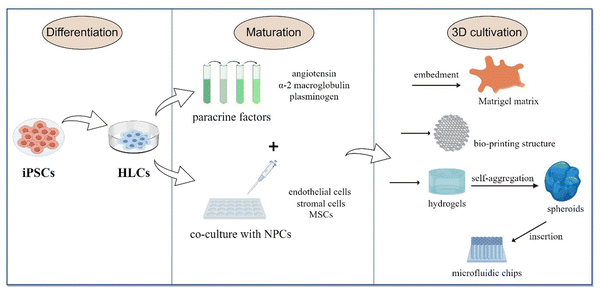
The culture process of iPSCs to LOs is simple. The cells are cultured in a medium with added growth factors; they first proliferate and differentiate to a specific mature state at the 2D level. Then, they are collected and embedded in a matrix or scaffold material to form a 3D structure and maintain continuous growth (Fig. 3). LOs can be derived either from the culture of a single cell type or a co-culture system containing multiple cell types. A single-type cell culture ensures the proliferation and self-organization of homogenous cells, while a co-culture of multiple cell types can better mimic the liver organ structure.47
Currently, LO cultures mainly adopt a gradually induced differentiation scheme. After being differentiated into HLCs, the organoids continue to mature in the medium supplemented with paracrine factors, such as angiotensin, α-2 macroglobulin and plasminogen, which jointly simulate the signaling environment for maturation to transform single cells into organs.48 Then they are embedded in the Matrigel matrix, which is a gelatinous protein mixture derived from the extracellular matrix (ECM) of Engelbreth-Holm-Swarm mouse sarcoma and is mainly composed of laminin, collagen IV and entactin.49 This matrix is extremely bioactive and supports the structure and proliferation of different organoid types. Currently, most of the organoid generation field relies on Matrigel to provide structural support and enable them to achieve 3D suspension growth.50 Hence, the generation of human LOs from iPSCs is realized by adding different growth factors or chemicals at the corresponding stages and with the structural and functional support provided by the Matrigel matrix.
3D systems involving multiple cell types can produce more functional in-vivo-like liver models than traditional single-cell types. In the liver, non-parenchymal cells are also an important part of maintaining function. The major trend in organoid development is co-culturing iPSC-HLCs with endothelial cells, stromal cells and mesenchymal stem cells (MSCs) in 3D cultures using various new biomaterials. The available 3D culture models include bio-printed structures, microfluidic chips, and hydrogels.51–53 Hydrogels mimic ECM properties and are thus ideal scaffolds for in-vitro 3D liver models. They can produce organoids with the same efficiency as Matrigel without the possible immunogenic disadvantages.54 For example, Schoonjans and team utilized a polyethylene glycol hydrogel, which was complemented with the key ECM proteins of the native liver, such as laminin-111, collagen IV and fibronectin. This way, they produced LOs with improved hepatic maturation and preserved differentiation capacity.55 Another group used hydrogels with iPSC-HLCs to form self-aggregated spheroids, which were then inserted on microfluidic chips and co-cultured with human umbilical vein endothelial cells and adipose tissue-derived MSCs.56 They provided terminal proof that iPSC-HLCs can successfully be cultured in 3D as spheroids and on microfluidic chips, and co-culturing iPSC-HLCs with non-parenchymal cells enhances their functionality. Besides, some non-parenchymal cells may provide the capability to differentiate into supportive lineages, such as pro-fibrotic cells, hepatic stellate cells, and liver resident macrophages, Kupffer cells.57 Using iPSC-HLCs composed of these cells to derive multi-cellular human liver organoids that can provide an overview of steatosis, inflammation, fibrosis and other liver disease phenotypes more completely.
LOs derived from iPSCs show regenerative properties similar to those of the human liver.58 The in-vitro expansion of iPSCs is almost efficient and unlimited. Therefore, LOs can achieve long-term expansion in vitro, maintain genetic stability, and provide the possibility of large-scale production to form biobanks. Meanwhile, they can eliminate the limitation of dependence on primary tissues. Once iPSC cell lines are established, these cells can be repeatedly grown indefinitely to produce HLCs and organoids.59 The 3D structure can significantly improve LO maturation and make them closely resemble an adult liver. For example, their functions in albumin secretion, glycogen storage, drug metabolism and lipid transport have been well expressed.60 LOs derived from iPSCs have been applied to study oncogenic mutations, based on which host–pathogen relationships can be studied in vitro. There have been some studies on the application of liver organoids to discern the mechanism of host-HBV/HCV interactions that lead to primary liver cancer.39,61 Altogether, LOs offer a promising tool for various liver-related studies. Therefore, LO-based disease modeling and drug development will be their important biomedical applications in the field of liver diseases.
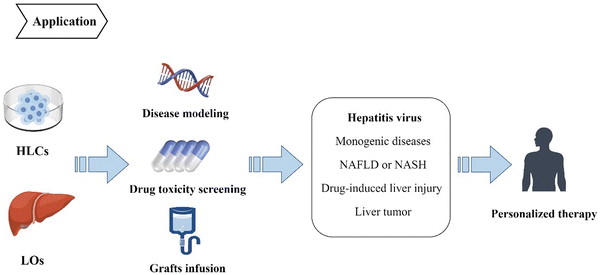
4. Application of iPSC-derived HLCs and LOs
HLCs and LOs derived from iPSCs have been widely used in preclinical studies of multiple liver diseases, including viral hepatitis, inherited monogenic diseases, non-alcoholic fatty liver disease, drug-induced liver injury, and liver tumors (Fig. 4). It is a cutting-edge method to simulate disease pathophysiology in vitro, and the CRISPR/Cas9 gene editing technology can be used to introduce or replicate specific mutations in the iPSCs cell lines.62 However, on the other hand, gene-editing modeling has obvious limitations the mutation sites related to the disease must be known. Therefore, other techniques are utilized for some diseases with unknown pathogenesis to build disease models, such as simulating the disease environment to induce morbidity. Successful in-vitro disease simulation and the establishment of disease models support the possibility of screening therapeutic targets and drugs. Disease models can also be used for experimentation and analysis of human intervention factors to provide a good theoretical basis for clinical strategies, which is of great significance to clinical treatment.63,64 Besides, HLCs and liver organoids can be used directly as grafts, especially organoids. They can also be transplanted to implant bio-artificial liver devices or can form functional units of implantable devices to provide temporary liver function, thereby serving as compensations to delay disease progression or enable repair.65,664.1. Hepatitis virus
Hepatitis virus infection is not only a common cause of most liver diseases but also the main factor that induces primary liver cancer. HBV and HCV are the most common pathogens, with about 170 million HBV infections and 350 million HCV infections reported worldwide.67 In the past, the mechanism of interaction between the virus and host was not clear due to the lack of reproducible and amplifiable in-vitro modeling systems that could precisely recapitulate the virus life cycle. With the development of iPSCs, Schwartz et al. found that when infected by HCV, iPSC-HLCs can replicate the whole infection cycle of HCV to establish models in infectious liver diseases.68 Sakurai et al. demonstrated that during the differentiation of iPSCs into HLCs, the expression levels of several genes involved in HBV infection were elevated.69 The sodium taurocholate co-transporting polypeptide (NTCP) is a functional receptor for HBV, and the overexpression of NTCP can improve the efficiency of HBV infection. Li et al. reported the enhanced expression of NTCP and proved that the medium containing iPSC-HLCs made the cells highly susceptible to HBV infection.70 As a result, iPSC-HLCs can hopefully become a platform for studying host inflammatory responses to pathogens and eventually provide opportunities to find new drug targets.Moreover, LOs with components derived from human iPSCs can provide new guidelines for developing models of hepatitis virus in vitro, owing to their prominent susceptibility and capability to precisely recapitulate the virus life cycle.71 Elisa De Crignis et al. infected liver organoids with HBV and found that the model successfully expressed HBV RNA and proteins and produced HBV with infectious activity.61 This organoid platform can be used to screen drugs that inhibit HBV replication and test drug toxicity. In another paper, the researchers established a functional organoid by culturing iPSC-derived endodermal, mesenchymal, and endothelial cells in a three-dimensional microwell culture system. This liver organoid was susceptible to HBV and could sustain HBV replication for a prolonged duration, reproducing the viral life cycle and virus-induced liver dysfunction.72 The above results indicate that iPSC-derived organoids can provide in-vitro infection systems for research on viral hepatitis.
4.2. Monogenic diseases
The use of iPSC-HLCs or organoids as novel models of several monogenic diseases is attracting increasing attention. The most common approach is to introduce known mutations through CRISPR/Cas9 technology to induce differentiation into the desired phenotype.Alagille syndrome (ALGS) is a multi-organ disorder caused by mutations in the Notch ligand JAG1 gene, which manifests as chronic cholestasis.73 Yuan et al. exploited a method for differentiating human iPSCs into LOs. During organoid development, the JAG1 mutation in ALGS was simulated, and the expression of JAG1 mRNA was regulated in the in-vitro model. The features of ALGS liver disease were recapitulated in the ALGS organoids.74,75 In another work, LOs were successfully produced to reproduce the cystic fibrosis process, and as a result, they mimicked disease phenotypes, such as impaired activity of the transmembrane conduction regulator channel.76,77 Moreover, the R778L mutation was introduced into iPSCs or embryonic stem cells from patients with Wilson's disease to acquire modified HLCs. The results showed that compared with the wild group, mutational HLCs had more cell death and higher sensitivity to copper ions, closer to the phenotype of Wilson's disease.78 In 2022, AAT deficiency patient-specific iPSCs were engineered, aiming to recapitulate cellular phenotypes that result from the expression of mutant ZAAT alleles and thereby determine the impact of ZAAT alleles on hepatocyte biology.79
As evidenced by the above studies, iPSCs are in-vitro research tools for multiple monogenic liver diseases. The successful replication of the phenotypes of monogenic diseases provides the possibility of establishing disease models in vitro. We can try to screen targets and therapeutic drugs based on these models, which is of great significance to clinical treatment.
4.3. MASLD or MASH
MASLD is the second most common cause of liver transplantation, next to liver tumors. The incidence and severity of MASLD are increasing, and it poses a risk of progression to end-stage liver diseases, such as cirrhosis and hepatocellular carcinoma.80 Additionally, it has been demonstrated that MASLD is associated with type 2 diabetes and can increase cardiovascular risk.81 In spite of this, its pathophysiological mechanism is not clear. Research has found that the I148M variant of patatin-like phospholipase domain-containing protein 3 (PNPLA3) is associated with MASLD.82 Tilson and team used CRISPR/Cas9 to modify iPSCs by knocking out the PNPLA3 gene. Then, the iPSCs were differentiated into HLCs and treated with either oleic acid or palmitic acid to induce MASLD-like phenotypes. It was found that PNPLA3 loss induced lipid accumulation, predisposition to steatosis, and increased sensitivity to ethanol- or methotrexate-induced hepatotoxicity.30 In another study, scientists combined stem cell biology with bioengineering principles; they used free fatty acid to induce MASLD pathological changes in iPSC organoids. It showed increased lipid accumulation and the expression of markers related to lipid metabolism. In addition, the expression of inflammatory and fibrotic markers was also increased.83 MASH is a severe form of MASLD. The organoids generated from MASH patient livers functionally manifested decreased albumin secretion, increased lipid storage and CYP450 metabolism. The transcriptome analysis of these liver organoids showed significantly increased expression of markers of liver fibrosis and tumor.84 This study opened up a new way for studying MASH. Elbadawy et al. created organoids of various stages of MASH in mice.85 The pathological effects of the model exhibited increased expression levels of liver fibrosis markers collagen I and α-SMA, demonstrating that these 3D cultures can reflect the degree of fibrosis in MASH. The above disease models summarize the key pathological features of MASLD or MASH and may provide tools for studying their underlying mechanisms, opening up a new path for their application in disease exploration and drug discovery.Intrasplenic transplantation of iPSC-HLCs has been shown to alleviate excessive accumulation of triglycerides and hepatitis symptoms, reduce the production of inflammatory cytokines and oxides, and reduce the production of apoptotic cells in MASLD mouse models.86 A high-throughput analysis based on iPSCs can be employed to screen potential therapeutic drugs. The technology of chemogenomic library and human iPSCs were used to screen 13,000 bioactive compounds and identify compounds that can significantly reduce the intracellular neutral lipid content, thus promoting the development of novel drugs for fatty liver disease.87 These results suggested that iPSCs may provide an alternative treatment and drug screening pathways for MASLD- and MASH-related end-stage liver diseases.
Organ crosstalk is a significant phenomenon that can affect the development and prognosis of MASLD. It has been reported that multiple tissues can influence MASLD development and correlate with MASLD severity, such as adipose tissues, pancreas, muscle, brain, and thyroid.88 Various cells or tissues can be co-cultured with iPSC-HLCs to provide different immune microenvironments and induce various phenotypes. Thus, in the future, organoid models will likely need to be combined with these cells and tissues to overcome multiple-organ crosstalk.
4.4. DILI
Drug hepatotoxicity is a major problem in all stages of clinical trials. It is the most common cause of post-marketing warnings and withdrawals. So far, establishing an in-vitro model that can predict drug hepatotoxicity, remains a significant challenge in drug development. With the development of stem cells, iPSC-derived hepatocytes or organoids are gradually being used as in-vitro tools for DILI detection. These technologies provide predictive models for novel drugs before they enter clinical trials and serve as potential in-vitro diagnostic tools.89IPSC-HLCs or organoids can be applied to predict DILI. They are also characterized by increased drug-related sensitivity. A study confirmed that iPSC-HLCs have increased time-dependent sensitivity to amiodarone, aflatoxin B1, and troglitazone.90 The results were also beneficial in predicting the degree of drug damage. Another study generated liver organoids from a 3D system on a chip, and they showed time-dependent and dose-dependent changes related to liver toxicity after exposure to acetaminophen.91 Researchers co-cultured HLCs with 47 known toxic drugs for 6 days and evaluated their toxicity by determining albumin, urea and ATP. The HLCs successfully detected 24 of 37 toxic drugs with a sensitivity of 65%, while 10 non-toxic drugs were detected with 100% accuracy.92 Shinozawa and team established a high-throughput toxicity screening model based on liver organoids, which tested 238 marketed drugs by measuring the transport activity of bile acids.93 Charles J et al. dispersed LOs derived from three separate iPSC lines in 384-well plates and exposed them to known hepatoxic drugs. The IC50 cytotoxicity values of the compounds were measured through this high-throughput screening format, showing the potential utility of iPSCs in DILI risk assessment.94 Wu et al. further used LOs derived from human iPSCs to model liver fibrogenesis induced by TGFβ or LPS treatment and established a high-throughput anti-fibrosis drug screening system, which demonstrated that SD208 and imatinib could significantly suppress fibrogenesis.95
In conclusion, prediction and screening platforms based on iPSCs have high potential in liver toxicology research, promoting drug screening applications and personalized medicine. However, the reliability and safety of the test have not been verified. It is also clear whether tissue heterogeneity will affect the effect of the drug, and whether the drug toxicity tests passed in mouse models can be safely applied to patients. Therefore, there are still many problems to be resolved for employing organoids to treat drug-induced liver injury from trial to the clinic.
4.5. Liver tumor
Tumor heterogeneity accounts for the difference in drug response among patients, which emphasizes the importance of precision medicine.96 Personalized strategies related to individual characteristics have crucial clinical relevance. Culture protocols for 3D tumor models have been established successfully in recent years, including those of the lungs,97 pancreas,98 colon99 and liver.100 Patient-derived models of tumors provide an in-vitro working object to study the molecular mechanism of tumorigenesis, seek therapeutic targets, test drug effects and formulate personalized strategies. For primary liver cancer, models based on patient-derived iPSCs or organoids are widely used as preclinical models to study precision medicine. They not only preserve the tumor microenvironment but also the tumor structure, and at the same time, can better recapitulate tumor heterogeneity.101 Here, we have summarized the immense progress made in establishing liver tumor models (Table 2). Based on tumor models, we can study the molecular mechanism of liver tumors, as well as guide patients toward early prevention and treatment.| Cell derivation | 3D cultivation method | Applications | Advantages | Ref. |
|---|---|---|---|---|
| Abbreviations: iPSC-EC, iPSC-derived endothelial cells; iPSC-MC, iPSC-derived mesenchymal cells; MASLD, metabolic dysfunction-associated steatotic liver disease; TME, tumor microenvironment; HCC, hepatocellular carcinoma; PLC, primary liver cancer | ||||
| iPSC-ECs, iPSC-MCs with Luciferase-expressing Huh7 cells | Ultra-purified alginate gel | • Induce the liver cancer mouse model | • High engraftment rate | 102 |
| • Study the mechanism of fibrosis or MASLD TME in HCC tumor development | • Controlled tumor size | |||
| • Simulation of the original TEM | ||||
| Hepatobiliary tumor tissue of patients | Suspended in Matrigel drops | • Assess the effect of neoantigen-directed therapy | • Recapitulate genetic complexity of original tumors | 103 |
| • Screen personalized immunotherapy targets | • Replication mutation | |||
| Hepatocellular carcinoma patients who underwent surgical resection | Seeded in spheroid formation medium | • Evaluate the treatment efficacy of candidate agents | • Maintain the histological features and expression profiles of the derived tumors | 104 |
| • Expandable in vitro | ||||
| • High generation efficiency | ||||
| Liver cancer tissues | Mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 with Matrigel 2 with Matrigel |
• Further pharmaco-proteogenomic analysis in models | • Retention of morphology, multiomics characteristics, and tumor heterogeneity | 105 |
| • Recapitulated previous cancer tissue–based molecular subtypes | ||||
| Tumor specimens | Solidification of Matrigel | • Screen sensitive drugs | • Exactly recapitulate the histopathological features of the original PLC | 106 |
| • Evaluate the efficacy of drugs directly targeting tumor cells | • Enrich aggressive PLC subpopulations | |||
| Liver tumors from patients who had no history of virus-meditated hepatitis | Cultured in organoids expansion medium | • Biomarker identification | • Recapitulate the histological architecture and expression profiles of the parent tumor | 107 |
| • Large-scale drug testing | • Retain the specific differences between patients as well as between tumor subtypes | |||
| • Long-term expansion in vitro | ||||
As mentioned above, tumor organoids have shown the potential to model tumor environments and facilitate basic research on cancers. In the field of personalized drug selection, tumor organoids have been widely used for high-throughput drug screening for hepatocellular carcinoma patients. Ji and team established a biobank of 65 human liver cancer organoids and screened a series of drugs and compounds that are used in clinical practice or under development, including sorafenib and lenvatinib, to achieve high prediction accuracy of drug responses.105 Furthermore, they also assessed potential drug combination therapies and predicted that lenvatinib may have better combinatorial effects with temsirolimus. Yuan Zhang et al. also evaluated the in-vitro drug response of hepatocellular carcinoma using a tumor organoid model.108 To aid this cause, single-cell RNA sequencing of liver tumor models may be helpful for dissecting drug resistance mechanisms.109 In the future, research on liver cancer organoids will be focused on searching for more biomarkers that facilitate early diagnosis and patient stratification.101
5. Summary and Limitations
After years of unremitting efforts by researchers, iPSC derivants have gradually expanded from 2-dimensional space to 3-dimensional space and from basic research to preclinical studies, with wide application prospects in treating various types of liver diseases.HLCs or LOs derived from iPSCs serve as objects of in-vitro research toward addressing the limitations of primary cells related to sources, amplification, and storage. HLCs and cholangiocytes differentiated from iPSCs can be applied to modeling molecular mechanisms at the cellular level, toxicity testing of drugs, and cell therapy in regenerative medicine in vitro. Liver organoids have emerged as potential tools in the drug development phase. Currently, the main research topics related to organoids are disease modeling and prediction of drug hepatotoxicity to develop individualized treatment strategies.110–112 In addition, the transplantation of iPSC derivatives as grafts has shown good results in animal models. A study has shown that when cell patches of iPSC-HLCs were attached to the injured liver surfaces, they successfully ameliorated lethal acute liver injury induced by the infusion of carbon tetrachloride, suggesting the high potential of grafting iPSC-HLCs.113 Recent studies on transplantation protocols of organoids have demonstrated LO transplantation rates of 80% and survival times of up to 90 days.114
However, there are some limitations to iPSC application scenarios that remain to be addressed. First and foremost, the main safety issue regarding iPSCs is the risk of tumorigenicity.115 This is because all four reprogramming factors employed for iPSC differentiation have been associated with tumor development, especially c-Myc, which is one of the most frequently mutated genes in human cancers.116 Furthermore, some of the added cytokines also play a role in tumor development, such as the dominant negative mutation of p53. Researchers should strictly control the differentiation conditions to support the aberrant expression of tumor-related mutant genes. Secondly, the amplification and culturing of iPSCs in vitro inevitably cause genetic alterations, chromosomal abnormalities, gene mutations and other uncontrollable and unpredictable problems. Uncontrolled safety issues in clinical applications pose a problem for cell therapy with iPSCs. Thirdly, immune rejection is another critical issue. Early studies have shown that undifferentiated iPSCs were rejected in syngeneic recipients of cell therapy.117 Moreover, this reflects another issue that the differentiation scheme needs for further maturation. HLCs and LOs are composed of cells with different degrees of differentiation. The immature cells, on the one hand, will become confounding factors in the culture system and produce unknown effects. On the other hand, the disease models cannot fully reproduce the disease characteristics, which limits their applicability in areas, such as drug testing and cell replacement.118,119 In other words, the liver is an organ with a highly complex composition and function that cannot be understood by analyzing a single type of cell or the pathogenesis of liver-related genetic metabolic diseases cannot be fully reproduced by a single cell. The reprogramming of iPSCs is expected to be the main means of promoting maturity in the future. In addition, the extracellular matrix components required for LO cultures are not clearly defined, which means there are many variables.120 Fourthly, so far, the vast majority of iPSC-based disease modeling studies performed involve comparative evaluation of a few patient-derived iPSC lines. This approach makes it easier to learn about disease-associated variants and phenotypes, while not the actual disease-causing variants. In the future, the biobank needs to be expanded to include enormous disease-related iPSC cell lines to obtain more disease information.121 Last, but not least, there is a lack of large-scale clinical trials to validate the precision and safety of iPSCs. Interestingly, the number of clinical trials on hepatocytes and LOs is large (Table 3). In the future, the stability and safety of iPSCs should be further clarified; HLCs and liver organoids should be tested in clinical trials and a standardized training model must be developed to exert their clinical application value.
| NCT Number | Title | Year | Country | Disease | Status | Results |
|---|---|---|---|---|---|---|
| Abbreviations: iPSC, induced pluripotent stem cells; ACLF, acute-on-chronic liver failure; CLF, chronic liver failure; NGS, next-generation sequencing; PDO, patient-derived tumor organoids; HCC, hepatocellular carcinoma; HAI, hepatic artery infusion; NA, not available. | ||||||
| IPSCs | ||||||
| NCT03867526 | Establishment of human cellular disease models for Wilson disease (IPSWILSON) | 2018 | Pakistan | Wilson disease | Completed | NA |
| NCT0095369 | Patient-specific induced pluripotency stem cells (PSiPS) | 2009 | Iran | Hepatic disorders | Completed | NA |
| Hepatocytes | ||||||
| NCT04806581 | Clinical study of hepatocyte transplantation for liver cirrhosis | 2021 | China | Liver cirrhosis | Not yet recruiting | NA |
| NCT04496479 | Allogenic hepatocyte transplantation into periduodenal lymph nodes | 2022 | United States | End stage liver disease | Recruiting | NA |
| NCT05727722 | Micro-encapsulated hepatocyte intraperitoneal transplantation in liver failure adults | 2023 | China | ACLF, CLF | Not yet recruiting | NA |
| Organoids | ||||||
| NCT06077591 | Prospective clinical calidation of NGS and PDO guided therapy in patients with advanced/inoperable solid tumors (PDO) | 2024 | China | HCC, colorectal cancer | Not yet recruiting | NA |
| NCT05932836 | An organoid-on-chips technique based on biopsy samples and its efficacy in predicting the response to HAI in HCC | 2023 | China | HCC | Recruiting | NA |
| NCT05183425 | Patient-derived organoids predicts the clinical efficiency of colorectal liver metastasis | 2022 | China | Colorectal liver metastasis | Recruiting | NA |
To sum up, hepatocytes and LOs derived from iPSCs represent two dimensions of regeneration methods and are used for disease modeling, drug screening, personalized medicine and other applications. Although some challenges remain to be sorted, iPSCs show immense capacity and superiority in liver disease treatment.
Abbreviations
| iPSCs | Induced pluripotent stem cells |
| MASLD | Metabolic dysfunction-associated steatotic liver disease |
| DILI | Drug-induced liver injury |
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| HLCs | Hepatocyte-like cells |
| LOs | Liver organoids |
| MASH | Metabolic dysfunction-associated steatohepatitis |
| FGF2 | Fibroblast growth factor 2 |
| BMP4 | Bone morphogenetic protein 4 |
| HGF | Hepatocyte growth factor |
| OSM | Oncostatin M |
| DEX | Dexamethasone |
| AAT | Alpha-1 antitrypsin |
| CYP450 | Cytochrome P450 |
| CRISPR | Clustered, regularly interspaced, short palindromic repeats |
| ECM | Extracellular matrix |
| NTCP | Sodium taurocholate co-transporting polypeptide |
| ALGS | Alagille syndrome |
| PNPLA3 | Phospholipase domain-containing protein 3 |
Author contributions
Conceptualization, Yulong Shang, Lina Cui and Xia Zhou, Ying Han; writing—original draft preparation, Ruobing Ju, Xia Zhou and Siyuan Tian; writing—review and editing, Yulong Shang, Lina Cui, Xia Zhou and Ying Han; visualization, Ruobing Ju, Siyuan Tian, Shuoyi Ma, Miao Zhang, Jingyi Liu and Keshuai Sun. All authors have read and agreed to the published version of the manuscript.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank the Figdraw (https://www.figdraw.com) for providing graphic elements in creating the figures in this review. This study was funded by National Key Research and Development Program of China (No. 2020YFA0710803 and 2017YFA0105704), National Natural Science Foundation of China (No. 81820108005, 82173241, 82200680, 82270551, and 81900502), and Key Research and Development Program of Shaanxi province, China (No. 2021ZDLSF02-07 and 2022ZDLSF03-03).References
- J. Xiao, F. Wang, N. K. Wong, J. He, R. Zhang, R. Sun, Y. Xu, Y. Liu, W. Li, K. Koike, W. He, H. You, Y. Miao, X. Liu, M. Meng, B. Gao, H. Wang and C. Li, J. Hepatol., 2019, 71, 212–221 CrossRef PubMed.
- S. Ling, G. Jiang, Q. Que, S. Xu, J. Chen and X. Xu, Liver Int., 2022, 42, 2110–2116 CrossRef PubMed.
- B. J. Dwyer, M. T. Macmillan, P. N. Brennan and S. J. Forbes, J. Hepatol., 2021, 74, 185–199 CrossRef CAS PubMed.
- A. Giancotti, V. D'Ambrosio, S. Corno, C. Pajno, G. Carpino, G. Amato, F. Vena, A. Mondo, L. Spiniello, M. Monti, L. Muzii, D. Bosco, E. Gaudio, D. Alvaro and V. Cardinale, Cytotherapy, 2022, 24, 376–384 CrossRef CAS PubMed.
- C. Busletta, E. Novo and M. Parola, Hepatol Int., 2013, 7, 299–305 CrossRef CAS PubMed.
- K. Takahashi and S. Yamanaka, Cell, 2006, 126, 663–676 CrossRef CAS PubMed.
- K. Takahashi, K. Tanabe, M. Ohnuki, M. Narita, T. Ichisaka, K. Tomoda and S. Yamanaka, Cell, 2007, 131, 861–872 CrossRef CAS PubMed.
- J. Yu, M. A. Vodyanik, K. Smuga-Otto, J. Antosiewicz-Bourget, J. L. Frane, S. Tian, J. Nie, G. A. Jonsdottir, V. Ruotti, R. Stewart, I. I. Slukvin and J. A. Thomson, Science, 2007, 318, 1917–1920 CrossRef CAS PubMed.
- S. Yamanaka, Cell Stem Cell, 2010, 7, 1–2 CrossRef CAS PubMed.
- N. Maherali, T. Ahfeldt, A. Rigamonti, J. Utikal, C. Cowan and K. Hochedlinger, Cell Stem Cell, 2008, 3, 340–345 CrossRef CAS PubMed.
- T. Zhou, C. Benda, S. Dunzinger, Y. Huang, J. C. Ho, J. Yang, Y. Wang, Y. Zhang, Q. Zhuang, Y. Li, X. Bao, H. F. Tse, J. Grillari, R. Grillari-Voglauer, D. Pei and M. A. Esteban, Nat. Protoc., 2012, 7, 2080–2089 CrossRef CAS PubMed.
- J. Staerk, M. M. Dawlaty, Q. Gao, D. Maetzel, J. Hanna, C. A. Sommer, G. Mostoslavsky and R. Jaenisch, Cell Stem Cell, 2010, 7, 20–24 CrossRef CAS PubMed.
- G. Liu, B. T. David, M. Trawczynski and R. G. Fessler, Stem Cell Rev. Rep., 2020, 16, 3–32 CrossRef PubMed.
- J. T. Dimos, K. T. Rodolfa, K. K. Niakan, L. M. Weisenthal, H. Mitsumoto, W. Chung, G. F. Croft, G. Saphier, R. Leibel, R. Goland, H. Wichterle, C. E. Henderson and K. Eggan, Science, 2008, 321, 1218–1221 CrossRef CAS PubMed.
- Y. Hirami, F. Osakada, K. Takahashi, K. Okita, S. Yamanaka, H. Ikeda, N. Yoshimura and M. Takahashi, Neurosci. Lett., 2009, 458, 126–131 CrossRef CAS PubMed.
- J. Zhang, G. F. Wilson, A. G. Soerens, C. H. Koonce, J. Yu, S. P. Palecek, J. A. Thomson and T. J. Kamp, Circ. Res., 2009, 104, e30–41 CAS.
- D. Taura, M. Sone, K. Homma, N. Oyamada, K. Takahashi, N. Tamura, S. Yamanaka and K. Nakao, Arterioscler., Thromb., Vasc. Biol., 2009, 29, 1100–1103 CrossRef CAS PubMed.
- Z. Song, J. Cai, Y. Liu, D. Zhao, J. Yong, S. Duo, X. Song, Y. Guo, Y. Zhao, H. Qin, X. Yin, C. Wu, J. Che, S. Lu, M. Ding and H. Deng, Cell Res., 2009, 19, 1233–1242 CrossRef PubMed.
- Y. Z. Xie and R. X. Zhang, J. Neurol. Sci., 2015, 36, 21–27 CrossRef CAS PubMed.
- C. Yang, J. Al-Aama, M. Stojkovic, B. Keavney, A. Trafford, M. Lako and L. Armstrong, Stem Cells, 2015, 33, 2643–2651 CrossRef PubMed.
- G. Song, X. Li, Y. Shen, L. Qian, X. Kong, M. Chen, K. Cao and F. Zhang, Cell Biochem. Biophys., 2015, 71, 1463–1473 CrossRef CAS PubMed.
- G. Roman, B. Stavik, K. H. Lauritzen, P. M. Sandset, S. P. Harrison, G. J. Sullivan and M. E. Chollet, Front. Physiol., 2023, 14, 1094249 CrossRef PubMed.
- D. Dutta, I. Heo and H. Clevers, Trends Mol. Med., 2017, 23, 393–410 CrossRef CAS PubMed.
- N. Graffmann, B. Scherer and J. Adjaye, Stem Cell Res., 2022, 61, 102763 CrossRef CAS PubMed.
- K. Si-Tayeb, F. K. Noto, M. Nagaoka, J. Li, M. A. Battle, C. Duris, P. E. North, S. Dalton and S. A. Duncan, Hepatology, 2010, 51, 297–305 CrossRef CAS PubMed.
- Q. Luo, N. Wang, H. Que, E. Mai, Y. Hu, R. Tan, J. Gu and P. Gong, Int. J. Mol. Sci., 2023, 24 Search PubMed.
- J. Blaszkiewicz and S. A. Duncan, Genes, 2022, 13 CrossRef CAS PubMed.
- Z. Ai, B. Niu, K. Duan, C. Si, S. Wang, L. Xiang, X. Zhu, Q. Zhu, C. Feng, Y. Yin, S. Zhao, R. Kong, W. Ji and T. Li, Biomaterials, 2020, 249, 120015 CrossRef CAS PubMed.
- T. Fukushima, S. Uchiyama, H. Tanaka and H. Kataoka, Int. J. Mol. Sci., 2018, 19, 3435 CrossRef PubMed.
- S. G. Tilson, C. M. Morell, A. S. Lenaerts, S. B. Park, Z. Hu, B. Jenkins, A. Koulman, T. J. Liang and L. Vallier, Hepatology, 2021, 74, 2998–3017 CrossRef CAS PubMed.
- J. Guo, L. Duan, X. He, S. Li, Y. Wu, G. Xiang, F. Bao, L. Yang, H. Shi, M. Gao, L. Zheng, H. Hu and X. Liu, Adv. Sci., 2021, 8, 2004680 CrossRef CAS PubMed.
- R. Boon, M. Kumar, T. Tricot, I. Elia, L. Ordovas, F. Jacobs, J. One, J. De Smedt, G. Eelen, M. Bird, P. Roelandt, G. Doglioni, K. Vriens, M. Rossi, M. A. Vazquez, T. Vanwelden, F. Chesnais, A. El Taghdouini, M. Najimi, E. Sokal, D. Cassiman, J. Snoeys, M. Monshouwer, W.-S. Hu, C. Lange, P. Carmeliet, S.-M. Fendt and C. M. Verfaillie, Nat. Commun., 2020, 11, 1393 CrossRef CAS PubMed.
- A. Laemmle, M. Poms, B. Hsu, M. Borsuk, V. Rüfenacht, J. Robinson, M. C. Sadowski, J. M. Nuoffer, J. Häberle and H. Willenbring, Hepatology, 2022, 76, 646–659 CrossRef CAS PubMed.
- R. Li, Y. Zhao, J. J. Yourick, R. L. Sprando and X. Gao, in Stem Cell Assays: Methods and Protocols, ed. N. Kannan and P. Beer, Springer US, New York, NY, 2022, pp. 127–142 DOI:10.1007/978-1-0716-1979-7_9.
- E. Zahmatkesh, N. Khoshdel Rad, N. Hossein-Khannazer, M. Mohamadnejad, R. Gramignoli, M. Najimi, R. Malekzadeh, M. Hassan and M. Vosough, Expert Rev. Gastroenterol. Hepatol., 2023, 17, 237–249 CrossRef CAS PubMed.
- H. Li, Y. Yang, W. Hong, M. Huang, M. Wu and X. Zhao, Signal Transduction Targeted Ther., 2020, 5, 1 CrossRef PubMed.
- Y. Xu and Z. Li, Comput. Struct. Biotechnol. J., 2020, 18, 2401–2415 CrossRef CAS PubMed.
- A. Marsee, F. J. M. Roos, M. M. A. Verstegen, H. P. B. O. Consortium, H. Gehart, E. de Koning, F. Lemaigre, S. J. Forbes, W. C. Peng, M. Huch, T. Takebe, L. Vallier, H. Clevers, L. J. W. van der Laan and B. Spee, Cell Stem Cell, 2021, 28, 816–832 CrossRef CAS PubMed.
- C. Olgasi, A. Cucci and A. Follenzi, Int. J. Mol. Sci., 2020, 21 CAS.
- G. S. P. Hsia, J. Esposito, L. A. da Rocha, S. L. G. Ramos and O. K. Okamoto, Stem Cells Int., 2021, 2021, 6632160 Search PubMed.
- L. Chen, X. Wei, D. Gu, Y. Xu and H. Zhou, Cancer Lett., 2023, 555, 216048 CrossRef CAS PubMed.
- S. Suominen, T. Hyypijev, M. Venäläinen, A. Yrjänäinen, H. Vuorenpää, M. Lehti-Polojärvi, M. Räsänen, A. Seppänen, J. Hyttinen, S. Miettinen, K. Aalto-Setälä and L. E. Viiri, Cells, 2023, 12, 2368 CrossRef CAS PubMed.
- A. Ootani, X. Li, E. Sangiorgi, Q. T. Ho, H. Ueno, S. Toda, H. Sugihara, K. Fujimoto, I. L. Weissman, M. R. Capecchi and C. J. Kuo, Nat. Med., 2009, 15, 701–706 CrossRef CAS PubMed.
- M. A. Lancaster and M. Huch, Dis. Models & Mech., 2019, 12 Search PubMed.
- M. Huch, C. Dorrell, S. F. Boj, J. H. van Es, V. S. Li, M. van de Wetering, T. Sato, K. Hamer, N. Sasaki, M. J. Finegold, A. Haft, R. G. Vries, M. Grompe and H. Clevers, Nature, 2013, 494, 247–250 CrossRef CAS PubMed.
- T. Takebe, K. Sekine, M. Enomura, H. Koike, M. Kimura, T. Ogaeri, R. R. Zhang, Y. Ueno, Y. W. Zheng, N. Koike, S. Aoyama, Y. Adachi and H. Taniguchi, Nature, 2013, 499, 481–484 CrossRef CAS PubMed.
- C. Nikokiraki, A. Psaraki and M. G. Roubelakis, Cells, 2022, 11 CrossRef CAS PubMed.
- C. Caiazza, S. Parisi and M. Caiazzo, Biology, 2021, 10 CrossRef CAS PubMed.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS PubMed.
- A. Messina, E. Luce, N. Benzoubir, M. Pasqua, U. Pereira, L. Humbert, T. Eguether, D. Rainteau, J. C. Duclos-Vallée, C. Legallais and A. Dubart-Kupperschmitt, Cells, 2022, 11 CAS.
- H. K. Kang, M. Sarsenova, D.-H. Kim, M. S. Kim, J. Y. Lee, E.-A. Sung, M. G. Kook, N. G. Kim, S. W. Choi, V. Ogay and K.-S. Kang, Cells, 2021, 10, 1268 CrossRef CAS PubMed.
- G. Lee, H. Kim, J. Y. Park, G. Kim, J. Han, S. Chung, J. H. Yang, J. S. Jeon, D.-H. Woo, C. Han, S. K. Kim, H.-J. Park and J.-H. Kim, Biomaterials, 2021, 269, 120529 CrossRef CAS PubMed.
- M. Danoy, Y. Tauran, S. Poulain, R. Jellali, J. Bruce, M. Leduc, M. Le Gall, Y. Koui, H. Arakawa, F. Gilard, B. Gakiere, Y. Kato, C. Plessy, T. Kido, A. Miyajima, Y. Sakai and E. Leclerc, APL Bioeng., 2021, 5 CAS.
- M. R. Poorna, R. Jayakumar, J. P. Chen and U. Mony, Colloids Surf., B, 2021, 207, 111991 CrossRef CAS PubMed.
- G. Sorrentino, S. Rezakhani, E. Yildiz, S. Nuciforo, M. H. Heim, M. P. Lutolf and K. Schoonjans, Nat. Commun., 2020, 11, 3416 CrossRef CAS PubMed.
- S. Suominen, T. Hyypijev, M. Venäläinen, A. Yrjänäinen, H. Vuorenpää, M. Lehti-Polojärvi, M. Räsänen, A. Seppänen, J. Hyttinen, S. Miettinen, K. Aalto-Setälä and L. E. Viiri, Cells, 2023, 12 CrossRef CAS PubMed.
- R. Ouchi, S. Togo, M. Kimura, T. Shinozawa, M. Koido, H. Koike, W. Thompson, R. A. Karns, C. N. Mayhew, P. S. McGrath, H. A. McCauley, R.-R. Zhang, K. Lewis, S. Hakozaki, A. Ferguson, N. Saiki, Y. Yoneyama, I. Takeuchi, Y. Mabuchi, C. Akazawa, H. Y. Yoshikawa, J. M. Wells and T. Takebe, Cell Metab., 2019, 30, 374–384 CrossRef CAS PubMed .e376.
- Y. Guan, D. Xu, P. M. Garfin, U. Ehmer, M. Hurwitz, G. Enns, S. Michie, M. Wu, M. Zheng, T. Nishimura, J. Sage and G. Peltz, JCI Insight, 2023, 8 CrossRef PubMed.
- R. Nguyen, S. Da Won Bae, L. Qiao and J. George, Cancer Lett., 2021, 508, 13–17 CrossRef CAS PubMed.
- L. Sun, Y. Wang, J. Cen, X. Ma, L. Cui, Z. Qiu, Z. Zhang, H. Li, R. Z. Yang, C. Wang, X. Chen, L. Wang, Y. Ye, H. Zhang, G. Pan, J. S. Kang, Y. Ji, Y. W. Zheng, S. Zheng and L. Hui, Nat. Cell Biol., 2019, 21, 1015–1026 CrossRef CAS PubMed.
- E. De Crignis, T. Hossain, S. Romal, F. Carofiglio, P. Moulos, M. M. Khalid, S. Rao, A. Bazrafshan, M. M. Verstegen, F. Pourfarzad, C. Koutsothanassis, H. Gehart, T. W. Kan, R. J. Palstra, C. Boucher, I. J. JN, M. Huch, S. F. Boj, R. Vries, H. Clevers, L. J. van der Laan, P. Hatzis and T. Mahmoudi, eLife, 2021, 10 CrossRef CAS PubMed.
- D. Hockemeyer and R. Jaenisch, Cell Stem Cell, 2016, 18, 573–586 CrossRef CAS PubMed.
- C. Günther, T. Brevini, F. Sampaziotis and M. F. Neurath, Dig. Liver Dis., 2019, 51, 753–760 CrossRef PubMed.
- L. Zhang, X. J. Ma, Y. Y. Fei, H. T. Han, J. Xu, L. Cheng and X. Li, Pharmacol. Ther., 2022, 232, 108004 CrossRef CAS PubMed.
- C. T. Nicolas, R. D. Hickey, H. S. Chen, S. A. Mao, M. Lopera Higuita, Y. Wang and S. L. Nyberg, Stem Cells, 2017, 35, 42–50 CrossRef PubMed.
- S. P. Harrison, S. F. Baumgarten, R. Verma, O. Lunov, A. Dejneka and G. J. Sullivan, Front. Med., 2021, 8, 574047 CrossRef PubMed.
- Y. Hutin, M. Nasrullah, P. Easterbrook, B. D. Nguimfack, E. Burrone, F. Averhoff and M. Bulterys, Morb. Mortal. Wkly. Rep., 2018, 67, 773–777 CrossRef PubMed.
- R. E. Schwartz, K. Trehan, L. Andrus, T. P. Sheahan, A. Ploss, S. A. Duncan, C. M. Rice and S. N. Bhatia, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2544–2548 CrossRef CAS PubMed.
- F. Sakurai, S. Mitani, T. Yamamoto, K. Takayama, M. Tachibana, K. Watashi, T. Wakita, S. Iijima, Y. Tanaka and H. Mizuguchi, Sci. Rep., 2017, 7, 45698 CrossRef CAS PubMed.
- X. Li, Z. Xu, B. Mitra, M. Wang, H. Guo and Z. Feng, Cell Biosci., 2021, 11, 123 CrossRef CAS PubMed.
- D. Cao, J. Y. Ge, Y. Wang, T. Oda and Y. W. Zheng, World J. Gastroenterol., 2021, 27, 4784–4801 CrossRef CAS PubMed.
- Y.-Z. Nie, Y.-W. Zheng, K. Miyakawa, S. Murata, R.-R. Zhang, K. Sekine, Y. Ueno, T. Takebe, T. Wakita, A. Ryo and H. Taniguchi, EBioMedicine, 2018, 35, 114–123 CrossRef PubMed.
- R. G. Rowe and G. Q. Daley, Nat. Rev. Genet., 2019, 20, 377–388 CrossRef CAS PubMed.
- Y. Guan, D. Xu, P. M. Garfin, U. Ehmer, M. Hurwitz, G. Enns, S. Michie, M. Wu, M. Zheng, T. Nishimura, J. Sage and G. Peltz, JCI Insight, 2017, 2 CrossRef PubMed.
- J. Roper and Ö. H. Yilmaz, Cell Stem Cell, 2019, 24, 841–842 CrossRef CAS PubMed.
- M. M. A. Verstegen, F. J. M. Roos, K. Burka, H. Gehart, M. Jager, M. de Wolf, M. J. C. Bijvelds, H. R. de Jonge, A. I. Ardisasmita, N. A. van Huizen, H. P. Roest, J. de Jonge, M. Koch, F. Pampaloni, S. A. Fuchs, I. F. Schene, T. M. Luider, H. P. J. van der Doef, F. Bodewes, R. H. J. de Kleine, B. Spee, G. J. Kremers, H. Clevers, I. J. JNM, E. Cuppen and L. J. W. van der Laan, Sci. Rep., 2020, 10, 21900 CrossRef CAS PubMed.
- R. Fiorotto, M. Amenduni, V. Mariotti, L. Fabris, C. Spirli and M. Strazzabosco, Hepatology, 2018, 67, 972–988 CrossRef CAS PubMed.
- D. Kim, S. B. Kim, J. L. Ryu, H. Hong, J. H. Chang, T. J. Yoo, X. Jin, H. J. Park, C. Han, B. H. Lee, J. H. Choi, H. W. Yoo, J. H. Kim and D. H. Woo, Cells, 2020, 9 Search PubMed.
- J. E. Kaserman, R. B. Werder, F. Wang, T. Matte, M. I. Higgins, M. Dodge, J. Lindstrom-Vautrin, P. Bawa, A. Hinds, E. Bullitt, I. S. Caballero, X. Shi, R. E. Gerszten, N. Brunetti-Pierri, M. Liesa, C. Villacorta-Martin, A. N. Hollenberg, D. N. Kotton and A. A. Wilson, Cell Rep., 2022, 41, 111775 CrossRef CAS PubMed.
- L. Zhang, K. Pu, X. Liu, S. D. W. Bae, R. Nguyen, S. Bai, Y. Li and L. Qiao, Front. Med., 2021, 8, 644594 CrossRef PubMed.
- S. Yang, H. Hu, H. Kung, R. Zou, Y. Dai, Y. Hu, T. Wang, T. Lv, J. Yu and F. Li, MedComm, 2023, 4, e274 CrossRef CAS PubMed.
- T. C. Yip, H. W. Lee, W. K. Chan, G. L. Wong and V. W. Wong, J. Hepatol., 2022, 76, 726–734 CrossRef CAS PubMed.
- Y. Wang, H. Wang, P. Deng, T. Tao, H. Liu, S. Wu, W. Chen and J. Qin, ACS Biomater. Sci. Eng., 2020, 6, 5734–5743 CrossRef CAS PubMed.
- S. McCarron, B. Bathon, D. M. Conlon, D. Abbey, D. J. Rader, K. Gawronski, C. D. Brown, K. M. Olthoff, A. Shaked and T. D. Raabe, Hepatology, 2021, 74, 1825–1844 CrossRef CAS PubMed.
- M. Elbadawy, M. Yamanaka, Y. Goto, K. Hayashi, R. Tsunedomi, S. Hazama, H. Nagano, T. Yoshida, M. Shibutani, R. Ichikawa, J. Nakahara, T. Omatsu, T. Mizutani, Y. Katayama, Y. Shinohara, A. Abugomaa, M. Kaneda, H. Yamawaki, T. Usui and K. Sasaki, Biomaterials, 2020, 237, 119823 CrossRef CAS PubMed.
- Y. Chien, C. S. Huang, H. C. Lin, K. H. Lu, P. H. Tsai, Y. H. Lai, K. H. Chen, S. D. Lee, Y. H. Huang and C. Y. Wang, Oncotarget, 2018, 9, 18594–18606 CrossRef PubMed.
- M. Parafati, S. H. Bae, R. J. Kirby, M. Fitzek, P. Iyer, O. Engkvist, D. M. Smith and S. Malany, Int. J. Mol. Sci., 2020, 21, 9557 CrossRef CAS PubMed.
- S.-X. Wang, J.-S. Yan and Y.-S. Chan, Int. J. Mol. Sci., 2022, 23, 11850 CrossRef CAS PubMed.
- S. Kammerer, Int. J. Mol. Sci., 2021, 22, 10214 CrossRef CAS PubMed.
- G. Holmgren, A. K. Sjögren, I. Barragan, A. Sabirsh, P. Sartipy, J. Synnergren, P. Björquist, M. Ingelman-Sundberg, T. B. Andersson and J. Edsbagge, Drug Metab. Dispos., 2014, 42, 1401–1406 CrossRef PubMed.
- Y. Wang, H. Wang, P. Deng, W. Chen, Y. Guo, T. Tao and J. Qin, Lab Chip, 2018, 18, 3606–3616 RSC.
- B. R. Ware, D. R. Berger and S. R. Khetani, Toxicol. Sci., 2015, 145, 252–262 CrossRef CAS PubMed.
- T. Shinozawa, M. Kimura, Y. Cai, N. Saiki, Y. Yoneyama, R. Ouchi, H. Koike, M. Maezawa, R.-R. Zhang, A. Dunn, A. Ferguson, S. Togo, K. Lewis, W. L. Thompson, A. Asai and T. Takebe, Gastroenterology, 2021, 160(831–846), e810 Search PubMed.
- C. J. Zhang, S. R. Meyer, M. J. O’Meara, S. Huang, M. M. Capeling, D. Ferrer-Torres, C. J. Childs, J. R. Spence, R. J. Fontana and J. Z. Sexton, J. Hepatol., 2023, 78, 998–1006 CrossRef CAS PubMed.
- X. Wu, D. Jiang, Y. Yang, S. Li and Q. Ding, Cell Regener., 2023, 12, 6 CrossRef CAS PubMed.
- A. Kashyap, M. A. Rapsomaniki, V. Barros, A. Fomitcheva-Khartchenko, A. L. Martinelli, A. F. Rodriguez, M. Gabrani, M. Rosen-Zvi and G. Kaigala, Trends Biotechnol., 2022, 40, 647–676 CrossRef CAS PubMed.
- J. R. Rock, M. W. Onaitis, E. L. Rawlins, Y. Lu, C. P. Clark, Y. Xue, S. H. Randell and B. L. M. Hogan, Proc. Natl. Acad. Sci., 2009, 106, 12771–12775 CrossRef CAS PubMed.
- Sylvia F. Boj, C.-I. Hwang, Lindsey A. Baker, Iok In. C. Chio, Dannielle D. Engle, V. Corbo, M. Jager, M. Ponz-Sarvise, H. Tiriac, Mona S. Spector, A. Gracanin, T. Oni, Kenneth H. Yu, R. van Boxtel, M. Huch, Keith D. Rivera, John P. Wilson, Michael E. Feigin, D. Öhlund, A. Handly-Santana, Christine M. Ardito-Abraham, M. Ludwig, E. Elyada, B. Alagesan, G. Biffi, Georgi N. Yordanov, B. Delcuze, B. Creighton, K. Wright, Y. Park, Folkert H. M. Morsink, I. Q. Molenaar, Inne H. Borel Rinkes, E. Cuppen, Y. Hao, Y. Jin, Isaac J. Nijman, C. Iacobuzio-Donahue, Steven D. Leach, Darryl J. Pappin, M. Hammell, David S. Klimstra, O. Basturk, Ralph H. Hruban, George J. Offerhaus, Robert G. J. Vries, H. Clevers and David A. Tuveson, Cell, 2015, 160, 324–338 CrossRef CAS PubMed.
- T. Sato, D. E. Stange, M. Ferrante, R. G. J. Vries, J. H. van Es, S. van den Brink, W. J. van Houdt, A. Pronk, J. van Gorp, P. D. Siersema and H. Clevers, Gastroenterology, 2011, 141, 1762–1772 CrossRef CAS PubMed.
- M. Huch, H. Gehart, R. van Boxtel, K. Hamer, F. Blokzijl, Monique M. A. Verstegen, E. Ellis, M. van Wenum, Sabine A. Fuchs, J. de Ligt, M. van de Wetering, N. Sasaki, Susanne J. Boers, H. Kemperman, J. de Jonge, Jan N. M. Ijzermans, Edward E. S. Nieuwenhuis, R. Hoekstra, S. Strom, Robert R. G. Vries, Luc J. W. van der Laan, E. Cuppen and H. Clevers, Cell, 2015, 160, 299–312 CrossRef CAS PubMed.
- K. Chen, Y. Li, B. Wang, X. Yan, Y. Tao, W. Song, Z. Xi, K. He and Q. Xia, Front. Immunol., 2023, 14, 1101324 CrossRef CAS PubMed.
- R. Qiu, S. Murata, C. Cheng, A. Mori, Y. Nie, S. Mikami, S. Hasegawa, T. Tadokoro, S. Okamoto and H. Taniguchi, Cancers, 2021, 13, 3997 CrossRef CAS PubMed.
- W. Wang, T. Yuan, L. Ma, Y. Zhu, J. Bao, X. Zhao, Y. Zhao, Y. Zong, Y. Zhang, S. Yang, X. Qiu, S. Shen, R. Wu, T. Wu, H. Wang, D. Gao, P. Wang and L. Chen, Adv. Sci., 2022, 9, 2105810 CrossRef CAS PubMed.
- S. Wang, Y. Wang, X. Xun, C. Zhang, X. Xiang, Q. Cheng, S. Hu, Z. Li and J. Zhu, J. Exp. Clin. Cancer Res., 2020, 39, 22 CrossRef CAS PubMed.
- S. Ji, L. Feng, Z. Fu, G. Wu, Y. Wu, Y. Lin, D. Lu, Y. Song, P. Cui, Z. Yang, C. Sang, G. Song, S. Cai, Y. Li, H. Lin, S. Zhang, X. Wang, S. Qiu, X. Zhang, G. Hua, J. Li, J. Zhou, Z. Dai, X. Wang, L. Ding, P. Wang, D. Gao, B. Zhang, H. Rodriguez, J. Fan, H. Clevers, H. Zhou, Y. Sun and Q. Gao, Sci. Transl. Med., 2023, 15, eadg3358 CrossRef CAS PubMed.
- L. Xian, P. Zhao, X. Chen, Z. Wei, H. Ji, J. Zhao, W. Liu, Z. Li, D. Liu, X. Han, Y. Qian, H. Dong, X. Zhou, J. Fan, X. Zhu, J. Yin, X. Tan, D. Jiang, H. Yu and G. Cao, Cell. Oncol., 2022, 45, 1019–1036 CrossRef CAS PubMed.
- L. Broutier, G. Mastrogiovanni, M. M. A. Verstegen, H. E. Francies, L. M. Gavarró, C. R. Bradshaw, G. E. Allen, R. Arnes-Benito, O. Sidorova, M. P. Gaspersz, N. Georgakopoulos, B.-K. Koo, S. Dietmann, S. E. Davies, R. K. Praseedom, R. Lieshout, J. N. M. Ijzermans, S. J. Wigmore, K. Saeb-Parsy, M. J. Garnett, L. J. W. van der Laan and M. Huch, Nature Medicine, 2017, 23, 1424–1435 CrossRef CAS PubMed.
- Y. Zhang, Z.-Y. Wang, H.-S. Jing, H.-D. Zhang, H.-X. Yan, J.-X. Fan and B. Zhai, Int. J. Mol. Med., 2022, 49, 51 CrossRef PubMed.
- Y. Zhao, Z.-X. Li, Y.-J. Zhu, J. Fu, X.-F. Zhao, Y.-N. Zhang, S. Wang, J.-M. Wu, K.-T. Wang, R. Wu, C.-J. Sui, S.-Y. Shen, X. Wu, H.-Y. Wang, D. Gao and L. Chen, Adv. Sci., 2021, 8, 2003897 CrossRef CAS PubMed.
- E. Driehuis, A. van Hoeck, K. Moore, S. Kolders, H. E. Francies, M. C. Gulersonmez, E. C. A. Stigter, B. Burgering, V. Geurts, A. Gracanin, G. Bounova, F. H. Morsink, R. Vries, S. Boj, J. van Es, G. J. A. Offerhaus, O. Kranenburg, M. J. Garnett, L. Wessels, E. Cuppen, L. A. A. Brosens and H. Clevers, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 26580–26590 CrossRef CAS PubMed.
- Y. Kita, A. Hamada, R. Saito, Y. Teramoto, R. Tanaka, K. Takano, K. Nakayama, K. Murakami, K. Matsumoto, S. Akamatsu, T. Yamasaki, T. Inoue, Y. Tabata, Y. Okuno, O. Ogawa and T. Kobayashi, Br. J. Cancer, 2019, 121, 1027–1038 CrossRef CAS PubMed.
- A. Lampis, P. Carotenuto, G. Vlachogiannis, L. Cascione, S. Hedayat, R. Burke, P. Clarke, E. Bosma, M. Simbolo, A. Scarpa, S. Yu, R. Cole, E. Smyth, J. F. Mateos, R. Begum, B. Hezelova, Z. Eltahir, A. Wotherspoon, N. Fotiadis, M. A. Bali, C. Nepal, K. Khan, M. Stubbs, J. C. Hahne, P. Gasparini, V. Guzzardo, C. M. Croce, S. Eccles, M. Fassan, D. Cunningham, J. B. Andersen, P. Workman, N. Valeri and C. Braconi, Gastroenterology, 2018, 154, 1066–1079 CrossRef CAS PubMed .e1065.
- Y. Nagamoto, K. Takayama, K. Ohashi, R. Okamoto, F. Sakurai, M. Tachibana, K. Kawabata and H. Mizuguchi, J. Hepatol., 2016, 64, 1068–1075 CrossRef PubMed.
- W. C. Peng, C. Y. Logan, M. Fish, T. Anbarchian, F. Aguisanda, A. Álvarez-Varela, P. Wu, Y. Jin, J. Zhu, B. Li, M. Grompe, B. Wang and R. Nusse, Cell, 2018, 175(1607–1619), e1615 Search PubMed.
- V. Volarevic, B. S. Markovic, M. Gazdic, A. Volarevic, N. Jovicic, N. Arsenijevic, L. Armstrong, V. Djonov, M. Lako and M. Stojkovic, Int. J. Med. Sci., 2018, 15, 36–45 CrossRef CAS PubMed.
- S. Yamanaka, Cell Stem Cell, 2020, 27, 523–531 CrossRef CAS PubMed.
- M. X. Doss and A. Sachinidis, Cells, 2019, 8, 403 CrossRef CAS PubMed.
- J. L. Corbett and S. A. Duncan, Front. Med., 2019, 6, 265 CrossRef PubMed.
- N. Roy-Chowdhury, X. Wang, C. Guha and J. Roy-Chowdhury, Hepatol. Int., 2017, 11, 54–69 CrossRef PubMed.
- R. Fiorotto and M. Strazzabosco, Cell. Mol. Gastroenterol. Hepatol., 2019, 8, 197–207 CrossRef PubMed.
- K. Musunuru, F. Sheikh, R. M. Gupta, S. R. Houser, K. O. Maher, D. J. Milan, A. Terzic and J. C. Wu, Circ.: Genomic Precis. Med., 2018, 11, e000043 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |