 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In silico identification of SARS-CoV-2 spike (S) protein–ACE2 complex inhibitors from eight Tecoma species and cultivars analyzed by LC-MS†
Seham S. El Hawarya,
Amira R. Khattab *b,
Hanan S. Marzoukc,
Amira S. El Senousya,
Mariam G. A. Alexc,
Omar M. Alyd,
Mohamed Telebe and
Usama Ramadan Abdelmohsen
*b,
Hanan S. Marzoukc,
Amira S. El Senousya,
Mariam G. A. Alexc,
Omar M. Alyd,
Mohamed Telebe and
Usama Ramadan Abdelmohsen *fg
*fg
aPharmacognosy Department, Faculty of Pharmacy, Cairo University, Egypt
bPharmacognosy Department, College of Pharmacy, Arab Academy for Science, Technology and Maritime Transport, Alexandria 1029, Egypt. E-mail: Dr_amira_khattab@aast.edu
cPharmacognosy Department, Faculty of Pharmacy, Pharos University in Alexandria, Egypt
dMedicinal Chemistry Department, Faculty of Pharmacy, Minia University, Minia, 61519, Egypt
eDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Alexandria University, Alexandria, 21521, Egypt
fPharmacognosy Department, Faculty of Pharmacy, Deraya University, Minia 61111, Egypt. E-mail: usama.ramadan@mu.edu.eg
gPharmacognosy Department, Faculty of Pharmacy, Minia University, Minia, 61519, Egypt
First published on 26th November 2020
Abstract
Coronavirus (CoV) is a positive RNA genome virus causing a global panic nowadays. Tecoma is a medicinally-valuable genus in the Bignoniaceae family, with some of its species exhibiting anti-HIV activity. This encouraged us to conduct an in silico exploration of some phytocompounds in Tecoma species cultivated in Egypt, namely Tecoma capensis and its four varieties i.e. yellow, harmony, pink and red, T. grandiflora Loisel., T. radicans L., and one hybrid i.e. Tecoma × smithii W. Watson. LC/MS-based metabolite profiling of the studied Tecoma plants resulted in the dereplication of 12 compounds (1–12) belonging to different phytochemical classes viz. alkaloids, iridoids, flavonoids and fatty acid esters. The in silico inhibitory action of these compounds against SARS-CoV-2 spike protein C-terminal domain in complex with human ACE2 was assessed via molecular docking. Succinic acid decyl-3-oxobut-2-yl ester (10), a fatty acid ester, possessed the best binding affinity (−6.77 kcal mol−1), as compared to hesperidin (13) (−7.10 kcal mol−1).
1. Introduction
Coronaviruses (CoVs) are positive RNA genome viruses, belonging to the Coronaviridae family of the Nidovirales order, which is divided into four genera (A, B, C and D). SARS-CoV-2 belongs to the B genus. CoV possess four structural proteins: spike protein, envelope protein, membrane protein, and nucleocapsid protein.1 Spike protein promotes host attachment and viral cell membrane fusion during virus infection.2 Potential anti-coronavirus treatments can be divided into two main categories, one operating on the human immune system or human cells, and the other on the coronavirus itself.3 Viruses often bind to receptor proteins on the surface of cells in order to enter human cells, for example, the SARS virus links with human angiotensin-converting enzyme 2 (hACE2) receptor.4 Protein–protein docking showed that SARS-CoV-2 spike proteins have a strong affinity for hACE2.5 Through virtual screening many compounds could be identified as inhibitors of hACE2, however these potential hACE2 inhibitors might not be beneficial for the management of SARS-CoV-2 infection due to the protective role of hACE2 in lung injury. Hesperidin was the only compound reported till now that could target the binding interface between spike protein and hACE2. It was reported that hesperidin can lie on the middle shallow pit of the surface of the receptor-binding domain (RBD) of spike protein.6 Shang J. et al., determined the crystal structure of RBD of the spike protein of SARS-CoV-2 (engineered to facilitate crystallization) in complex with hACE2. In comparison with the SARS-CoV RBD, an hACE2-binding ridge in SARS-CoV-2 RBD has a more compact conformation; moreover, several residues changes in the SARS-CoV-2 RBD stabilize two virus-binding hotspots at the RBD–hACE2 interface. These structural features of SARS-CoV-2 RBD increase its hACE2 binding affinity.7Tecoma genus is one of the medicinally-valuable members in Bignoniaceae family, embracing fourteen species of shrubs and small trees, of which twelve are native to America and two to Africa. The leaves were traditionally used by people in Latin America for diabetes management.8 Later on, several Tecoma species were reported to possess a wide variety of pharmacological actions viz. anti-inflammatory, antipyretic, analgesic, antimicrobial, antioxidant, hepatoprotective and cytotoxic actions.9 Tecoma stans (L.) Juss. ex Kunth, Tecoma capensis and Tecoma undulata possessed anti-diabetic action. T. sambucifolia H.B.K. had anti-inflammatory and antinociceptive actions as well as cytotoxicity against human hepatoma cell line. T. undulata Seem. showed hepatoprotective actions.10 Tecoma species are reported to be enriched in a diverse array of phytochemicals viz. alkaloids, flavonoids, iridoids, naphthoquinones, coumarins, chromones and steroids.10
Many research endeavors are now directed towards finding an effective therapy against COVID 19.11–18 As an extension from our previous research work,2,19,20 we conducted a screening for natural products that could possess potential anti-SARS actions, however with more focus on the plants reported to possess antiviral as well as antimalarial activities (compared to the antimalarial medication “chloroquine” with approved anti SARS-CoV activity21). We found that among Tecoma species, T. undulata was reported to exhibit anti-HIV activity22 and T. mollis possessed anti-malarial activity using chloroquine sensitive clones of Plasmodium falciparum.23 These reports encouraged us to conduct an in silico exploration of some representative members of Tecoma species cultivated in Egypt namely i.e. Tecoma capensis Lindl., T. capensis var. yellow, T. capensis var. harmony, T. grandiflora Loisel., T. radicans (L.) Juss., T. capensis var. pink, T. capensis var. red, and one hybrid Tecoma × smithii W. Watson.
2. Materials and methods
2.1. Plant material
Fresh samples of eight Tecoma species and cultivars were collected during the years 2016 and 2017, which are enlisted in ESI (Table S1)† along with their collection geographical locations and voucher specimen codes. The botanical specimens were identified by Professor Selim Zidan (Head of Botany Department, Faculty of Science, Alexandria University, Egypt), Mrs Therese Labib (Mazhar Botanical Garden, Egypt) and Mr Mohamed El-Gebaly (Plant Taxonomists in El-Orman Botanical Garden, Egypt).2.2. Preparation of plant extract and LC-MS metabolic profiling
Leaves of Tecoma species and cultivars under study (1600 g) were exhaustively extracted with methanol by cold maceration in 10 L methanol (2 L each × 5 times). The total methanolic extract was evaporated under reduced pressure by rotary evaporator at a temperature not exceeding 45 °C, yielding 180–200 mg of total methanolic extract of each studied plants. About 2 mg of each crude methanolic extract was dissolved separately in 1 ml MeOH and filtered using 0.2 μm membrane filter and then subjected to LC-HRESIMS analysis as previously reported in ref. 24. An Acquity Ultra-Performance Liquid Chromatography (UPLC) system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, MA, USA). Chromatographic separation was performed using a BEH C18 column (2.1 × 100 mm, 1.7 mm particle size) and a guard column (2.1 × 5 mm, 1.7 mm particle size) using the method previously described in ref. 19. Fig. S1† depicts the total ion chromatograms of the eight studied plants. MZmine 2.12 was used for processing the obtained raw data by the negative ionization mode. The processed data set was then subjected to molecular formula prediction and peak identification via dereplication using online METLIN25 and Dictionary of Natural Products (DNP)26 databases.2.3. Docking studies
3. Results and discussion
3.1. LC-HRESIMS based metabolic profiling
Twelve phytocompounds belonging to different phytochemical classes viz. alkaloids, iridoids, flavonoids, and fatty acid esters, have been identified by dereplication of the obtained LC-HRESIMS derived metabolite profiles of the eight Tecoma species and cultivars (Table 1). These metabolites have been annotated using databases (METLIN and DNP databases). From these databases, the molecular ion peaks appeared at m/z 325.1835, 349.1141, 327.2163 and 311.1672, with their predicted molecular formulas of C21H26O3, C19H26O6, C18H32O5 and C20H40O2 were dereplicated to be corresponding to four fatty acid esters viz. octanoic acid, 4-benzyloxyphenyl ester (1), fumaric acid, 3,4-dimethoxyphenyl heptyl ester (2), succinic acid, decyl 3-oxobut-2-yl ester (10) and valeric acid, pentadecyl ester (12), respectively.| # | Compound name | RT (min) | [M − H]− | Molecular formula | Error (0.1 m/z or 5 ppm) | T. smithii Wil. Wats. | T. radicans (L.) Juss. | T. capensis var. harmony | T. capensis var. yellow | T. capensis var. pink | T. capensis var. red | T. grandiflora (Thunb.) Loisel. | T. capensis (Thunb.) Lindl. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Octanoic acid, 4-benzyloxyphenyl ester | 1.32 | 325.1835 | C21H26O3 | 0.0134 | + | − | + | + | − | + | + | + |
| 2 | Fumaric acid, 3,4-dimethoxyphenyl heptyl ester | 1.82 | 349.1141 | C19H26O6 | −0.0495 | + | + | − | + | − | + | + | + |
| 3 | Boschniakine | 2.07 | 160.5217 | C10H11NO | 0.0446 | + | + | + | + | + | − | − | + |
| 4 | Luteolin 7-O-D-glucopyranoside | 2.18 | 447.1537 | C21H20O11 | 0.0604 | − | − | − | + | + | − | − | + |
| 5 | Actinidine | 2.89 | 147.9783 | C10H13N | 0.0109 | + | + | + | + | + | − | + | + |
| 6 | Skytanthine | 3.16 | 166.3757 | C11H21N | 0.02156 | + | − | + | + | + | + | + | + |
| 7 | Tecomanine (syn. tecomine) | 3.35 | 178.0705 | C11H17NO | 0.0013 | + | − | − | + | + | − | + | + |
| 8 | 7-O-(p-OH)cinnamoyltecomoside | 3.38 | 521.1938 | C25H30O12 | 0.0973 | − | − | + | + | − | + | − | + |
| 9 | 7-O-(p-MeO)cinnamoyltecomoside | 3.60 | 535.2093 | C26H32O12 | 0.0122 | − | − | + | + | + | + | − | + |
| 10 | Succinic acid, decyl 3-oxobut-2-yl ester | 3.78 | 327.2163 | C18H32O5 | 0.0134 | + | − | + | + | − | + | + | + |
| 11 | 7-O-(p-OH)benzoyltecomoside | 4.96 | 495.2212 | C23H28O12 | 0.0267 | − | − | + | + | + | − | − | − |
| 12 | Valeric acid, pentadecyl ester | 6.78 | 311.1672 | C20H40O2 | 0.0537 | + | + | − | + | − | + | + | + |
Besides, four alkaloids were dereplicated; boschniakine (3) with its molecular ion peak detected at m/z 160.5217, and a predicted molecular formula of C10H11NO, which was formerly identified in T. stans.28,29 Besides, actinidine (5), skytanthine (6), and tecomanine (7) were identified based on their molecular ion peaks appearing at m/z 147.9783, 166.3757 and 178.0705 and possessing predicted molecular formulas of C10H13N, C11H21N and C11H17NO which were previously isolated from several Tecoma spp. i.e. T. stans and T. arequipensis.28,30 Three iridoid glucosides were tentatively identified as being 7-O-(p-OH)cinnamoyltecomoside (8), 7-O-(p-MeO)cinnamoyltecomoside (9) and 7-O-(p-OH)benzoyl tecomoside (11) based on their respective molecular ion peaks at m/z 521.1938, 535.2093 and 495.2212, and molecular formulas of C25H30O12, C26H32O12 and C23H28O12. These compounds were previously isolated from T. capensis,31 however, they are reported here in most of the studied Tecoma species for the first time. One flavonoid was identified as being luteolin 7-O-β-D-glucopyranoside (4) by its molecular ion peak depicted at m/z 447.1537, corresponding to the suggested molecular formula (C21H20O11) which was formerly detected in T. stans fruits.32
3.2. Molecular docking analysis of selected phytocompounds from Tecoma species and cultivars
Docking simulations results (Table 2) showed that most of the studied phytocompounds displayed moderate to promising binding affinities compared to hesperidin (13). Succinic acid, decyl-3-oxobut-2-yl ester (10) came at the top of the list due to its best binding affinity (−6.77 kcal mol−1) among the studied phytocompounds. 7-O-(p-OH)cinnamoyltecomoside (8), valeric acid, pentadecyl ester (12), octanoic acid, 4-benzyloxyphenyl ester (1), 7-O-(p-OH)benzoyl tecomoside (11) showed slightly less binding affinities ranging from −6.73 to −6.48 kcal mol−1. Moderate fitting was observed in case of the two phytocompounds viz. 7-O-(p-MeO)cinnamoyltecomoside (9), and fumaric acid, 3,4-dimethoxyphenyl heptyl ester (2) with binding affinities of −5.44 and −5.28 kcal mol−1, respectively. The remaining phytocompounds showed relatively low binding affinities.| No. | Name of phytoligands | ΔGa (kcal mol−1) | Interactions at the binding interfaceb | |
|---|---|---|---|---|
| hACE2 residues | SARSCoV-2-CTD residues | |||
| a The ligand–receptor complex binding free energy at RMSD < 2 Å.b The key residues involved in the SARS-CoV-2-CTD–2hACE complex formation are listed in bold. | ||||
| 1 | Octanoic acid, 4-benzyloxyphenyl ester | −6.66 | No interaction | Gln493 |
| 2 | Fumaric acid, 3,4-dimethoxyphenyl heptyl ester | −5.28 | No interaction | Glu406, Arg408, Lys417 |
| 3 | Boschniakine | −3.76 | Lys353 | Gln493 |
| 4 | Luteolin 7-O-D-glucopyranoside | −4.84 | Glu37, Lys353 | Glu406, Gln493 |
| 5 | Actinidine | −4.80 | No interaction | No interaction |
| 6 | Skytanthine | −4.47 | No interaction | Arg403 |
| 7 | Tecomanine (syn. tecomine) | −4.64 | Lys353 | Gly496 |
| 8 | 7-O-(p-OH)cinnamoyltecomoside | −6.73 | No interaction | Glu406, Gln409 |
| 9 | 7-O-(p-MeO)cinnamoyltecomoside | −5.44 | His34, Ala389, Arg393 | Arg403 |
| 10 | Succinic acid, decyl 3-oxobut-2-yl ester | −6.77 | His34, Glu35 | Gln493 |
| 11 | 7-O-(p-OH)benzoyl tecomoside | −6.48 | Glu37, Lys353 | Asp405, Arg408, Gly496 |
| 12 | Valeric acid, pentadecyl ester | −6.73 | Lys353 | Gly496 |
| 13 | Hesperidin | −7.10 | His34, Ala387 | Gln409, Lys417, Ser494 |
Inspecting the binding modes of the promising phytocompounds revealed that most of them were able to accommodate into the interface and interact with the key amino acids, in most cases, through hydrogen bonds (Fig. 1 and 2). Thus, they may destabilize or even prevent the virus–receptor engagement that is generally dominated by polar contacts mediated by these key hydrophilic amino acid residues.27 Succinic acid, decyl-3-oxobut-2-yl ester (10), showing the best binding affinity in the current study, exhibited H–π interactions with His34 of the hACE-2 involved in the complex formation (Fig. 1Q and R). Among the studied phytocompounds, the second most promising in silico activity was recorded by valeric acid, pentadecyl ester (12) which interacted with SARS-CoV-2-CTD Gly496 and hACE2 Lys353 (Fig. 1U and V). Similarly, 7-O-(p-OH)benzoyl tecomoside (11) exhibited these key interactions in addition to binding hACE-2 Glu37 (Fig. 1S and T). It is worth mentioning that a single Lys353 mutation was reported to be sufficient to abolish the interactions at the interface. Interestingly, boschniakine (3; Fig. 1E and F), luteolin 7-O-D-glucopyranoside (4; Fig. 1G and H), tecomanine (7; Fig. 1K and L) were able to interact with Lys353 despite their relatively low binding affinities to the complex. Notably, fumaric acid, 3,4-dimethoxyphenyl heptyl ester (2) showed interactions with SARS-CoV-2 CTD through its key residue Lys417 as well as Glu406 with no interactions with the hACE-2 side (Fig. 1C and D). This observation was also recorded in case of octanoic acid, 4-benzyloxyphenyl ester (1; Fig. 1A and B), skytanthine (6; Fig. 1I and J) and 7-O-(p-OH)cinnamoyltecomoside (8; Fig. 1M and N). On the other hand, actinidine (5) did not exhibit any considerable interactions with its residues.
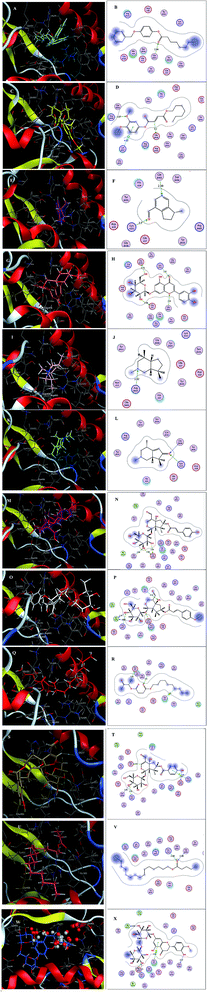 | ||
| Fig. 1 (A) 3D binding mode of 1 (cyan sticks), (B) 2D binding mode of 1, (C) 3D binding mode of 2 (yellow sticks), (D) 2D binding mode of 2, (E) 3D binding mode of 3 (magenta sticks), (F) 2D binding mode of 3, (G) 3D binding mode of 4 (orange sticks), (H) 2D binding mode of 4, (I) 3D binding mode of 6 (light pink sticks), (J) 2D binding mode of 6, (K) 3D binding mode of 7 (green sticks), (L) 2D binding mode of 7, (M) 3D binding mode of 8 (deep pink sticks), (N) 2D binding mode of 8, (O) 3D binding mode of 9 (white sticks), (P) 2D binding mode of 9, (Q) 3D binding mode of 10 (red sticks), (R) 2D binding mode of 10, (S) 3D binding mode of 11 (deep yellow sticks), (T) 2D binding mode of 11, (U) 3D binding mode of 12 (pink sticks), (V) 2D binding mode of 12 (W) 3D binding mode of 13 (blue sticks), (X) 2D binding mode of 13 in the binding interface of SARS-CoV-2-CTD in complex with hACE2 (PDB ID 6LZG). | ||
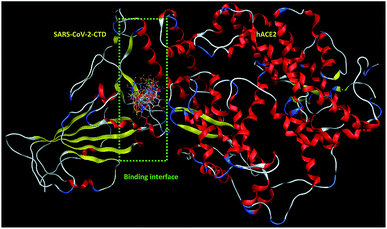 | ||
| Fig. 2 Overlay of 1 (cyan sticks), 2 (yellow sticks), 3 (magenta sticks), 4 (orange sticks), 6 (light pink sticks), 7 (green sticks), 8 (deep pink sticks), 9 (white sticks), 10 (red sticks), 11 (deep yellow sticks), 12 (pink sticks) and 13 (blue sticks) in the binding interface of SARS-CoV-2-CTD in complex with hACE2 (PDB ID 6LZG). | ||
4. Conclusions
In the current study, in silico exploration was conducted for some of Tecoma phytocompounds that could inhibit SARS-CoV entry to its host cells. Twelve compounds from eight Tecoma plants belonging to different phytochemical classes viz. alkaloids, iridoids, flavonoids, and fatty acid esters were dereplicated using LC-HRESIMS, among which a fatty acid ester named succinic acid, decyl-3-oxobut-2-yl ester (10) was reported to possess the best binding affinity (−6.77 kcal mol−1), followed by 7-O-(p-OH)cinnamoyltecomoside (8), valeric acid, pentadecyl ester (12), octanoic acid, 4-benzyloxyphenyl ester (1), 7-O-(p-OH)benzoyl tecomoside (11) which showed slightly less binding affinities ranging from −6.73 to −6.48 kcal mol−1, as compared to hesperidin (13). These phytocompounds could serve as potential candidates for the discovery of anti-SARS drugs that possess a preventive potential, however the therapeutic potential is yet to be validated using in vitro and in vivo studies.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank Prof. Tobias Gulder (University of Dresden) for LC-MS measurement.References
- B. J. Bosch, R. van der Zee, C. A. M. de Haan and P. J. M. Rottier, J. Virol., 2003, 77, 8801–8811 CrossRef CAS.
- A. M. Sayed, A. R. Khattab, A. M. AboulMagd, H. M. Hassan, M. E. Rateb, H. Zaid and U. R. Abdelmohsen, RSC Adv., 2020, 10, 19790–19802 RSC.
- A. S. Omrani, M. M. Saad, K. Baig, A. Bahloul, M. Abdul-Matin, A. Y. Alaidaroos, G. A. Almakhlafi, M. M. Albarrak, Z. A. Memish and A. M. Albarrak, Lancet Infect. Dis., 2014, 14, 1090–1095 CrossRef CAS.
- X. Y. Ge, J. L. Li, X. Lou Yang, A. A. Chmura, G. Zhu, J. H. Epstein, J. K. Mazet, B. Hu, W. Zhang, C. Peng, Y. J. Zhang, C. M. Luo, B. Tan, N. Wang, Y. Zhu, G. Crameri, S. Y. Zhang, L. F. Wang, P. Daszak and Z. L. Shi, Nature, 2013, 503, 535–538 CrossRef CAS.
- X. Xu, P. Chen, J. Wang, J. Feng, H. Zhou, X. Li, W. Zhong and P. Hao, Sci. China: Life Sci., 2020, 63, 457–460 CrossRef CAS.
- C. Wu, Y. Liu, Y. Yang, P. Zhang, W. Zhong, Y. Wang, Q. Wang, Y. Xu, M. Li, X. Li, M. Zheng, L. Chen and H. Li, Acta Pharm. Sin. B, 2020, 10(5), 766–788 CrossRef.
- J. Shang, G. Ye, K. Shi, Y. Wan, C. Luo, H. Aihara, Q. Geng, A. Auerbach and F. Li, Nature, 2020, 581, 221–224 CrossRef CAS.
- L. Aguilar-Santamaría, G. Ramírez, P. Nicasio, C. Alegría-Reyes and A. Herrera-Arellano, J. Ethnopharmacol., 2009, 124, 284–288 CrossRef.
- S. Verma, Phytochemical and pharmacological review study on Tecoma stans Linn, 2016, vol. 4 Search PubMed.
- M. Rahmatullah, W. Samarrai, R. Jahan, S. Rahman, Z. U. M. E. U. Miajee, M. H. Chowdhury, S. Bari, A. B. M. A. Bashar, A. K. Azad and S. Ahsan, DNA Repair, 2010, 4, 236–253 Search PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS.
- A. K. Ghosh, M. Brindisi, D. Shahabi, M. E. Chapman and A. D. Mesecar, ChemMedChem, 2020, 15, 907–932 CrossRef CAS.
- R. Alexpandi, J. F. De Mesquita, S. K. Pandian and A. V. Ravi, Front. Microbiol., 2020, 11 DOI:10.3389/fmicb.2020.01796.
- W. R. Ferraz, R. A. Gomes, A. L. S Novaes and G. H. Goulart Trossini, Future Med. Chem., 2020, 12, 1815–1828 CrossRef CAS.
- D. Gentile, V. Patamia, A. Scala, M. T. Sciortino, A. Piperno and A. Rescifina, Mar. Drugs, 2020, 18(4) DOI:10.3390/md18040225.
- S. Shahinshavali, K. A. Hossain, A. V. D. N. Kumar, A. G. Reddy, D. Kolli, A. Nakhi, M. V. B. Rao and M. Pal, Tetrahedron Lett., 2020, 61(40) DOI:10.1016/j.tetlet.2020.152336.
- M. Hagar, H. A. Ahmed, G. Aljohani and O. A. Alhaddad, Int. J. Mol. Sci., 2020, 21(11) DOI:10.3390/ijms21113922.
- S. T. Ngo, N. Quynh Anh Pham, L. Thi Le, D.-H. Pham and V. V. Vu, J. Chem. Inf. Model., 2020 DOI:10.1021/acs.jcim.0c00491.
- A. I. Owis, M. S. El-Hawary, D. El Amir, O. M. Aly, U. R. Abdelmohsen and M. S. Kamel, RSC Adv., 2020, 10, 19570–19575 RSC.
- F. M. Abd El-Mordy, M. M. El-Hamouly, M. T. Ibrahim, G. A. El-Rheem, O. M. Aly, A. M. Abd El-Kader, K. A. Youssif and U. R. Abdelmohsen, RSC Adv., 2020, 10, 32148–32155 RSC.
- A. S. Achutha, V. L. Pushpa and S. Suchitra, J. Proteome Res., 2020, 19(11), 4706–4717, DOI:10.1021/acs.jproteome.0c00683.
- R. Kumawat, S. Sharma and S. Kumar, Acta Pol. Pharm., 2012, 69, 993–996 CAS.
- W. M. Abdel-Mageed, E. Y. Backheet, A. A. Khalifa, Z. Z. Ibraheim and S. A. Ross, Fitoterapia, 2012, 83, 500–507 CrossRef CAS.
- A. H. Elmaidomy, R. Mohammed, H. M. Hassan, A. I. Owis, M. E. Rateb, M. A. Khanfar, M. Krischke, M. J. Mueller and U. R. Abdelmohsen, Metabolites, 2019, 17(8) DOI:10.3390/md17080465.
- C. Guijas, J. R. Montenegro-Burke, X. Domingo-Almenara, A. Palermo, B. Warth, G. Hermann, G. Koellensperger, T. Huan, W. Uritboonthai, A. E. Aisporna, D. W. Wolan, M. E. Spilker, H. P. Benton and G. Siuzdak, Anal. Chem., 2018, 90, 3156–3164 CrossRef CAS.
- M. Whittle, P. Willett, W. Klaffke and P. Van Noort, J. Chem. Inf. Comput. Sci., 2003, 43, 449–457 CrossRef CAS.
- Q. Wang, Y. Zhang, L. Wu, S. Niu, C. Song, Z. Zhang, G. Lu, C. Qiao, Y. Hu, K. Y. Yuen, Q. Wang, H. Zhou, J. Yan and J. Qi, Cell, 2020, 181(4), 894–904 CrossRef CAS.
- L. Costantino, L. Raimondi, R. Pirisino, T. Brunetti, P. Pessotto, F. Giannessi, A. P. Lins, D. Barlocco, L. Antolini and S. A. El-Abady, Farmaco, 2003, 58, 781–785 CrossRef CAS.
- A. Al-Azzawi, A. Al-Guboori, A. Abdul-Sada and M. Al-Azzawi, Planta Med., 2010, 76(12) DOI:10.1055/s-0030-1264698.
- G. H. Harris, E. C. Fixman, F. R. Stermitz and L. Castedo, J. Nat. Prod., 1988, 51, 543–548 CrossRef CAS.
- M. Guiso, C. Marra, F. Piccioni and M. Nicoletti, Phytochemistry, 1997, 45, 193–194 CrossRef CAS.
- M. Marzouk, A. Gamal-Eldeen, M. Mohamed and M. El-Sayed, Z. Naturforsch., C: J. Biosci., 2006, 61, 783–791 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra08997d |
| This journal is © The Royal Society of Chemistry 2020 |
