Open Access Article
Open Access ArticleCarbon dots for highly effective photodynamic inactivation of multidrug-resistant bacteria†
Dina I.
Abu Rabe
a,
Oluwayemisi O.
Mohammed
a,
Xiuli
Dong
a,
Amankumar K.
Patel
b,
Christopher M.
Overton
b,
Yongan
Tang
c,
Sophia
Kathariou
d,
Ya-Ping
Sun
 *b and
Liju
Yang
*b and
Liju
Yang
 *a
*a
aDepartment of Pharmaceutical Sciences, Biomanufacturing Research Institute and Technology Enterprise (BRITE), North Carolina Central University, Durham, NC 27707, USA. E-mail: lyang@nccu.edu
bDepartment of Chemistry and Laboratory for Emerging Materials and Technology, Clemson University, Clemson, South Carolina 29634, USA. E-mail: syaping@clemson.edu
cDepartment of Mathematics and Physics, North Carolina Central University, Durham, NC 27707, USA
dDepartment of Food, Bioprocessing & Nutrition Sciences, North Carolina State University, Raleigh, NC 27695, USA
First published on 5th May 2020
Abstract
For addressing the ever increasing challenge of multidrug-resistant (MDR) bacterial infections, specifically designed and prepared carbon dots (CDots) of small carbon nanoparticles with surface functionalization–passivation by oligomeric polyethylenimine were found to be readily activated by visible light to effectively and efficiently inactivate MDR bacterial strains. The inactivation was evaluated under various combinations of experimental conditions (dot concentrations, light intensities, and treatment times), with the results collectively suggesting CDots as a new class of promising agents for combating MDR bacteria. Mechanistic origins and implications of the observed strong antibacterial actions as relevant to the photoexcited state processes in CDots and the photodynamically induced cellular damages leading to the death of the bacterial cells were explored, with the results discussed.
Infectious diseases caused by ever increasing antibiotic-resistant bacteria represent one of the most serious threats to public health globally.1,2 Among the multidrug resistant (MDR) pathogens that have emerged over the recent decades, MDR Enterococci are ranked second only to Staphylococci as aetiological agents for hospital acquired infections (HAI).3,4Enterococci are the leading causes for bacteremia, urinary tract infections, and surgical wound infections,4–6 with E. faecium and E. faecalis responsible for the majority of enterococcal infections.2,7–10 These strains have acquired strong resistance capabilities through multiple mechanisms, and their resistance to virtually all existing antibiotics in clinic9,11 presents particularly tough challenges for countermeasure strategies. In fact, not only are Enterococci able to recruit and maintain a variety of gene clusters for MDR, but also serve as donors of the resistance gene clusters to other pathogenic microorganisms,12,13 thus driving the spread and evolution of antibiotic resistance.
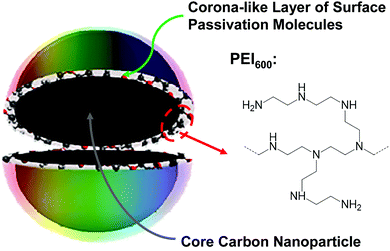
In the fight to control MDR pathogens, alternative to the development of new drugs and therapeutic paradigms have been approaches based on antimicrobial agents and mechanisms that are beyond the bacterial resistance. One approach proven effective has been photodynamic inactivation (PDI), in which a photo-sensitizer is employed to generate reactive oxygen species (ROS) under light illumination for their damaging multiple cellular components and ultimately killing the bacterial cells.14,15 Organic dyes have traditionally been used as photo-sensitizers, but more recently nanoscale materials have shown great promise as effective PDI agents for their many unique and advantageous characteristics, such as the robust photoexcited state processes for more effective and efficient ROS generation, photostability for repeated PDI actions, among others. In fact, recent reports on the PDI activities of semiconductor quantum dots (QDs) against MDR bacteria have generated some excitements in the research communities and also in the news media,16,17 despite the fact that the reported results for the QDs were generally suggesting the toughness of MDR bacterial strains in comparison with their non-resistant counterparts.16,17 For example, in the well publicized study of CdTe QDs for PDI against drug-resistant Staphylococcus aureus, Escherichia coli, Klebsiella pneumonia, and Salmonella typhimurium, only a maximum of 80% or 92% (less than one log) of the bacterial cells were killed with 35 nM CdTe QDs after 6 hours of illumination with the normal light intensity or 8 hours of illumination with triple the normal light intensity, respectively.16 The inactivation was attributed to the light-activated redox species (LARS) interfering with the redox homeostatsis of the bacterial cells. Competing with semiconductor QDs are the structurally rather simple carbon nanomaterials dubbed carbon dots (CDots, Fig. 1),18,19 which have recently emerged to represent a new class of highly effective PDI agents under visible/natural light against a number of bacterial strains.20–24 In the work reported here, specifically designed and prepared CDots with visible-light activation were found to be able to kill drug resistant E. faecium and E. faecalis efficiently and quantitatively.
CDots are generally defined as small carbon nanoparticles with various surface passivation schemes,19,25 and the dot structure may be viewed as a special core–shell configuration, each with a “hard” carbon nanoparticle core and a “soft’ shell of organic species for the passivation function (Fig. 1). In this work, oligomeric polyethylenimine (molecular weight ∼600, PEI600) was used for the surface functionalization–passivation of pre-processed and selected small carbon nanoparticles, thus the resulting dots denoted as PEI600–CDots. The small carbon nanoparticles of averaging around 5 nm in diameter were harvested from commercially acquired carbon nanopowders (purity >99%), which according to the supplier were produced by the high-temperature carbonization of plant materials. The protocol for the harvesting included the oxidative acid treatment of the as-supplied carbon nanopowders, followed by a combination of solvent-dispersing and centrifuging steps and then solvent removal to obtain the targeted small carbon nanoparticles.26 The nanoparticles thus harvested were then dispersed in PEI600 melt for microwave energy-induced thermal reactions, in which the nanoparticles were functionalized by PEI600 through radical addition27 and/or other surface attachment modes.23 The PEI600–CDots prepared in the thermally induced functionalization reactions were found to be highly stable in terms of their optical properties remaining unchanged after their exposure to visible light for many hours or storage under ambient conditions for many months.
The sample characterization by using a series of instrumental methods yielded results that confirmed the dot structure as defined (Fig. 1), namely carbon nanoparticle cores of around 5 nm in average diameter surface-passivated by PEI moieties in the CDots (sizes mostly 4–8 nm, Fig. 2). The observed UV/vis absorption spectrum of PEI600–CDots in an aqueous solution (Fig. 2) is similar to that of the aqueous suspended precursor carbon nanoparticles, characterized by significant photon-harvesting in the visible spectrum, and the observed fluorescence emission spectra and quantum yields (∼20% at 400 nm excitation) are comparable to those of other CDots reported previously.26
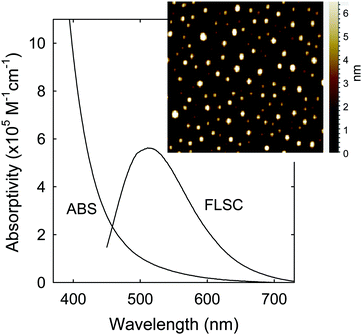 | ||
| Fig. 2 Optical absorption (ABS) and fluorescence (FLSC) spectra of PEI600–CDots in aqueous solution. Inset: Representative AFM images for the CDots. | ||
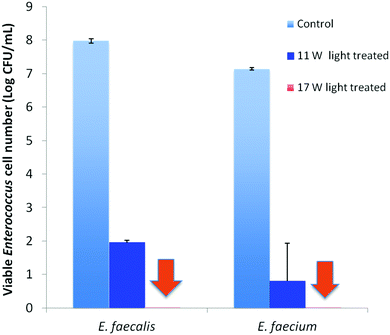
Visible light-induced PDI activities of PEI600–CDots against MDR E. faecalis (resistant to tetracycline, nalidixic acid, and low-level streptomycin and erythromycin) and E. faecium (resistant to tetracycline, erythromycin, nalidixic acid, and low-level ciprofloxacin) were evaluated, with excellent outcomes. Experimentally, each strain in PBS buffer at a concentration of 107–108 CFU mL−1 was treated with 1.9 μM PEI600–CDots under visible light of 400–800 nm (11 W LED – light intensity ∼4.8 mW cm−2) for 30 min, resulting in average viable cell reductions of ∼6.0 log for E. faecalis and ∼6.3 log for E. faecium (Fig. 3), close to complete inactivation. In the dark controls, the same treatment of PEI600–CDots without light exposure had no effects on the bacterial cells. Also no effects were found for the exposure of E. faecalis and E. faecium to the visible light in the absence of any CDots.
The evaluation results also suggested dependencies of the PDI effectiveness on various combinations of parameters and conditions, including the dot concentration, light intensity, and exposure time. Fig. 3 shows the PDI results on MDR E. faecalis and E. faecium by PEI600–CDots at dot concentration of 1.9 μM with 30 min exposure of visible light at two levels of intensity. The treatment at higher light intensity of ∼7.8 mW cm−2 (17 W LED) resulted in the complete inactivation of both E. faecalis and E. faecium, achieving ∼8.0 log and ∼7.1 log viable cell reduction, respectively (Fig. 3), while at the lower light intensity of ∼4.8 mW cm−2 (11 W LED), ∼6.0 log and ∼6.3 log viable cell reduction for E. faecalis and E. faecium was achieved, respectively (Fig. 3). In another combination to keep the light intensity at ∼4.8 mW cm−2, a much lower dot concentration of 0.62 μM could be coupled with a longer light exposure time of 90 min to achieve the similarly effective inactivation, with viable cell reductions of ∼7.0 log for E. faecalis and ∼7.1 log for E. faecium.
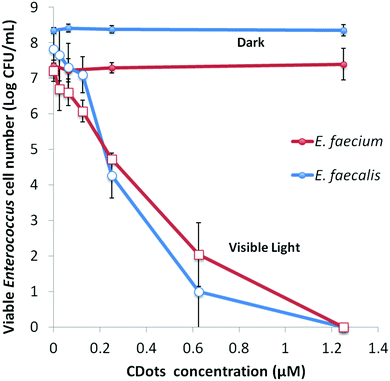
Under the same light intensity and exposure time (∼4.8 mW cm−2 for 60 min), the evaluations on variable dot concentrations show that the meaningful inactivation of more than 1 log could be achieved with the dot concentration as low as 0.12 μM in the samples containing 107–108 CFU mL−1 of E. faecalis or E. faecium, while complete eradication of bacterial cells in the samples was achieved at dot concentration of 1.25 μM (Fig. 4). And the concentration dependency of CDots’ PDI was clearly observed for both strains (Fig. 4).
The results presented above collectively suggest that the CDots of relatively simple chemical compositions and nanoscale structures could readily be activated under visible light for highly effective killing of MDR bacteria, specifically the highly ranked MDR E. faecium and E. faecalis. While there are no comparable results available on PDI actions of conventional semiconductor QDs and related nanomaterials against the same MDR bacterial strains, the high performances of CDots are apparent, with the quantitative inactivation achieved at relatively low concentrations coupled with the exposure to simple household light sources. Among similar uses of conventional semiconductor QDs, the recently reported CdTd QDs for PDI against several drug-resistant bacterial strains resulted in only about one log bacterial cell reduction.16 The obviously high performance levels of CDots demonstrated in the PDI of the MDR Enterococci strains make these simple carbon nanomaterials extremely promising in the development of countermeasures to control MDR bacteria. Beyond the high performance, the CDots platform also offers other major advantages. Unlike many popular yet highly toxic conventional semiconductor QDs due to their containing Cd (cadmium) or other heavy metals, CDots are known as being benign and nontoxic in vitro and in vivo,28–30 thus offering excellent opportunities for broad applications in many healthcare settings and human living environments.
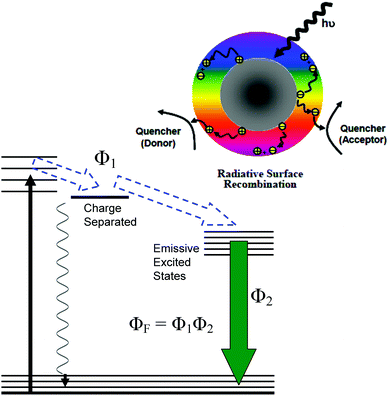
For PDI, the strong optical absorptions of CDots for photon-harvesting in the visible spectrum represent a major advantage. The estimated molar absorptivities for CDots with core carbon nanoparticles in sizes of 5 nm in diameter are around 5 × 105 M−1 cm−1 at 400–450 nm (Fig. 2). Mechanistically, upon photoexcitation of CDots there are rapid charge transfer and separation in the dots (Fig. 5), and the electrons and holes thus produced are trapped at various surface defect sites that are stabilized by the passivation of the organic moieties (Fig. 1).25 The redox pairs likely serve the antibacterial function that is similar to or even more potent than that of the ROS produced in the traditional photosensitization, responsible in a major part for the observed high inactivation performance of the CDots. There have been experimental results suggesting that radiative recombinations of the electrons and holes (the population of the emissive excited states, Fig. 5) for the observed bright and colorful fluorescence emissions in CDots are rapid, namely the redox pairs are short-lived.31 Thus, their potent antibacterial activities must be on a fast time scale, probably requiring the dots next to the targeted bacterial cells. In this regard, the PEI moieties with abundant amino groups on the dot surface (Fig. 1) could be protonated at near neutral pH in the buffer solution and positively charged to facilitate the “binding” of PEI600–CDots to the Enterococci cells, similar to what were found in the interactions of CDots with other bacteria.20
The emissive excited states in CDots (Fig. 5) are relatively longer lived, averaging on the order of 5–10 ns,31 capable of the same ROS generation as that with traditional photosensitization. The high fluorescence quantum yields of PEI600–CDots suggest relatively inefficient competing processes for the excited state energies, which must have benefited the ROS generation by the CDots (essentially bound to the targeted bacterial cells) to be more competitive and efficient. The combination of the ROS-like (by the initially formed redox pairs from the rapid charge separation following photoexcitation) and classical ROS actions on different time scales may be credited for the observed uniquely effective PDI against the MDR Enterococci.
The photo-generated ROS are known to damage various cell components such as carbohydrates, DNA, and/or lipids and proteins, leading to the cell death.32–34 For CDots, their photodynamic killing of cancer cells has been demonstrated.35–37 In this work, the elevated intracellular ROS level was detected in both E. faecalis and E. faecium cells upon their treatment with PEI600–CDots, increasing 3.8 and 3.2 fold in comparison to control samples, respectively, after the cells were treated with 1.2 μM CDots for 1 h.
Both intracellular and extracellular ROS are capable of disrupting cell membranes,38,39 and the resulting cell structural damages can be visualized by using electron microscope. In the transmission electron microscopy (TEM) imaging experiment, the structural changes in the CDots-treated E. faecium cells were probed in reference to the untreated control cells. As compared in Fig. 6, while the treated cells still maintained their shape, without obvious disruption in cell morphology, there were apparently significant changes in the cytoplasm with the distinct condensed areas (the light area mostly in the center of cell, Fig. 6). The condensation of the cytoplasm must be due to the denaturing and precipitation of proteins and other cytoplasmic constituents induced by the PDI treatment with CDots, as similar condensed cytoplasm was observed in cells treated by other bactericidal agents,40–42 for which an example is the condensation in Listeria innocua cells treated with electrolyzed water and acidic sanitizers.41 Also consistent with the cell membrane damages and increased permeability was the observed release of more extracellular vesicles by the treated cells than the control cells.
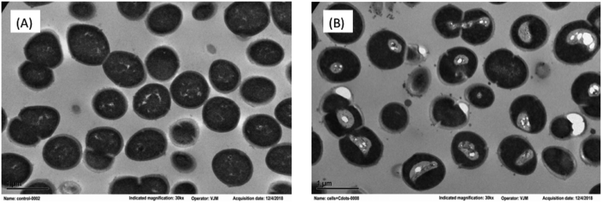 | ||
| Fig. 6 TEM images of E. faecium cells (A) without treatment, and (B) post-treatment with 1.2 μM PEI600–CDots under the 11 W visible LED for 1 h. | ||
In conclusion, CDots as structurally and composition-wise simple carbon-based nanomaterials could readily be activated by visible light to inactivate effectively and efficiently different strains of MDR Enterococcus. Mechanistically the inactivation is credited to the photodynamic effect, for which a special advantage of CDots is for the initially produced redox pairs upon photoexcitation to contribute substantially to the killing of the targeted bacterial cells (in addition to the actions of ROS generated in processes similar to those found in traditional photosensitization). The reported results further establish CDots as a new class of promising visible/natural light-activated broad spectral antibacterial agents, including especially their being capable of killing MDR bacterial pathogens in addition to various non-resistant bacterial species previously reported. With their simple structure and composition and their preparation from benign and abundant precursors, CDots may find broad applications in combating MDR bacterial infections through effective inactivation of MDR pathogens at various contaminated sites in their spread/transmission routes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from NSF grants 1701399, 1701424 and 1855905, and USDA grant 2019-67018-29689 is gratefully acknowledged.References
- W. H. Organization, Antimicrobial Resistance: Global Report on Surveillance, the WHO Document Production Services, Geneva, Switzerland, 2014.
- L. B. Rica, J. Infect. Dis., 2008, 197, 1079–1081 CrossRef PubMed.
- A. I. Hidron, J. R. Edwards, J. Patel, T. C. Horan, D. M. Sievert, D. A. Pollock, S. K. Fridkin, T. National Healthcare Safety Network and F. Participating National Healthcare Safety Network, Infect. Control Hosp. Epidemiol., 2008, 29, 996–1011 CrossRef.
- D. R. Schaberg, D. H. Culver and R. P. Gaynes, Am. J. Med., 1991, 91, 72S–75S CrossRef CAS.
- T. G. Emori and R. P. Gaynes, Clin. Microbiol. Rev., 1993, 6, 428–442 CrossRef CAS PubMed.
- W. R. Jarvis, CDC, 1996, 1–7 CAS.
- C. A. Arias, G. A. Contreras and B. E. Murray, Clin. Microbiol. Infect., 2010, 16, 555–562 CrossRef CAS.
- B. Hota, Clin. Infect. Dis., 2004, 39, 1182–1189 CrossRef PubMed.
- M. M. Huycke, D. F. Sahm and M. S. Gilmore, Emerging Infect. Dis., 1998, 4, 239–249 CrossRef CAS.
- R. M. van Harten, R. J. L. Willems, N. I. Martin and A. P. A. Hendrickx, Trends Microbiol., 2017, 25, 467–479 CrossRef CAS PubMed.
- B. L. Hollenbeck and L. B. Rice, Virulence, 2012, 3, 421–433 CrossRef PubMed.
- S. Chang, D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin and T. Vancomycin-Resistant Staphylococcus aureus Investigative, N. Engl. J. Med., 2003, 348, 1342–1347 CrossRef PubMed.
- A. J. Ray, N. J. Pultz, A. Bhalla, D. C. Aron and C. J. Donskey, Clin. Infect. Dis., 2003, 37, 875–881 CrossRef PubMed.
- D. E. J. G. J. Dolmans, D. Fukumura and R. K. Jain, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef CAS PubMed.
- L. B. Josefsen and R. W. Boyle, Met.-Based Drugs, 2008, 2008, 276109 Search PubMed.
- C. M. Courtney, S. M. Goodman, J. A. McDaniel, N. E. Madinger, A. Chatterjee and P. Nagpal, Nat. Mater., 2016, 15, 529–534 CrossRef CAS PubMed.
- C. M. Courtney, S. M. Goodman, T. A. Nagy, M. Levy, P. Bhusal, N. E. Madinger, C. S. Detweiler, P. Nagpal and A. Chatterjee, Sci. Adv., 2017, 3, e1701776 CrossRef PubMed.
- Y. P. Sun, US Pat. 7,829,772 B2, 2010 Search PubMed.
- Y. P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. F. Wang, P. J. G. Luo, H. Yang, M. E. Kose, B. L. Chen, L. M. Veca and S. Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- D. I. Abu Rabe, M. M. Al Awak, F. Yang, P. A. Okonjo, X. Dong, L. R. Teisl, P. Wang, Y. Tang, N. Pan, Y. P. Sun and L. Yang, Int. J. Nanomed., 2019, 14, 2655–2665 CrossRef CAS PubMed.
- M. M. Al Awak, P. Wang, S. Wang, Y. Tang, Y. P. Sun and L. Yang, RSC Adv., 2017, 7, 30177–30184 RSC.
- X. Dong, A. E. Bond, N. Pan, M. Coleman, Y. Tang, Y. P. Sun and L. Yang, Int. J. Nanomed., 2018, 13, 8025–8035 CrossRef CAS PubMed.
- Y. Hu, M. M. Al Awak, F. Yang, S. Yan, Q. Xiong, P. Wang, Y. Tang, L. Yang, G. E. LeCroy, X. Hou, C. E. Bunker, L. Xu, N. Tomlinson and Y. P. Sun, J. Mater. Chem. C, 2016, 4, 10554–10561 RSC.
- M. J. Meziani, X. Dong, L. Zhu, L. P. Jones, G. E. LeCroy and F. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 10761–10766 CrossRef CAS PubMed.
- L. Cao, M. J. Meziani, S. Sahu and Y. P. Sun, Acc. Chem. Res., 2013, 46, 171–180 CrossRef CAS PubMed.
- G. E. LeCroy, S. K. Sonkar, F. Yang, L. M. Veca, P. Wang, K. N. Tackett, J. J. Yu, E. Vasile, H. J. Qian, Y. M. Liu, P. Luo and Y. P. Sun, ACS Nano, 2014, 8, 4522–4529 CrossRef CAS PubMed.
- X. Y. Ren, W. X. Liang, P. Wang, C. E. Bunker, M. Coleman, L. R. Teisl, L. Cao and Y. P. Sun, Carbon, 2019, 141, 553–560 CrossRef CAS.
- J.-H. Liu, Y. Wang, G.-H. Yan, F. Yang, H. Gao, Y. Huang, H. Wang, P. Wang, L. Yang, Y. Tang, L. R. Teisl and Y.-P. Sun, J. Nanosci. Nanotechnol., 2019, 19, 2130–2137 CrossRef CAS PubMed.
- Y. Wang, P. Anilkumar, L. Cao, J. H. Liu, P. G. Luo, K. N. Tackett, 2nd, S. Sahu, P. Wang, X. Wang and Y. P. Sun, Exp. Biol. Med., 2011, 236, 1231–1238 CrossRef CAS PubMed.
- S. T. Yang, X. Wang, H. Wang, F. Lu, P. G. Luo, L. Cao, M. J. Meziani, J. H. Liu, Y. Liu, M. Chen, Y. Huang and Y. P. Sun, J. Phys. Chem. C, 2009, 113, 18110–18114 CrossRef CAS PubMed.
- W. Liang, C. E. Bunker and Y.-P. Sun, ACS Omega, 2020, 5, 965–971 CrossRef CAS PubMed.
- L. J. Marnett, Carcinogenesis, 2000, 21, 361–370 CrossRef CAS PubMed.
- E. R. Stadtman and R. L. Levine, Ann. N. Y. Acad. Sci., 2000, 899, 191–208 CrossRef CAS PubMed.
- S. Yla-Herttuala, Ann. N. Y. Acad. Sci., 1999, 874, 134–137 CrossRef CAS PubMed.
- J. Ge, M. Lan, B. Zhou, W. Liu, L. Guo, H. Wang, Q. Jia, G. Niu, X. Huang, H. Zhou, X. Meng, P. Wang, C. S. Lee, W. Zhang and X. Han, Nat. Commun., 2014, 5, 4596 CrossRef CAS PubMed.
- P. Juzenas, A. Kleinauskas, P. G. Luo and Y. P. Sun, Appl. Phys. Lett., 2013, 103, 063701 CrossRef.
- Z. M. Markovic, B. Z. Ristic, K. M. Arsikin, D. G. Klisic, L. M. Harhaji-Trajkovic, B. M. Todorovic-Markovic, D. P. Kepic, T. K. Kravic-Stevovic, S. P. Jovanovic, M. M. Milenkovic, D. D. Milivojevic, V. Z. Bumbasirevic, M. D. Dramicanin and V. S. Trajkovic, Biomaterials, 2012, 33, 7084–7092 CrossRef CAS PubMed.
- Y. N. Slavin, J. Asnis, U. O. Häfeli and H. Bach, J. Nanobiotechnol., 2017, 15, 65 CrossRef PubMed.
- L. Wang, H. He, Y. Yu, L. Sun, S. Liu, C. Zhang and L. He, J. Inorg. Biochem., 2014, 135, 45–53 CrossRef CAS PubMed.
- T. Dai, A. Gupta, Y.-Y. Huang, R. Yin, C. K. Murray, M. S. Vrahas, M. E. Sherwood, G. P. Tegos and M. R. Hamblin, Antimicrob. Agents Chemother., 2013, 57, 1238–1245 CrossRef CAS PubMed.
- L. Feliciano, J. Lee and M. A. Pascall, J. Food Sci., 2012, 77, M182–187 CrossRef CAS PubMed.
- B. Sampathkumar, G. G. Khachatourians and D. R. Korber, Appl. Environ. Microbiol., 2003, 69, 122–129 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ma00078g |
| This journal is © The Royal Society of Chemistry 2020 |