Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Photoenzymatic epoxidation of styrenes†
Morten M. C. H.
van Schie
 a,
Caroline E.
Paul
a,
Caroline E.
Paul
 b,
Isabel W. C. E.
Arends
b,
Isabel W. C. E.
Arends
 c and
Frank
Hollmann
c and
Frank
Hollmann
 *a
*a
aDepartment of Biotechnology, Delft University of Technology, van der Maasweg 9, 2629 HZ Delft, The Netherlands. E-mail: f.hollmann@tudelft.nl
bLaboratory of Organic Chemistry, Wageningen University & Research, Stippeneng 4, 6708WE Wageningen, The Netherlands
cFaculty of Science, University of Utrecht, The Netherlands
First published on 22nd January 2019
Abstract
Two-component-diffusible-flavomonooxygenases are versatile biocatalysts for selective epoxidation-, hydroxylation- or halogenation reactions. Their complicated molecular architecture can be simplified using photochemical regeneration of the catalytically active, reduced FADH2 prosthetic group. In this contribution we provide the proof-of-concept and characterization for the direct regeneration of the styrene monooxygenase from Pseudomonas.
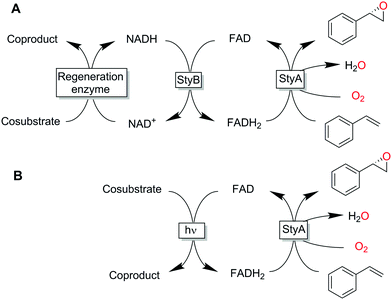
So-called two-component, diffusible flavin monooxygenases (2CDFMOs) are a diverse and preparatively highly interesting class of enzymes. For example, 2CDFMs catalyse regioselective aromatic hydroxylations and halogenation reactions as well as stereoselective epoxidation reactions.1–4
For this, 2CDFMOs rely on the reductive activation of molecular oxygen mediated by an enzyme-bound, reduced flavin cofactor (generally the reduced form of flavin adenine dinucleotide, FADH2). FADH2 itself is regenerated by a NAD(P)H-dependent reductase. There is an ongoing debate on the mechanism on how FADH2 reaches the monooxygenase subunit. Some studies suggest a freely diffusible FADH25 while others found indications for a complex between the reductase- and monooxygenase subunits thereby channelling the reduced flavin and protecting it from spontaneous aerobic reoxidation.6–8 The complicated molecular architecture of 2CDFMOs poses a challenge for their preparative application, which is mostly addressed by whole-cell systems.9–12 More recently, also fusion proteins combining the reductase- and monooxygenase subunits in one polypeptide chain are moving into the research focus.13,14 Reactions utilising isolated enzymes require the entire cascade outlined in Scheme 1.15–17 Hence, it is no surprise that alternative, more direct and simple regeneration systems for the reduced flavin cofactor have been evaluated. Examples include, transition metal-catalysed reduction of FAD,18,19 direct electrochemical regeneration20,21 or using chemical reductants.22
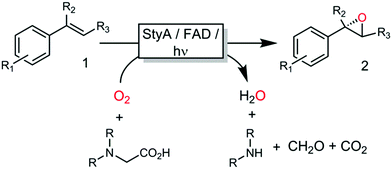
All these methods, however, despite significantly simplifying the reaction scheme, exhibited drawbacks such as reliance on specialized equipment or dependency on costly and enzyme-inactivating transition metal complexes. Instead, direct photochemical regeneration of reduced enzyme prosthetic groups is gaining increasing attention.23,24 We therefore set out to explore the possibility of direct, photocatalytic regeneration of FADH2 to promote StyA-catalysed epoxidations of styrene and its derivatives (Scheme 1, bottom). Very recently, Kottke and coworkers reported the successful application of this approach to promote a 2CDFM-driven halogenation reaction.25
The biocatalyst used in our study was styrene monooxygenase from Pseudomonas sp. VLB120 (StyA).5,26 The enzyme was recombinantly expressed in Escherichia coli and purified in one step yielding approximately 0.11 g of technically pure StyA per gram of cell free extract. Crude cell free extracts (CFE) were used for the first experiments (Fig. S1, ESI†). Even though the CFE contained significant catalase activity (Fig. S12, ESI†), we routinely added catalase externally to circumvent any possible negative effect of stemming from the spontaneous aerobic reoxidation of FADH2.27 Pleasingly, already in a first experiment under arbitrary reaction conditions, catalytic turnover and production of enantiopure (S)-styrene oxide was observed (Fig. S3, ESI†). It is worth mentioning here that all negative control experiments (i.e. performing the reaction under identical conditions while leaving out one of the reaction components StyA, FAD, EDTA or light) did not yield detectable product formation (data not shown).
Encouraged by these results, the substrate scope of this photoenzymatic reaction system was investigated. As shown in Table 1, both the relative reaction rates and the enantioselectivity of the photoenzymatic epoxidation reaction are comparable to results reported previously.11
| Product | R1 | R2 | R3 | [Product] [mM] | ee [%] |
|---|---|---|---|---|---|
| Conditions: [substrate]0 = 5 mM, [StyA] = 5.3 μM, [FAD] = 200 μM, [EDTA] = 20 mM, [catalase] = 600 U ml−1, [DMSO] = 1.25% (v/v), 100 mM KPi buffer pH 7, 35 °C, stirring at 300 rpm, light intensity of 40% for 1 h. The standard deviations represent those for three independent experiments. | |||||
| 2a | H | H | H | 0.60 ± 0.04 | >99 |
| 2b | m-CH3 | H | H | 2.12 ± 0.11 | 98 |
| 2c | p-F | H | H | 0.86 ± 0.13 | >99 |
| 2d | m-Cl | H | H | 3.10 ± 0.19 | 95 |
| 2e | p-F | H | H | 1.06 ± 0.14 | 97 |
| 2f | p-Br | H | H | 0.65 ± 0.15 | >99 |
| 2g | H | H | CH3 | 1.45 ± 0.09 | >99 |
| 2h | H | CH3 | H | 0.40 ± 0.12 | >99 |
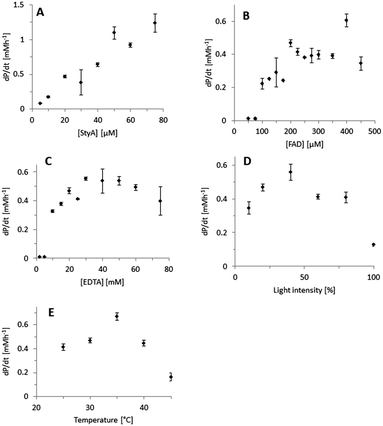
Indeed, excellent enantiomeric excess was achieved (95 to >99%). In order to further understand this system, we further characterised the influence of the single reaction components on the efficiency of the overall reaction using the technically pure StyA (Fig. 1). The product formation rate correlated linearly with the biocatalyst concentration applied (Fig. 1A), indicating that the biocatalytic step was overall rate-limiting.
Also increasing the concentration of the photocatalyst increased the overall product formation reaction (Fig. 1B) indicating that the concentration of the photoexcited FAD was rate-limiting as well. Below an FAD concentration of 100 μM no product formation was detectable, which may be attributed to the corresponding low concentration of FADH2 and inefficient utilisation by StyA.28,29 Above approximately 200 μM FAD, no further acceleration of the reaction rate was observed. Possibly this can be attributed to the decreasing optical transparency of the reaction mixture at elevated FAD concentration resulting in a complete utilization of all photons offered to the reaction system. Alternatively, elevated FAD concentrations may also favour the (undesired) futile oxidation of FADH2 to FAD and H2O2 as observed in previous experiments.27 Similar observations have been made previously.19,20,30 Variation of the concentration of the sacrificial electron donor (EDTA) had a similar effect (Fig. 1C). Additionally, the light intensity significantly influenced the overall reaction (Fig. 1D). To a certain extent, brighter reaction conditions favoured increased product formation. However, at very high light intensities also a dramatic reduction of the reaction rate was observed. We attribute this to an increased photobleaching of FAD leading to flavin degradation products,31 which are not accepted by StyA as prosthetic group. Finally, it is worth mentioning that an apparent optimal temperature of ca. 35 °C was observed (Fig. 1E). This is perfectly in line with the mesophilic character of the original host of StyA.
One shortcoming of the current photoenzymatic reaction setup, however, is the comparably poor robustness of the reactions. In general, after reaction times of ca. 1 h, no further conversion could be detected. In order to shed light on the reason for this limitation, a range of control experiments was conducted (Fig. S13, ESI†). The biocatalyst (StyA) itself was stable under the reaction conditions (Fig. S13, ESI,† column 2). However, when illuminating the photocatalyst (FAD) for 30 min prior start of the reaction (Fig. S13, ESI,† columns 3 and 4) significantly reduced styrene oxide accumulation was observed. This inactivation was almost complete illuminating FAD alone and was somewhat less pronounced in the presence of EDTA (serving as reducing agent for photoexcited FAD; the resulting reduced flavin being less photoactive). Clearly, the photoinstability of FAD represents the major limitation of the current reaction setup. This is also supported by the changes of the FAD spectrum upon illumination (Fig. S14, ESI†) and is in line with the well-known photodegradation of FAD to lumichrome.32 While the latter still possesses the desired photochemical properties it is not accepted by StyA as a prosthetic group, thereby explaining the poor robustness of the current reaction setup.
Overall, in the present study, we have provided a proof-of-concept for the direct, photochemical regeneration of styrene monooxygenase for the generation of enantiomerically pure epoxides. The photoinstability of the photocatalyst/prosthetic group FAD was identified as the major limitation of the current setup. Even though at the present stage of development this system is not suitable for preparative application we are convinced that it's conceptual simplicity will convince others to further improve the system.
We thank the Netherlands Organization for Scientific Re-search for financial support through a VICI grant (No. 724.014.003). CEP acknowledges a VENI grant (No. 722.015.011). Furthermore, we would like to thank Maarten Gorseling and Remco van Oosten for excellent technical support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. M. E. Huijbers, S. Montersino, A. H. Westphal, D. Tischler and W. J. H. van Berkel, Arch. Biochem. Biophys., 2014, 544, 2–17 CrossRef CAS PubMed.
- T. Heine, W. van Berkel, G. Gassner, K.-H. van Pée and D. Tischler, Biology, 2018, 7, 42 CrossRef PubMed.
- R. D. Ceccoli, D. A. Bianchi and D. V. Rial, Front. Microbiol., 2014, 5, 25 Search PubMed.
- W. Dijkman, G. Gonzalo, A. Mattevi and M. Fraaije, Appl. Microbiol. Biotechnol., 2013, 97, 5177–5188 CrossRef CAS PubMed.
- K. Otto, K. Hofstetter, M. Rothlisberger, B. Witholt and A. Schmid, J. Bacteriol., 2004, 186, 5292–5302 CrossRef CAS PubMed.
- A. Kantz, F. Chin, N. Nallamothu, T. Nguyen and G. T. Gassner, Arch. Biochem. Biophys., 2005, 442, 102–116 CrossRef CAS PubMed.
- A. Kantz and G. T. Gassner, Biochemistry, 2011, 50, 523–532 CrossRef CAS PubMed.
- E. Morrison, A. Kantz, G. T. Gassner and M. H. Sazinsky, Biochemistry, 2013, 52, 6063–6075 CrossRef CAS PubMed.
- B. Buehler, J. B. Park, L. M. Blank and A. Schmid, Appl. Environ. Microbiol., 2008, 74, 1436–1446 CrossRef CAS PubMed.
- J. B. Park, B. Buehler, T. Habicher, B. Hauer, S. Panke, B. Witholt and A. Schmid, Biotechnol. Bioeng., 2006, 95, 501–512 CrossRef CAS PubMed.
- A. Schmid, K. Hofstetter, H.-J. Feiten, F. Hollmann and B. Witholt, Adv. Synth. Catal., 2001, 343, 732–737 CrossRef CAS.
- H. Toda, T. Ohuchi, R. Imae and N. Itoh, Appl. Environ. Microbiol., 2015, 81, 1919–1925 CrossRef CAS PubMed.
- T. Heine, K. Tucker, N. Okonkwo, B. Assefa, C. Conrad, A. Scholtissek, M. Schlomann, G. Gassner and D. Tischler, Appl. Biochem. Biotechnol., 2017, 181, 1590–1610 CrossRef CAS PubMed.
- M. L. Corrado, T. Knaus and F. G. Mutti, ChemBioChem, 2018, 19, 679–686 CrossRef CAS PubMed.
- K. Hofstetter, J. Lutz, I. Lang, B. Witholt and A. Schmid, Angew. Chem., Int. Ed., 2004, 43, 2163–2166 CrossRef CAS PubMed.
- H. Toda, R. Imae and N. Itoh, Tetrahedron: Asymmetry, 2012, 23, 1542–1549 CrossRef CAS.
- H. Toda, R. Imae, T. Komio and N. Itoh, Appl. Microbiol. Biotechnol., 2012, 96, 407–418 CrossRef CAS PubMed.
- S. Unversucht, F. Hollmann, A. Schmid and K.-H. van Pée, Adv. Synth. Catal., 2005, 347, 1163–1167 CrossRef CAS.
- F. Hollmann, P. C. Lin, B. Witholt and A. Schmid, J. Am. Chem. Soc., 2003, 125, 8209–8217 CrossRef CAS PubMed.
- R. Ruinatscha, C. Dusny, K. Buehler and A. Schmid, Adv. Synth. Catal., 2009, 351, 2505–2515 CrossRef CAS.
- F. Hollmann, K. Hofstetter, T. Habicher, B. Hauer and A. Schmid, J. Am. Chem. Soc., 2005, 127, 6540–6541 CrossRef CAS PubMed.
- C. E. Paul, D. Tischler, A. Riedel, T. Heine, N. Itoh and F. Hollmann, ACS Catal., 2015, 5, 2961–2965 CrossRef CAS.
- W. Zhang and F. Hollmann, Chem. Commun., 2018, 54, 7281–7289 RSC.
- S. H. Lee, D. S. Choi, S. K. Kuk and C. B. Park, Angew. Chem., Int. Ed., 2018, 57, 7958–7985 CrossRef CAS PubMed.
- L. Schroeder, M. Frese, C. Müller, N. Sewald and T. Kottke, ChemCatChem, 2018, 10, 3336–3341 CrossRef CAS.
- S. Panke, B. Witholt, A. Schmid and M. G. Wubbolts, Appl. Environ. Microbiol., 1998, 64, 2032–2043 CAS.
- D. Holtmann and F. Hollmann, ChemBioChem, 2016, 17, 1391–1398 CrossRef CAS PubMed.
- D. Tischler, R. Kermer, J. A. D. Groning, S. R. Kaschabek, W. J. H. van Berkel and M. Schlomann, J. Bacteriol., 2010, 192, 5220–5227 CrossRef CAS PubMed.
- U. E. Ukaegbu, A. Kantz, M. Beaton, G. T. Gassner and A. C. Rosenzweig, Biochemistry, 2010, 49, 1678–1688 CrossRef CAS PubMed.
- F. Hollmann and A. Schmid, Biocatal. Biotransform., 2004, 22, 63–88 CrossRef CAS.
- W. Holzer, J. Shirdel, P. Zirak, A. Penzkofer, P. Hegemann, R. Deutzmann and E. Hochmuth, Chem. Phys., 2005, 308, 69–78 CrossRef CAS.
- P.-S. Song and D. E. Metzler, Photochem. Photobiol., 1967, 6, 691–709 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: For more details on the catalyst preparation, reaction procedure and GC chromatograms, etc. See DOI: 10.1039/c8cc08149b |
| This journal is © The Royal Society of Chemistry 2019 |