A review: the trend of progress about pH probes in cell application in recent years
Yongkang
Yue
a,
Fangjun
Huo
b,
Songyi
Lee
c,
Caixia
Yin
*a and
Juyoung
Yoon
*c
aKey Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education, Institute of Molecular Science, Key Laboratory of Materials for Energy Conversion and Storage of Shanxi Province, Shanxi University, Taiyuan 030006, China. E-mail: yincx@sxu.edu.cn
bResearch Institute of Applied Chemistry, Shanxi University, Taiyuan, 030006, China
cDepartment of Chemistry and Nano Science, Ewha Womans University, Seoul 120750, Korea. E-mail: jyoon@ewha.ac.kr
First published on 4th October 2016
Abstract
Intracellular pH values are some of the most important factors that govern biological processes and the acid–base homeostasis in cells, body fluids and organs sustains the normal operations of the body. Subcellular organelles including the acidic lysosomes and the alkalescent mitochondria undergo various processes such as intracellular digestion, ATP production and apoptosis. Due to their precise imaging capabilities, fluorescent probes have attracted great attention for the illustration of pH modulated processes. Furthermore, based on the unique acidic extracellular environment of acidic lysosomes, fluorescent probes can specifically be activated in cancer cells or tumors. In this review, recently reported lysosome and mitochondria specific pH imaging probes as well as pH-activatable cancer cell-targetable probes have been discussed.
Introduction
The hydronium ion is one of the most important species with crucial roles in biological systems. In organisms, the acid–base homeostasis in cells, body fluids and organs is a key factor for many processes such as proliferation and apoptosis, endocytosis, multidrug resistance, ion transport, and muscle contraction.1,2 The intracellular pH in a typical mammalian cell ranges from 4.5 in lysosomes to 8.0 in mitochondria.Acting as the peptic in cells, lysosomes contain a variety of hydrolytic enzymes which can degrade almost all kinds of endogenous and exogenous biomolecules.3,4 The acidic environment (pH 4.5–5.5) maintains the biological activity of these hydrolytic enzymes for digestion and degradation processes.5 During these metabolic processes, endogenous cell death effectors and reactive oxygen species stimulated by lysosomal membrane permeabilization (LMP) induce the release of hydrolytic enzymes in the cytosol and further initiate apoptosis.6 Alongside the degradation of cellular components by hydrolytic enzymes, cytosolic acidification further promotes cell death. For cancer cells, the pH values in both the cytosol and in lysosomes are lower than in normal cells.7,8 pH changes in the lysosomes and cytoplasm can reflect the cell status and metabolic processes.
Mitochondria are the main sites for aerobic respiration and the primary organelle for energy production. During the synthesis of adenosine 5′-triphosphate (ATP), proton extrusion provides an alkalescent system in mitochondria.9 Additionally, mitochondria participate in various metabolic processes including cell differentiation, calcium ion homeostasis and apoptosis.10,11
These pH dependent processes in the cytoplasm and in organelles sustain the operation of organisms and minor pH deviations can influence the physiological status. Diseases including cancer and Alzheimer's have been associated with abnormal pH values.12 Due to the momentous roles of pH values in cells, the ability to monitor and visualize pH values and distributions using non-invasive fluorescent probes is in high demand. Many studies have been conducted for this purpose in the past decade.13–19 Inchoate pH fluorescent probes with excellent cell membrane permeability and optical properties have been applied in cell imaging20–22 and further, to in vivo imaging using near infrared (NIR) fluorescence emission.23,24 In recent years, with the improvement of the resolution of confocal laser microscopes and fluorescent probe design, pH fluorescent probes for cellular imaging have provided deep insight into the more complicated and important subcellular structures like lysosomes and mitochondria, yielding a better understanding of pH related physiological and pathological processes.25,26 Furthermore, since the lysosome-targeting pH activatable fluorescent imaging probe was reported by Urano and co-workers in 2009,27 the pragmatic tumor cell specific labelling method has become increasingly attractive, especially in the past two years. In this review, we summarize the mitochondria and lysosome targetable pH probes reported in recent years and provide comprehensive details on their design strategies, sensing mechanisms and biological applications, especially in cancer cell imaging.
Non-specific fluorescent probes for pH detection
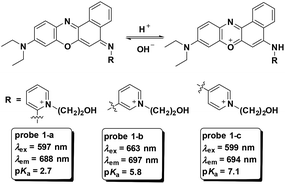
Fluorescent probes for pH detection are mainly based on two categories of design philosophy: (1) the acid–base neutralization reaction of the phenol, amine and N-heterocycles;1,17,28–30 and (2) nucleophilic addition reaction of the hydroxyl ion towards cyanine dyes.18,31 The optical responses in the above processes realized the detection of pH values. Particularly for those pH fluorescent probes with ratiometric fluorescence responses, the utilization of nigericin (K+/H+ ionophore) or monensin (Na+/H+ ionophore) to equilibrate the pH values in cells with the surrounding media promoted the quantitative detection of pH in living cells.32,33 In this section, we will discuss the representative cell imaging pH fluorescent probes reported in recent years.Based on the protonation of amine, three benzo[a]phenoxazine derivatives were developed as NIR fluorescent probes which featured pH decrease induced turn-on fluorescent responses for intracellular pH imaging by Liu and co-workers in 2013 (Fig. 1).29
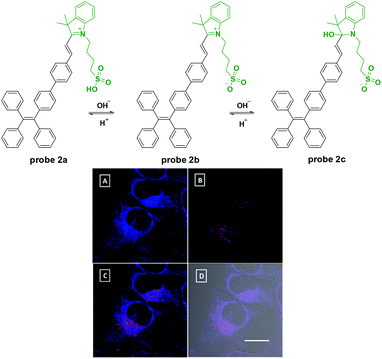
Tang's group reported a nucleophilic addition reaction based fluorescent probe for pH detection with AIE characteristics in 2013.18 After being modified with sulphonate, the tetraphenylethene cyanine derivative displayed two scope fluorescent responses with the altering of pH values. Probe 2 displays strong-to-medium red emissions at pH 5–7 (λex = 380 nm, λem = 615 nm), weak-to-no red emissions at pH 7–10, and no-to-strong blue emissions at pH 10–14 (λex = 380 nm, λem = 489 nm). In the acidic range, the increase of pH value would induce the equilibrium between sulfo acid and sulfonate in 2a which induced the fluorescence emission changes. Meanwhile, in the alkaline range, the pH higher than 10 would induce the nucleophilic addition reaction of the hydroxyl group and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N in the indolinium unit. This process broke the conjugation system in 2b and further induced the fluorescent responses. The pKa value of probe 2 in PBS buffer was 10. In further cell experiments with HeLa cells, the probe exhibits good cell-permeability and biocompatibility. And, interestingly, lipid compartments such as 1,2-dioleoyl-glycero-3-phosphocholine could facilitate the response process and shift its pKa to intracellular physiological pH range which promoted probe 2 to visualizing acidic and basic compartments with red and blue emissions in cell labeling, respectively. Furthermore, the ratiometric red blue emission signals can be employed as an indicator of local proton concentration. This probe has great potential for high-resolution and high throughput analysis of intracellular environments which may find an array of biomedical applications in cancer diagnosis and drug screening (Fig. 2).
N in the indolinium unit. This process broke the conjugation system in 2b and further induced the fluorescent responses. The pKa value of probe 2 in PBS buffer was 10. In further cell experiments with HeLa cells, the probe exhibits good cell-permeability and biocompatibility. And, interestingly, lipid compartments such as 1,2-dioleoyl-glycero-3-phosphocholine could facilitate the response process and shift its pKa to intracellular physiological pH range which promoted probe 2 to visualizing acidic and basic compartments with red and blue emissions in cell labeling, respectively. Furthermore, the ratiometric red blue emission signals can be employed as an indicator of local proton concentration. This probe has great potential for high-resolution and high throughput analysis of intracellular environments which may find an array of biomedical applications in cancer diagnosis and drug screening (Fig. 2).
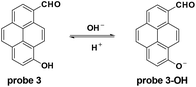
The Yang group modified pyrene to synthesize probe 3 which was enfolded with a polyurethane nanogel to improve the water solubility for pH detection in vitro and in vivo with ratiometric fluorescent responses.34 Accompanied by the pH increase from 4.0 to 10.0, the fluorescence emission changed from 510 nm to 610 nm and the pKa value was 7.8 ± 0.1. With good photostability and permeability, and nontoxicity, the probe was applied for intracellular pH detection in NIH/3T3 fibroblast cells with the culturing of nigericin in high K+ buffers. Furthermore, the probe could image the intracellular pH changes induced by ROS (Fig. 3).
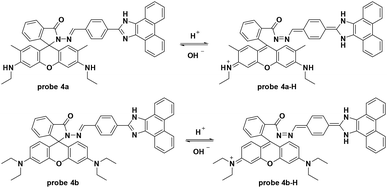
By modifying the rhodamine derivative, the Das group developed a new fluorescence resonance energy transfer (FRET) based fluorescent probe for pH detection.35 The pH decrease from 8.0 to 3.5 induced quenching of the original fluorescence emission at 493 nm and enhanced a new emission at 552 nm in PBS buffer–DMSO (98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). Probe 4a was inert towards various biothiols and metal ions including Na+, K+, Mg2+, Hg2+, Cu2+, Zn2+, Cr3+, Fe3+, Co2+, Cr3+, Pd2+, Pb2+, Ni2+, and Cd2+. Further cell experiments using Hct116 cells displayed low toxicity of probe 4a and co-imaging with Endoplasmic Reticulum Tracker Green demonstrated that the probe was mainly accumulated in the lipid dense region of the Hct116 cells. For further application, the probe could be used for pH imaging under the dexamethasone induced intracellular pH changes in Hct116 cells. Similar to probe 4a, the hydrazino containing probe 4b featured a hydrion induced ring-open process with turn-on fluorescence emission at 587 nm when excited at 530 nm (Fig. 4).36,37
2, v/v). Probe 4a was inert towards various biothiols and metal ions including Na+, K+, Mg2+, Hg2+, Cu2+, Zn2+, Cr3+, Fe3+, Co2+, Cr3+, Pd2+, Pb2+, Ni2+, and Cd2+. Further cell experiments using Hct116 cells displayed low toxicity of probe 4a and co-imaging with Endoplasmic Reticulum Tracker Green demonstrated that the probe was mainly accumulated in the lipid dense region of the Hct116 cells. For further application, the probe could be used for pH imaging under the dexamethasone induced intracellular pH changes in Hct116 cells. Similar to probe 4a, the hydrazino containing probe 4b featured a hydrion induced ring-open process with turn-on fluorescence emission at 587 nm when excited at 530 nm (Fig. 4).36,37
Mitochondria targeted fluorescent probes for pH detection
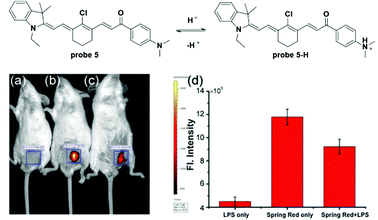
Mitochondria contain outer and inner membranes composed of phospholipid bilayers and proteins. The electrochemical potential across the mitochondrial inner membrane can accumulate lipophilic cations (as shown in Fig. 5) according to the Nernst equation.11,38 Applications of these lipophilic cations to fluorophores, various mitochondria-targetable fluorescent probes for reactive oxygen species (ROS),39–41 thiols42,43 and pH26,44 detection have been reported. For most of these probes, the cation moieties do not participate in the detection process and are used only for mitochondria localization. In this section, we discuss the fluorescent probes for mitochondria pH detection reported in the last two years.Li and co-workers utilized the cyanine moiety to synthesize a series of dyes for mitochondria labelling in 2014 (Fig. 6).44 The dimethylamino containing probe 5 displayed turn-on NIR fluorescent responses (λex = 530 nm, λem = 680 nm) to pH values ranging from 5.0 to 8.0, with large Stokes shifts owing to the asymmetric structure. The resonance structure of the cyanine moiety provided a lipophilic cation that improved the mitochondria-targetable properties of this series of fluorescent probes. Optical analysis demonstrated the excellent selectivity of probe 5 for various ions and ROS including K+, Na+, Ca2+, Mg2+, Zn2+, Cu2+, Mn2+, Co2+, NH4+, Cl−, SO42−, H2O2, ClO−, O2˙−, ONOO−, and ˙OH and the pKa value of probe 5 was determined to be 6.3. Co-localization imaging experiments using probe 5 with Mito-Tracker Green (MTG) demonstrated its mitochondria targetable property, which was further confirmed by the bioimaging of zebrafish models. Further cellular experiments with probe 5 for different pH values revealed the linear relationship of fluorescence intensities with pH values in HepG2 cells. The probe could be used for visualizing lactate and pyruvate induced mitochondria acidification, pH decrease during apoptosis and in lipopolysaccharide (LPS)-induced inflammatory tissues in mice models.
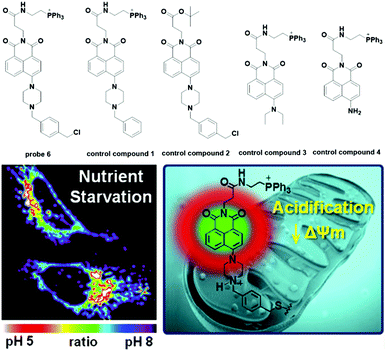
Later, Lee modified naphthalimide (fluorophore) with a triphenylphosphonium cation and benzyl chloride, which acted as the mitochondria targeting moiety and the immobility moiety respectively when reacted with mitochondrial proteins containing thiols, to synthesize probe 6 for mitochondria pH detection (Fig. 7).26 The naphthalimide-connected piperazine works as the pH sensitive switch through the photoinduced electron transfer (PET) process. Optical and cellular experiments with control compounds demonstrated the design strategies mentioned above. Control compound 1 displayed turn-on fluorescent responses for pH values in the 11 to 2 range and the pKa value was 6.18 ± 0.049. With inert responses to other metal ions, thiols and H2O2 and negligible cytotoxicity, probe 6 was successfully applied for quantitative measurement of pH changes in the mitochondria of HeLa cells through ratiometric imaging with Mito-Tracker Red, showing a pKa of 7.99 ± 0.49 in normal HeLa cells. Further experiments displayed that probe 6 could be used to identify mitochondrial heterogeneity. Interestingly, the probe could map the nutrient deprivation induced mitochondrial acidification and visualize the mitophagy process within autolysosomes when co-cultured with Lyso-Tracker Red (LTR).
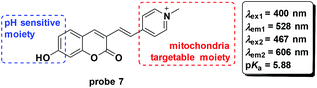
To develop fluorescent probes for ratiometric imaging of pH changes in mitochondria, Wu and co-workers connected the coumarin and pyridinium moieties to synthesize probe 7 based on pH controlled phenol and phenate transformation (Fig. 8).45 Optical experiments displayed a potential application in bioimaging and mitochondria pH detection. Co-culturing probe 7 and Mito-Tracker Deep Red with HeLa and 293T cells demonstrated the mitochondria labelling property. The probe could image the mitochondrial acidification induced by H2O2.
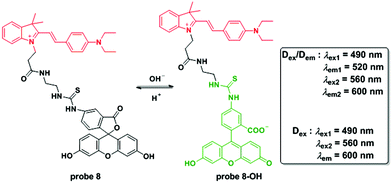
Chen and co-workers connected a cyanine group and fluorescein isothiocyanate to construct a ratiometric fluorescent system for pH detection through a dual excitation/dual emission mode (Dex/Dem) and a dual excitation (Dex) mode.46 The fluorescence ratios (F520/F600) of probe 8 for pH values ranging from 4.85 to 9.65 increase from 0.023 to 0.99 and the pKa value was determined to be 7.33 ± 0.03. The reversibility of the probe in the pH range of 5.00 to 9.00 persisted for 5 cycles. Cell experiments confirmed the mitochondria targetable property of probe 8 with the co-culturing of Mito-Tracker Deep Red 633 using MCF-7 cells. Further imaging upon the culture of probe 8-loaded MCF-7 cells with high K+ buffers at different pH values containing nigericin demonstrated the applicability of 8 for quantitative detection of pH changes in mitochondria. The results were further supported by ratiometric flow cytometry experiments. As a result of its excellent biological properties, probe 8 was used to monitor the pH fluctuation stimulated by carbonyl cyanide m-chlorophenylhydrazone (CCCP) used for mitochondrial proton gradient uncoupling across the inner membrane and the results were positive. Importantly, the probe could visualize the ROS induced mitochondrial acidification stimulated by phorbol myristate acetate (PMA), a stimulant of intracellular ROS generation (Fig. 9).
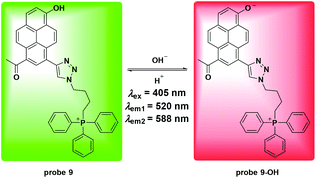
Similar to the above mentioned studies, Cao et al. utilized the pyrene moiety and triphenylphosphonium cation to synthesize probe 9 for mitochondria pH detection.47 As shown in Fig. 10, the probe displayed ratiometric fluorescent changes with pH values ranging from 4.0 to 11.0 and the pKa value was calculated to be 7.33 ± 0.04. Co-localizing experiments involving 9 and MTR with HeLa cells and NIH/3T3 cells revealed a mitochondria targeting property. The calibrated curve was further obtained to detect the pH values in mitochondria. Attractively, the probe displayed high thermostability under pH changes, which prompted the use of 9 for visualizing temperature induced pH fluctuation in mitochondria. The results showed that both cold stress and heat stress led to mitochondrial dysfunction. The application of H2O2 induced significant pH increase in both tumor and normal cells. However, tumor cells displayed faster increasing rates compared to normal cells, which might present a strategy for tumor cell discrimination from normal cells.
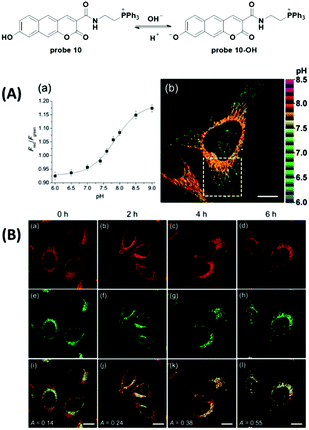
Recently, Sarkar et al. reported a two-photon fluorescent probe for ratiometric detection of pH values (Fig. 11).48 The increase of pH from 4.0 to 10.0 induced one-photon fluorescence emission changes in probe 10 from 542 nm to 604 nm with excitation at 370 nm and 453 nm, respectively. Co-localization experiments with 10 and mCherrymito-7, a red-emissive mitochondria labelling protein, demonstrated mitochondria specific accumulation. Two-photon ratiometric imaging (Fred/Fgreen) of 10 loaded HeLa cells was used to obtain the calibration curve and the in situ pKa value was 7.86 ± 0.05. Importantly, two-photon imaging of 10 loaded HeLa cells displayed distinct ratio differences within the whole cell as well as the heterogeneity of mitochondria pH values and further reflected the metabolic state. Treatment with CCCP induced prominent pH decrease originating from perinuclear mitochondria and extending to the periphery. The nutrient deprivation-induced mitochondria autophagy was observed with the co-culturing of the two-photon Lyso-Tracker and BLT-blue in HeLa cells. Furthermore, the probe could be used for pH detection in an astrocyte mitochondria model of Parkinson's disease and for rat hippocampus pH distribution imaging.
Lysosome targeted fluorescent probes for pH detection
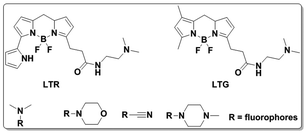
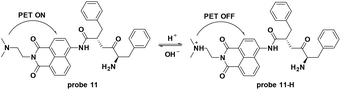
Lysosomes are membrane-bound vesicles found in almost all eukaryotic cells. Small amphiphilic molecules are lysosome membrane-permeable in the dissociative state but impermeable after protonation in acidic lysosomes. This promotes the accumulation of alkalescent moieties containing amphiphilic molecules like LTR and Lyso-Tracker Green (LTG).49–51 As shown in Fig. 12, structures including dimethylamino, cyan and morpholine have been applied to fluorescent probe design for lysosome specific labelling. In this section, these probes reported within the past 5 years are reviewed.To develop lysosome-targeted fluorescent probes with large Stokes shifts, Chen et al. modified naphthalimide to synthesize probe 11 for pH detection (Fig. 13).52 Low pH triggered PET off between the dimethylamino (lysosome labelling anchor) and naphthalimide, releasing fluorescence emission at 465 nm in Britton–Robinson buffer. The pKa value was 6.48 and the probe could be used for ES-2 cells pH imaging.
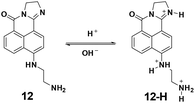
In 2013, the Zhou group utilized the naphthalimides as fluorophores to synthesize probe 12 for pH imaging in lysosomes.53 As shown in Fig. 14, the protonation process of 12 induced turn-off fluorescent responses with the excitation at 425 nm and ratiometric fluorescent responses with the excitation at 480 nm. The pKa value was calculated to be 5.88. With good water solubility and cell permeability, the probe was applied for lysosome pH imaging in HeLa cells.
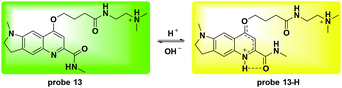
Li and co-workers utilized the quinoline moiety to develop a new ratiometric fluorescent probe 13 for lysosome pH detection (Fig. 15).54 The protonation activatable resonance charge transfer process induced fluorescence emission changes from 570 nm to 494 nm within the pH range of 3.0 to 6.0. The corresponding ratio of F494 nm/F570 nm changed 105-fold and the pKa value was 4.20 ± 0.015. Co-culturing 13 and LTR demonstrated the lysosome labelling property. Furthermore, NIH 3T3 cells were used to quantitatively detect the pH values of the lysosomes. The obtained pH for the lysosomes in NIH 3T3 cells was 4.25 ± 0.34 and both NH4Cl and chloroquine could induce lysosomal alkalization, which further supported the application of 13 for ratiometric detection of lysosomal pH fluctuation. Oxidative stress caused by exogenous H2O2 and endogenous GSH deficiency induced significant lysosomal alkalization whereas N-acetylcysteine caused a slight pH decrease. Treatment with exogenous NaClO did not induce significant pH changes in NIH 3T3 cells.
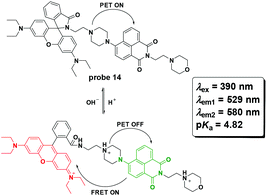
Zhang et al. reported probe 14 for lysosome pH imaging based on the protonation-induced PET OFF of the piperazine moiety to a naphthalimide derivative and further FRET from the naphthalimide moiety to the ring-open rhodamine moiety (Fig. 16).55 With excellent selectivity and low cytotoxicity, the probe was successfully used for pH imaging in HeLa cells.
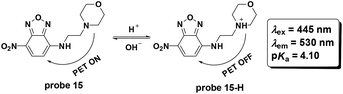
Later, Cao and co-workers utilized the fluorophore 4-nitro-2,1,3-benzoxadiazole to synthesize probe 15 for pH detection (Fig. 17).56 The PET based probe displayed turn-on fluorescent responses for pH values ranging from 9.0–2.0 and the pKa value was calculated to be 4.10. Co-localization with LTR displayed the lysosome labelling property. The probe could be used for imaging the lysosome pH changes induced by chloroquine and bafilomycin A1 in HeLa cells.
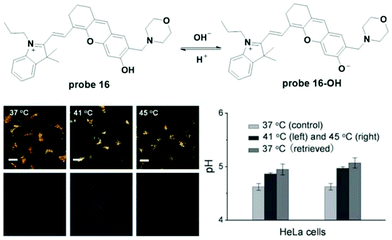
Wan et al. reported hemicyanine based ratiometric fluorescent probe 16 for lysosome pH detection in 2013.25 Deprotonation of the phenol moiety induced a decrease in the original fluorescence emission at 670 nm and the appearance of a new peak at 708 nm with a pH change from 4.0 to 7.4. The pKa value was 5.00 ± 0.01. With high selectivity, high thermostability and good biocompatibility, the probe was used for lysosome pH imaging in both HeLa and MCF-7 cells with the co-culturing of LTR. The probe was accurately accumulated in lysosomes. Further culturing of 16 loaded HeLa and MCF-7 cells in high K+ buffers at different pH values with nigericin was performed to obtain the calibration curve for quantitative detection of lysosome pH using the fluorescence intensity ratio of two channels (channel 1: 650–680; channel 2: 690–720). The normal lysosome pH values for HeLa and MCF-7 cells were calculated to be 4.58 ± 0.09 and 4.55 ± 0.08, respectively. Heat shock towards both HeLa and MCF-7 cells caused obvious lysosome pH increase and the stimulation was irreversible (Fig. 18).
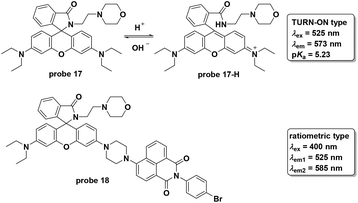
Shi and co-workers modified rhodamine B to develop a turn on fluorescent probe 17 for lysosome-specific pH imaging (Fig. 19).57 The probe displayed turn-on fluorescent responses with pH decrease from 7.4 to 4.5 with the pKa value of 5.23. With excellent selectivity, photostability, low cytotoxicity and lysosome localizability, the probe was used for visualizing chloroquine induced lysosomal alkalization and dexamethasone induced pH change during apoptosis in MCF-7 cells. Similar to probe 17, another rhodamine derivative, 18, has been reported recently.58 To connect to the naphthalimide moiety, the rhodamine derivative featured ratiometric fluorescent responses towards pH changes based on the FRET process from naphthalimide (donor) to the rhodamine (acceptor) moiety. Furthermore, the two-photon property of the naphthalimide derivative prompted the use of probe 18 to image lysosome pH changes in both HeLa cells and nude mice muscle tissue.
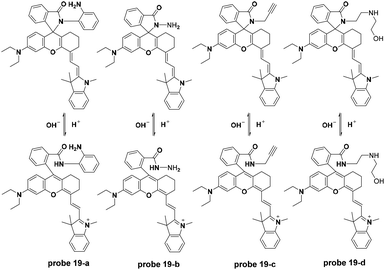
The Liu group synthesized a series of NIR fluorescent probes for lysosome imaging (Fig. 20).59 The four probes all displayed turn-on fluorescence emission at 743 nm with excitation at 670 nm when the pH decreased from 7.4 to 4.1. The corresponding pKa values for the four probes were calculated to be 5.8, 4.6, 4.9 and 5.4, respectively. However, peracid caused fluorescence quenching for the four probes. Co-localization experiments of the four probes with DND-189 showed that probes 19-b, c, and d could be used for lysosome imaging with negligible toxicity.
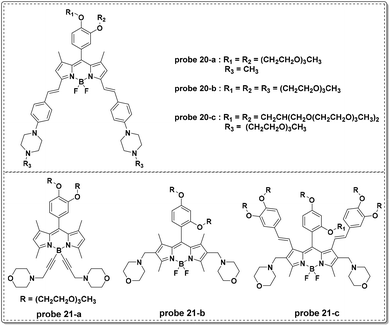
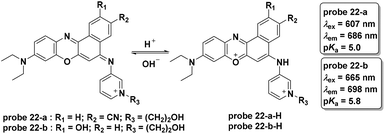
In 2015, the Liu group modified the BODIPY fluorophore to synthesize three NIR fluorescent probes (probe 20-a, b, c) for lysosome imaging (Fig. 21).60 Modulated by the protonation processes for piperazine moieties, which inhibited PET from the un-conjugated nitrogen atoms of piperazine moieties to BODIPY moieties, probes 20-a, b, and c all displayed turn-on fluorescent responses at 715 nm upon the decrease of pH from 9.98 to 2.2 in citrate–phosphate buffer solutions. The pKa values of the three probes were calculated to be 2.91, 3.19 and 3.57, respectively. Additionally, the three probes were inert towards various metal ions. Co-localization experiments with probe 20 and DND-189 using MDA-MB-231 and HUVEC-C cells revealed the acidic organelle activation properties of the three probes and they could be used for the imaging of pH changes in HUVEC-C cells.
In a follow-up study, the Liu group recently reported another series of BODIPY dyes (probe 21-a, b, c in Fig. 21).61 Unlike the previous probes involving PET processes from piperazine moieties to BODIPY moieties controlling fluorescent responses, the three new probes featured turn-on fluorescent responses with pH increase. The unique changes were further illustrated through DFT calculation and included d-PET processes from the fluorophores to the protonated morpholine moieties, further restrained by the increase of pH values. The use of morpholine as the lysosome anchor resulted in the use of the three probes for visualizing accumulation specifically in lysosomes. Probe 21-c could be used for intracellular pH imaging in HUVEC-C cells.
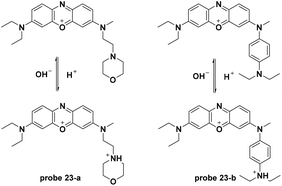
Sun and co-workers reported two NIR fluorescent probes for pH detection in aqueous solutions in 2013 (Fig. 22).62 The phenoxazine derivative probes 22-a and 22-b displayed turn-on fluorescent responses at 686 nm and 698 nm with pH values ranging from 7.0 to 3.4 and 7.8 to 3.4, respectively. The probes displayed high selectivity towards hydrion over other cations and amino acids. Co-culturing of probe 22-a, b and LTG with HeLa cells revealed the lysosome-specific imaging property of probe 22-a, which was attributed to the cyano moiety within the molecule.
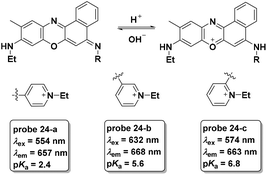
Similar to probe 22, two new phenoxazine derivatives were developed for pH detection in lysosomes.63 The pH controlled PET processes from the morpholine moiety and the aniline moiety to the phenoxazine fluorophore for probe 23-a and 23-b, respectively, demonstrated turn-on pH detection. The two probes displayed inert responses towards various metal ions and amino acids and could be used for lysosome pH imaging (Fig. 23).
As a follow-up, three new phenoxazine derivatives were further developed for lysosome pH imaging with turn-on NIR fluorescence emission (Fig. 24).8 Among the three probes, probe 24-a displayed the lowest pKa value and could be used for discrimination imaging of cancer cells including HeLa and KB cells over V97 cells (normal cells). Probes 24-b and 24-c did not exhibit the same property. The fluorescent responses of the three probes in cell experiments further demonstrated the lower lysosome pH values of cancer cells compared with normal cells.
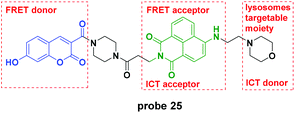
Recently, the Lin group reported a novel pH probe which contained two hydrion sensing sites, the hydroxy and the morpholine moiety, and featured ICT–PET–FRET during the detection process (Fig. 25).64 In the pH 3.0 system, the PET from morpholine to naphthalimide was inhibited and the FRET from coumarin to naphthalimide was activated. With pH increase, the PET process activated gradually and further, the ICT of the ionization of hydroxyl to coumarin increased. Theoretical calculations using the MarvinSketch software confirmed the above mentioned processes. The corresponding fluorescent responses changed from the original emission at 530 nm to 454 nm. Furthermore, the biological species including Ca2+, Mg2+, Cu2+, H2O2, Cys, GSH, H2S, SO2, and NO did not influence the detection of pH. Co-localization experiments displayed the lysosome labelling property of probe 25. Further cell experiments demonstrated that the probe could be used to monitor the chloroquine-induced lysosome pH changes in HeLa cells.
pH activatable probes for cancer cell specific imaging
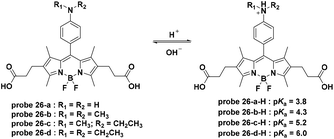
Small molecule pH-activatable fluorescent probes conjugated with active-targeting ligands for cancer specific receptors are some of the most promising cancer specific imaging systems with double security.65,66 The active-targeting ligands promote the accumulation of the fluorescent probes within the cancer cells or tissue. Furthermore, the pH activation process in the acidic organelles, such as lysosomes, of cancer cells activate fluorescence emission, leading to specific imaging. For these probes, exocytosis will induce fluorescence quenching during the pharmacokinetic process due to the reversible nature. Due to the high specific expression and quenching upon leakage from the cell, these probes provide a high signal to noise ratio (SNR).In 2009, Urano and co-workers synthesized a series of BODIPY dyes which featured off–on fluorescence emission with pH decrease based on the pH controlled PET process from the aniline moiety to the BODIPY fluorophore (Fig. 26).27 To create cancer cell targetable probes, trastuzumab, a monoclonal antibody for the human epidermal growth factor type 2 (HER2) receptor, was further conjugated to 26-b, c, and d through the amidation of the two carboxylic groups. The trastuzumab modified probes 26-b, c, and d all displayed lysosome-activatable fluorescent responses and exhibited almost no fluorescence emission immediately after addition whereas strong fluorescent spots were observed for 4 hours after addition within NIH3T3/HER2 cells. Further in vivo imaging experiments of probe 26-d-trastuzumab using a mouse model demonstrated the tumor specific imaging over normal tissues. Additionally, the signal was activated only in viable cancer cells and probe leakage from the damaged cells induced fluorescence signal quenching, providing a potential method to monitor therapy in living cancer cells.
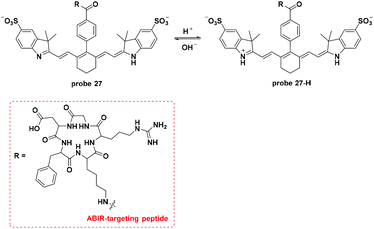
Lee and co-workers modified the Cy7 dye, which acted as the fluorophore and the hydrion bonding site, with a αvβ3 integrin receptor (ABIR, overexpressed in many tumor cells) targeting RGD peptides to develop a new pH activatable NIR fluorescent probe (Fig. 27).24 Co-culturing experiments with LTG revealed the lysosome accumulation property of probe 27. Probe 27 injected into 4T1/luc tumor-bearing mice accumulated in the tumor within one hour and peaked after 4 hours. Imaging in the orthotopic syngeneic breast tumor model further confirmed the tumor specific imaging of probe 27. The corresponding ex vivo imaging of the tissues presented an average tumor–muscle ratio value of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
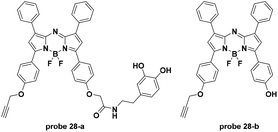
For another pH activatable cancer specific targeting probe, Han et al. connected aza-BODIPY dyes with a pH (low) insertion peptide (pHLIP) and a tumor membrane-targeting ligand, activated upon DGL loading (Fig. 28).67 The acidity of the extracellular environments of tumors induced the tumor specific accumulation of the pHLIP loaded probes. The pHLIP loaded probe 28-a and 28-b displayed turn-on fluorescence emission at 729 nm with pH decrease from 8 to 6.5 and the pKa values were 6.03 and 5.45, respectively. Both in vitro and in vivo experiments demonstrated the tumor (breast cancer and glioma) targetable properties of the two probes with relatively high tumor-background ratios.
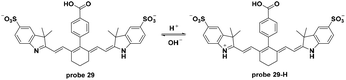
In 2015, Gilson and co-workers utilized Cy7 dyes in probe 29 as NIR fluorescent probes for tumor specific imaging via the pH activatable path.68 The decrease of pH induced turn-on fluorescent responses in probe 29 with the pKa value of 5.2. In vitro experiments using A431 or 4T1/luc cells in pH 7.4 and 6.4 systems demonstrated the internalization of the probe in cancer cells and, interestingly, illustrated how cell dying and death induced probe accumulation and fluorescence intensity enhancement. Both in vivo and ex vivo experiments demonstrated the specific imaging of tumors with a high tumor–muscle ratio after injection for 72 h and organ imaging further exhibited the excretion of the probe. Additionally, two-photon imaging of probe 29 in 4T1/luc tumors revealed the extracellular matrix of the tumors (Fig. 29).
Conclusions
Fluorescent probes for intracellular pH imaging and detection hold great potential to illustrate complex physiological and pathological processes. The non-invasive options afforded by fluorescent probes have brought unprecedented insight into cells, tissues and living bodies with the ability to visualize the pH distribution and changes, especially for subcellular specific imaging probes. Probes for the most acidic organelle (lysosomes) and the alkalescent organelle (mitochondria) have been widely reported based on the specific accumulation of alkalescent moiety-containing probes and cation-containing probes, respectively. Among these lysosome-targetable probes, probes 11, 15, 17, 19 and 24 featured lysosome-activatable turn-on fluorescence emission while probes 13, 14, 16, 18, and 25 displayed ratiometric fluorescent responses towards pH changes, which allowed the quantitative determination of the pH values of the lysosomes. For these lysosome-specific imaging probes, the mitochondria targetable probes (1–6) can achieve quantitative measurement of mitochondria pH values with high sensitivity. Importantly, most of these probes could image stimulations like ROS, chloroquine and starvation induced pH changes in lysosomes and mitochondria. Specifically, probes 6 and 10 could visualize the mitophagy process in autolysosomes with the co-culturing of lysosome-specific imaging dyes. Furthermore, the NIR probes (probes 1, 5, 19, 20, 22, 24, 27, 29), which feature minor photo-damage, lower background signal and deep tissue penetration, enable more promising in vivo imaging of pH changes.For cancer cells and tissues, specific tumor-targeting ligand oriented accumulation of pH-activatable fluorescent probes exhibits attractive potential for reversible and high SNR imaging of tumors. Hydrion activated fluorescence emission both in the acidic lysosomes and the extracellular environment of cancer cells provides a novel option for the visualization of tumor tissues.
Fluorescent probes for subcellular pH imaging have enabled the visualization of pH based metabolic processes. Based on the development of these imaging probes, a deeper understanding of pH related processes in cells and tissues can be obtained and will further provide guidance for pH-related disease diagnosis and treatment.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 21472118, 21672131), the Program for the Top Young and Middle-aged Innovative Talents of Higher Learning Institutions of Shanxi (2013802), the Talents Support Program of Shanxi Province (2014401), and Shanxi Province Outstanding Youth Fund (no. 2014021002). J. Y. is thankful for a grant from the National Creative Research Initiative Programs of the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIP) (no. 2012R1A3A2048814).Notes and references
- J. Yin, Y. Hu and J. Yoon, Chem. Soc. Rev., 2015, 44, 4619–4644 RSC.
- J. Han and K. Burgess, Chem. Rev., 2010, 110, 2709–2728 CrossRef PubMed.
- P. Boya and G. Kroemer, Oncogene, 2008, 27, 6434–6451 CrossRef PubMed.
- J. Ji, N. Rosenzweig, C. Griffin and Z. Rosenzweig, Anal. Chem., 2000, 72, 3497–3503 CrossRef PubMed.
- S. Wu, Z. Li, J. Han and S. Han, Chem. Commun., 2011, 47, 11276–11278 RSC.
- L. He, Y. Li, C.-P. Tan, R.-R. Ye, M.-H. Chen, J.-J. Cao, L.-N. Ji and Z.-W. Mao, Chem. Sci., 2015, 6, 5409–5418 RSC.
- E. J. Blott and G. M. Griffiths, Nat. Rev. Mol. Cell Biol., 2002, 3, 122–131 CrossRef PubMed.
- D. D. He, W. Liu, R. Sun, C. Fan, Y. J. Xu and J. F. Ge, Anal. Chem., 2015, 87, 1499–1502 CrossRef PubMed.
- X. Li, X. Guo, L. Cao, Z. Xun, S. Wang, S. Li, Y. Li and G. Yang, Angew. Chem., Int. Ed., 2014, 53, 7809–7813 CrossRef PubMed.
- P. Boya, R. A. Gonzalez-Polo, D. Poncet, K. Andreau, H. L. Vieira, T. Roumier, J. L. Perfettini and G. Kroemer, Oncogene, 2003, 22, 3927–3936 CrossRef PubMed.
- P. Bernardi, Physiol. Rev., 1999, 79, 1127–1155 Search PubMed.
- E. S. Trombetta, M. Ebersold, W. Garrett, M. Pypaert and I. Mellman, Science, 2003, 299, 1400–1403 CrossRef PubMed.
- D. Wu, L. Shao, Y. Li, Q. Hu, F. Huang, G. Yu and G. Tang, Chem. Commun., 2016, 52, 541–544 RSC.
- Y. I. Park, O. Postupna, A. Zhugayevych, H. Shin, Y. S. Park, B. Kim, H. J. Yen, P. Cheruku, J. S. Martinez, J. W. Park, S. Tretiak and H. L. Wang, Chem. Sci., 2015, 6, 789–797 RSC.
- C. Richter, C. Schneider, M. T. Quick, P. Volz, R. Mahrwald, J. Hughes, B. Dick, U. Alexiev and N. P. Ernsting, Phys. Chem. Chem. Phys., 2015, 17, 30590–30597 RSC.
- Q. A. Best, C. Liu, P. D. van Hoveln, M. E. McCarroll and C. N. Scott, J. Org. Chem., 2013, 78, 10134–10143 CrossRef PubMed.
- D. Asanuma, Y. Takaoka, S. Namiki, K. Takikawa, M. Kamiya, T. Nagano, Y. Urano and K. Hirose, Angew. Chem., Int. Ed., 2014, 53, 6085–6089 CrossRef PubMed.
- S. Chen, Y. Hong, Y. Liu, J. Liu, C. W. Leung, M. Li, R. T. Kwok, E. Zhao, J. W. Lam, Y. Yu and B. Z. Tang, J. Am. Chem. Soc., 2013, 135, 4926–4929 CrossRef PubMed.
- Y. Yue, F. Huo, S. Lee, C. Yin, J. Yoon, J. Chao, Y. Zhang and F. Cheng, Chem. – A Eur. J., 2016, 22, 1239–1243 CrossRef PubMed.
- B. Tang, F. Yu, P. Li, L. Tong, X. Duan, T. Xie and X. Wang, J. Am. Chem. Soc., 2009, 131, 3016–3023 CrossRef PubMed.
- T. Myochin, K. Kiyose, K. Hanaoka, H. Kojima, T. Terai and T. Nagano, J. Am. Chem. Soc., 2011, 133, 3401–3409 CrossRef PubMed.
- R. Pal and D. Parker, Chem. Commun., 2007, 474–476, 10.1039/b616665b.
- S. Luo, E. Zhang, Y. Su, T. Cheng and C. Shi, Biomater., 2011, 32, 7127–7138 CrossRef PubMed.
- H. Lee, W. Akers, K. Bhushan, S. Bloch, G. Sudlow, R. Tang and S. Achilefu, Bioconjugate Chem., 2011, 22, 777–784 CrossRef PubMed.
- Q. Wan, S. Chen, W. Shi, L. Li and H. Ma, Angew. Chem., Int. Ed., 2014, 53, 10916–10920 CrossRef PubMed.
- M. H. Lee, N. Park, C. Yi, J. H. Han, J. H. Hong, K. P. Kim, D. H. Kang, J. L. Sessler, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2014, 136, 14136–14142 CrossRef PubMed.
- Y. Urano, D. Asanuma, Y. Hama, Y. Koyama, T. Barrett, M. Kamiya, T. Nagano, T. Watanabe, A. Hasegawa, P. L. Choyke and H. Kobayashi, Nat. Med., 2009, 15, 104–109 CrossRef PubMed.
- T. Jokic, S. M. Borisov, R. Saf, D. A. Nielsen, M. Kuhl and I. Klimant, Anal. Chem., 2012, 84, 6723–6730 CrossRef PubMed.
- W. Liu, R. Sun, J.-F. Ge, Y.-J. Xu, Y. Xu, J.-M. Lu, I. Itoh and M. Ihara, Anal. Chem., 2013, 85, 7419–7425 CrossRef PubMed.
- H. J. Park, C. S. Lim, E. S. Kim, J. H. Han, T. H. Lee, H. J. Chun and B. R. Cho, Angew. Chem., Int. Ed., 2012, 51, 2673–2676 CrossRef PubMed.
- L. Fan, Q. Liu, D. Lu, H. Shi, Y. Yang, Y. Li, C. Dong and S. Shuang, J. Mater. Chem. B, 2013, 1, 4281–4288 RSC.
- M. Lee, N. G. Gubernator, D. Sulzer and D. Sames, J. Am. Chem. Soc., 2010, 132, 8828–8830 CrossRef PubMed.
- J. Llopis, J. M. McCaffery, A. Miyawaki, M. G. Farquhar and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6803–6808 CrossRef.
- L. Cao, X. Li, S. Wang, S. Li, Y. Li and G. Yang, Chem. Commun., 2014, 50, 8787–8790 RSC.
- G. U. Reddy, A. H. A. F. Ali, N. Taye, S. Chattopadhyay and A. Das, Org. lett., 2015, 17, 5532–5535 CrossRef PubMed.
- G. U. Reddy, V. Ramu, S. Roy, N. Taye, S. Chattopadhyay and A. Das, Chem. Commun., 2014, 50, 14421–14424 RSC.
- P. Mahato, S. Saha, P. Das, H. Agarwalla and A. Das, RSC Adv., 2014, 4, 36140–36174 RSC.
- G. R. Rosania, J. W. Lee, L. Ding, H. S. Yoon and Y. T. Chang, J. Am. Chem. Soc., 2003, 125, 1130–1131 CrossRef PubMed.
- J. Zhou, L. Li, W. Shi, X. Gao, X. Li and H. Ma, Chem. Sci., 2015, 6, 4884–4888 RSC.
- L. Yuan, L. Wang, B. K. Agrawalla, S. J. Park, H. Zhu, B. Sivaraman, J. Peng, Q. H. Xu and Y. T. Chang, J. Am. Chem. Soc., 2015, 137, 5930–5938 CrossRef CAS PubMed.
- Y. Wen, K. Liu, H. Yang, Y. Liu, L. Chen, Z. Liu, C. Huang and T. Yi, Anal. Chem., 2015, 87, 10579–10584 CrossRef CAS PubMed.
- C. S. Lim, G. Masanta, H. J. Kim, J. H. Han, H. M. Kim and B. R. Cho, J. Am. Chem. Soc., 2011, 133, 11132–11135 CrossRef CAS PubMed.
- C. Y. Kim, H. J. Kang, S. J. Chung, H. K. Kim, S. Y. Na and H. J. Kim, Anal. Chem., 2016, 88, 7178–7182 CrossRef CAS PubMed.
- P. Li, H. Xiao, Y. Cheng, W. Zhang, F. Huang, W. Zhang, H. Wang and B. Tang, Chem. Commun., 2014, 50, 7184–7187 RSC.
- M. Y. Wu, K. Li, Y. H. Liu, K. K. Yu, Y. M. Xie, X. D. Zhou and X. Q. Yu, Biomater., 2015, 53, 669–678 CrossRef CAS PubMed.
- Y. Chen, C. Zhu, J. Cen, Y. Bai, W. He and Z. Guo, Chem. Sci., 2015, 6, 3187–3194 RSC.
- L. Cao, Z. Zhao, T. Zhang, X. Guo, S. Wang, S. Li, Y. Li and G. Yang, Chem. Commun., 2015, 51, 17324–17327 RSC.
- A. R. Sarkar, C. H. Heo, L. Xu, H. W. Lee, H. Y. Si, J. W. Byun and H. M. Kim, Chem. Sci., 2016, 7, 766–773 RSC.
- A. C. Macintrye and D. J. Cutler, J. Pharm. Sci., 1988, 77, 196–199 CrossRef.
- B. Lemieux, M. D. Percival and J. P. Falgueyret, Anal. Biochem., 2004, 327, 247–251 CrossRef CAS PubMed.
- Z. Li, S. Wu, J. Han and S. Han, Analyst, 2011, 136, 3698–3706 RSC.
- L. Chen, J. Li, Z. Liu, Z. Ma, W. Zhang, L. Du, W. Xu, H. Fang and M. Li, RSC Adv., 2013, 3, 13412–13416 RSC.
- J. Hu, F. Wu, S. Feng, J. Xu, Z. Xu, Y. Chen, T. Tang, X. Weng and X. Zhou, Sens. Actuators, B, 2014, 196, 194–202 CrossRef CAS.
- G. Li, D. Zhu, L. Xue and H. Jiang, Org. lett., 2013, 15, 5020–5023 CrossRef CAS PubMed.
- X.-F. Zhang, T. Zhang, S.-L. Shen, J.-Y. Miao and B.-X. Zhao, J. Mater. Chem. B, 2015, 3, 3260–3266 RSC.
- X. J. Cao, L. N. Chen, X. Zhang, J. T. Liu, M. Y. Chen, Q. R. Wu, J. Y. Miao and B. X. Zhao, Anal. Chim. Acta, 2016, 920, 86–93 CrossRef CAS PubMed.
- X.-L. Shi, G.-J. Mao, X.-B. Zhang, H.-W. Liu, Y.-J. Gong, Y.-X. Wu, L.-Y. Zhou, J. Zhang and W. Tan, Talanta, 2014, 130, 356–362 CrossRef CAS PubMed.
- L. Zhou, D.-Q. Lu, Q. Wang, S. Hu, H. Wang, H. Sun and X. Zhang, Sens. Actuators, B, 2017, 238, 274–280 CrossRef CAS.
- G. K. Vegesna, J. Janjanam, J. Bi, F.-T. Luo, J. Zhang, C. Olds, A. Tiwari and H. Liu, J. Mater. Chem. B, 2014, 2, 4500–4508 RSC.
- J. Zhang, M. Yang, C. Li, N. Dorh, F. Xie, F.-T. Luo, A. Tiwari and H. Liu, J. Mater. Chem. B, 2015, 3, 2173–2184 RSC.
- J. Zhang, M. Yang, W. Mazi, K. Adhikari, M. Fang, F. Xie, L. Valenzano, A. Tiwari, F.-T. Luo and H. Liu, ACS Sens., 2016, 1, 158–165 CrossRef CAS PubMed.
- R. Sun, W. Liu, Y.-J. Xu, J.-M. Lu, J.-F. Ge and M. Ihara, Chem. Commun., 2013, 49, 10709–10711 RSC.
- X. L. Wang, X. J. Li, R. Sun, Y. J. Xu and J. F. Ge, Analyst, 2016, 141, 2962–2969 RSC.
- B. Dong, X. Song, C. Wang, X. Kong, Y. Tang and W. Lin, Anal. Chem., 2016, 88, 4085–4091 CrossRef CAS PubMed.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS PubMed.
- X. Hu, X. Guan, J. Li, Q. Pei, M. Liu, Z. Xie and X. Jing, Chem. Commun., 2014, 50, 9188–9191 RSC.
- L. Han, M. Liu, D. Ye, N. Zhang, E. Lim, J. Lu and C. Jiang, Biomater., 2014, 35, 2952–2960 CrossRef CAS PubMed.
- R. C. Gilson, R. Tang, A. Som, C. Klajer, P. Sarder, G. P. Sudlow, W. J. Akers and S. Achilefu, Mol. Pharmaceutics, 2015, 12, 4237–4246 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |