Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of a specific fluorescent probe to detect advanced glycation end products (AGEs)†
Heewon
Cho‡
a,
Na-Kyeong
Hong‡
a,
Insung
Yong
 b,
Haw-Young
Kwon
a,
Nam-Young
Kang
c,
Larissa Miasiro
Ciaramicoli
a,
Pilnam
Kim
b and
Young-Tae
Chang
b,
Haw-Young
Kwon
a,
Nam-Young
Kang
c,
Larissa Miasiro
Ciaramicoli
a,
Pilnam
Kim
b and
Young-Tae
Chang
 *a
*a
aDepartment of Chemistry, Pohang University of Science and Technology (POSTECH), Pohang, Gyeongsangbuk-do 37673, Republic of Korea. E-mail: ytchang@postech.ac.kr
bDepartment of Bio and Brain Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, 34141, Republic of Korea
cDepartment of Convergence IT Engineering, Pohang University of Science and Technology (POSTECH), Pohang, Gyeongsangbuk-do 37673, Republic of Korea
First published on 23rd May 2024
Abstract
Advanced glycation end products (AGEs) play a pivotal role in the aging process, regarded as a hallmark of aging. Despite their significance, the absence of adequate monitoring tools has hindered the exploration of the relationship between AGEs and aging. Here, we present a novel AGE-selective probe, AGO, for the first time. AGO exhibited superior sensitivity in detecting AGEs compared to the conventional method of measuring autofluorescence from AGEs. Furthermore, we validated AGO's ability to detect AGEs based on kinetics, demonstrating a preference for ribose-derived AGEs. Lastly, AGO effectively visualized glycation products in a collagen-based mimicking model of glycation. We anticipate that this study will enhance the molecular tool sets available for comprehending the physiological processes of AGEs during aging.
1. Introduction
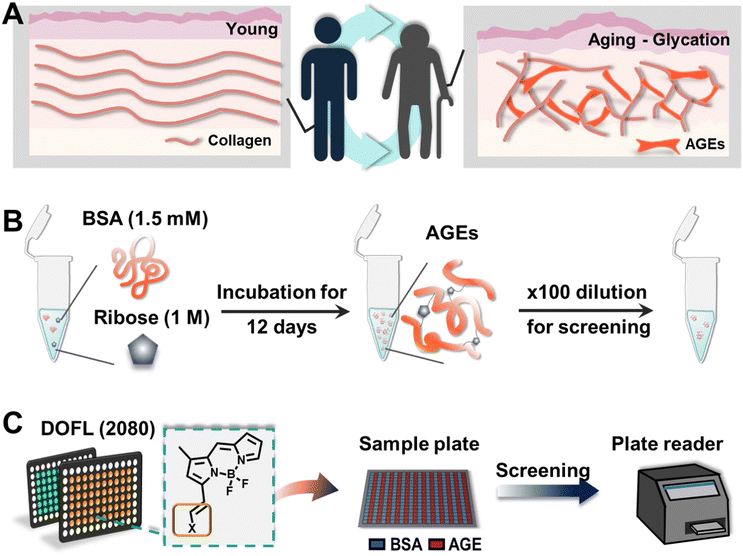
Advanced glycation end products (AGEs) are a diverse group of compounds formed through the non-enzymatic reaction between reducing sugars and proteins, lipids, or nucleic acids.1 This reaction, known as glycation or a Maillard reaction, occurs spontaneously under normal physiological conditions.2 AGEs accumulate over time in various tissues and organs, particularly in long-lived proteins like collagen and elastin (Fig. 1A).3 Recent research has highlighted AGEs as a molecular hallmark of aging, demonstrating their significant role in the aging process. Additionally, the accumulation of AGEs has been linked to age-related diseases such as diabetes, cardiovascular diseases, neurodegenerative disorders, and kidney dysfunction.4 Understanding the relationship between the accumulation of AGEs and aging is crucial for developing therapeutic strategies. Monitoring AGE levels in biological samples can provide valuable insights into the progression of age-related conditions. However, this task is complicated due to the lack of precise measurement techniques for specific AGEs and the absence of accurate AGE-mimicking models that replicate physiological conditions.4The most common method for detecting AGEs involves measuring their autofluorescence using fluorescence spectroscopy.5 However, this approach lacks specificity and sensitivity and can be influenced by other fluorescent compounds present in biological samples, resulting in high background interference. Given the potential of AGEs as biomarkers, it is essential to overcome these limitations and develop new molecular tools for sensitive detection.
Fluorescent small molecules have gained significant attention for their potential in sensing and visualization. Traditional probe development often relies on established molecular recognition processes for specific substrates, but their effectiveness in complex biological environments can vary. This has limited the pace and scope of probe development. To address these challenges, diverse fluorescence library methods have been employed to create novel fluorescent probes, even when the target recognition mechanism is not fully understood.6 Our group has developed extensive fluorescence libraries, such as the diversity oriented fluorescence library (DOFL) with over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds.7 Through a systematic approach, we have successfully identified more than 30 fluorescent small molecules applicable to various biological samples.8–12 In this study, we introduced the AGE-selective probe, AGO, which was developed through screening the DOFL. AGO demonstrated the highest selectivity among 2080 compounds and superior sensitivity to AGEs compared to their autofluorescence. Additionally, AGO effectively detected ribose-derived glycation products compared to other sugars and disaccharides. We also showed that AGO could visualize glycation products in a glycated collagen framework, simulating the natural glycation of the matrix.
000 compounds.7 Through a systematic approach, we have successfully identified more than 30 fluorescent small molecules applicable to various biological samples.8–12 In this study, we introduced the AGE-selective probe, AGO, which was developed through screening the DOFL. AGO demonstrated the highest selectivity among 2080 compounds and superior sensitivity to AGEs compared to their autofluorescence. Additionally, AGO effectively detected ribose-derived glycation products compared to other sugars and disaccharides. We also showed that AGO could visualize glycation products in a glycated collagen framework, simulating the natural glycation of the matrix.
2. Experimental section
2.1. Materials
D-(+)-Galactose (Sigma Aldrich; St. Louis, Mo, USA), D-(+)-mannose, D-(+)-ribose, sucrose, and bovine serum albumin (BSA) were purchased from Sigma Aldrich (St. Louis, Mo, USA). D-(+)-glucose was acquired from Alfa Aesar (Haverhill, MA, USA) and D-(+)-xylose was acquired from TCI (Tokyo, Japan).2.2. Glycation of BSA
BSA was dissolved in PBS containing 0.05% sodium azide to yield a stock solution of 3 mM. This solution was then mixed with carbohydrates, prepared in PBS with 0.05% sodium azide, resulting in final concentrations of 1.5 mM BSA and 1 M carbohydrates. The mixtures were incubated at 37 °C, and incubation days were dependent on each experiment. Before resuspension, all solutions were filtered with 0.45 μm and 0.22 μm membranes sequentially (Millipore, USA).2.3. DOFL screening
26 DOFLs (including 2080 compounds) were used for 384-well based screening. For the screening, compounds (100 μM in DMSO) were mixed with AGE solution to a final concentration of 5 μM. After 30 minutes of incubation at room temperature, plates were read using a plate reader (Molecular Devices, USA).2.4. Fluorescence microscopy
BSA (1.5 mM) and D-ribose (1 M) were incubated at 37 °C until 12 days at an interval of 2 days. After 12 days, samples were stained with AGO (5 μM) for 30 min at room temperature. The samples (50 μL) were transferred to a 384-well plate and then the stock solution was diluted 100 times. The fluorescence from solution was recorded by fluorescence microscopy using an Operetta CLS (PerkinElmer, Waltham, MA, USA) with a ×10 objective lens. AGE autofluorescence was recorded using a DAPI channel (excitation: 355–385/emission: 430–500), and AGO fluorescence was measured with a TRITC channel (excitation: 530–560/570–650). Each channel was set to give 20% power and expose to 10 ms.2.5. Assay of AGEs derived by different carbohydrate types
The mixtures of BSA (1.5 mM) and each carbohydrate (1 M), including glucose, galactose, mannose, ribose, xylose, and sucrose, were incubated at 37 °C for 24 days, and the mixtures were prepared at an interval of 6 days. To conduct the assay, the samples were made after 100× dilution and transferred into a 96-well plate. The fluorescence was measured using a plate reader (Molecular Devices, USA) after staining with AGO (5 μM) for 30 min at room temperature. The autofluorescence data were acquired by excitation at 370 nm and emission at 420 nm. The AGO graph was obtained by excitation at 490 nm and emission at 570 nm.2.6. Preparation of a glycated collagen hydrogel
To fabricate collagen microgels, the collagen type I solution (Corning, 354249) was neutralized to a physiological pH (approximately 7.0–7.4) utilizing a mixture composed of 1 N sodium hydroxide (NaOH), distilled water, and 10× phosphate-buffered saline (PBS), which was conducted on ice as per the manufacturer's instructions. Subsequently, the solution's concentration was meticulously adjusted to 3 mg mL−1. Following the neutralization process, 100 μL of the collagen mixture was carefully pipetted into each well of a 24-well plate (SPL, 30024). Gelation was then initiated by incubating the plates at 37 °C for 1 h, facilitating the polymerization of the collagen. Upon completion of the gelation process, the resulting hydrogel was immersed in a 500 mM ribose solution (Merck, R9629) prepared in PBS. This was incubated at 37 °C for a duration of one week to ensure adequate modification of the hydrogel properties.2.7. Immunofluorescence staining
After the glycation process, collagen microgels were washed three times with PBS for 10 minutes each to remove any residual ribose. Following this, they were fixed using 4% paraformaldehyde (PFA) for 1 h at room temperature to preserve their microstructures. After fixation, the samples were permeabilized with 0.15% Triton-X 100 in PBS for 30 min. To prevent non-specific antibody binding, the microgels were then blocked with 1% BSA in PBS for 1 h. Subsequently, they were incubated overnight at 4 °C with an anti-collagen I (Abcam, ab21286) antibody, followed by a 2-hour incubation with secondary antibodies (Merck, F0382) at room temperature. For the comparison of AGE selectivity in the glycated extracellular matrix (ECM), the hydrogels were stained with 10 μM AGO/AGN dyes for 1 h. Each step was followed by three 10-minute washes with PBS. Finally, fluorescence images were captured and analyzed using a confocal laser scanning microscope (Nikon, A1R).2.8. Chemical materials and general methods for AGO synthesis
All used compounds and solvents were purchased from Alfa Aesar (Haverhill, MA, USA), Sigma Aldrich (St. Louis, MO, USA), Combi-Blocks (San Diego, USA), TCI (Tokyo, Japan), and Samchun Chemicals (Seoul, Republic of Korea). All the chemicals were directly used without further purification. MERCK silica gel 60 (230–400 mesh, 0.040–0.063 mm) was used for normal-phase column chromatography. The optical properties were investigated with a SpectraMax M2e spectrophotometer (molecular devices) in a 96 well plate (clear bottom) and a QS high-precision cuvette. The relative fluorescence quantum yield method was selected, and rhodamine B (Φ = 0.49) was utilized as the standard. The quantum yield was calculated using eqn (1). For the analytical characterization of AGO, a HPLC (Agilent, 1260 series) system with a DAD (diode array detector) and a single quadrupole mass spectrometer (Agilent, 6100 series, ESI†) were used. Eluents (A: H2O with 0.1% formic acid (FA) and B: ACN with 0.1% FA) and a Zorbax SB-C18 column (2.1 × 50 mm, 1.8 μm particle size, 80 Å pore size) were used. High-performance liquid chromatography (HPLC) was performed using a Prep. HPLC (Shimadzu) with a PDA detector with a C18(2) Luna column (5 μm, 250 mm × 21.2 mm, 100 Å). A gradient elution of 60% B for 5 min, then 60% B to 100% B for 40 min and then 100% B for 55 min was performed at a flow rate of 8 mL min−1 (solvent A: H2O; B: ACN). 1H and 13C NMR spectra were obtained using a Bruker Avance III HD 850 spectrometer.| Φfi =(Fi/Fs)(fs/fi)(ni/ns)2·Φfs | (1) |
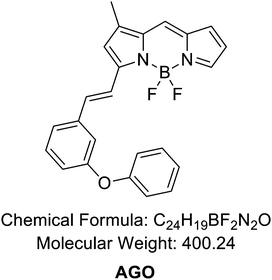
2.9. Synthesis of AGO
The reaction followed the below procedure. BD (4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-S-indacene) (30 mg, 136 μmole), 3-phenoxybenzaldehyde (32.3 mg, 164 μmole), acetic acid (47 μL, 818 μmole), and pyrrolidine (69 μL, 818 μmole) were dissolved in ACN (5 mL) and stirred at 70 °C for 30 min. The collected mixture was concentrated in vacuo. The residue was purified via high-performance liquid chromatography (HPLC) carried out using a Prep. HPLC (Shimadzu) with a PDA detector with a C18(2) Luna column (5 μm, 250 mm × 21.2 mm, 100 Å). A gradient elution of 60% B for 5 min, then 60% B to 100% B for 40 min, and then 100% B for 55 min was performed at a flow rate of 8 mL min−1 (solvent A: H2O; B: ACN). After purification, a purple solid was obtained (39.8 mg, 73%). 1H NMR (850 MHz, chloroform-d) δ 7.71 (s, 1H), 7.63 (d, J = 16.3 Hz, 1H), 7.43 (d, J = 7.74 Hz, 1H), 7.38 (q, J = 7.65 Hz, 3H), 7.33 (d, J = 16.3 Hz, 1H), 7.28 (s, 1H), 7.24 (t, J = 1.7 Hz), 7.21 (s, 1H), 7.16 (t, J = 7.40 Hz, 1H), 7.06 (d, J = 7.74 Hz, 2H), 7.01 (dd, J = 8.14 Hz, 1H), 6.97 (d, J = 3.83 Hz, 1H), 6.75 (s, 1H), 6.49 (q, J = 3.74 Hz, 3H), 2.33 (s, 3H). 13C NMR (214 MHz, chloroform-d) δ 158.22, 157.73, 156.95, 144.50, 139.55, 139.01, 137.75, 137.73, 133.30, 130.18, 129.88, 126.48, 123.57, 122.41, 120.09, 119.34, 118.97, 118.51, 117.17, 116.69, 11.51. LC-MS (ESI†) [M + H]+, m/z calcd for C24H19BF2N2O 400.2, found: 423.2.2.10. Synthesis of AGN
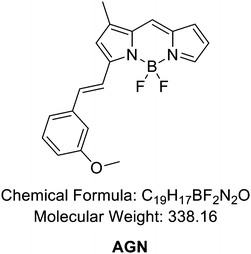
The reaction followed the below procedure. BD (100 mg, 454 μmole), 3-methoxybenzaldehyde (74.2 mg, 545 μmole), acetic acid (104 μL, 1820 μmole), and pyrrolidine (152 μL, 1820 μmole) were dissolved in ACN (10 mL) and stirred at 70 °C for 30 min. The collected mixture was concentrated in vacuo. The residue was purified via high-performance liquid chromatography (HPLC) carried out using a Prep. HPLC (Shimadzu) with a PDA detector with a C18(2) Luna column (5 μm, 250 mm × 21.2 mm, 100 Å). A gradient elution of 60% B for 5 min, then 60% B to 100% B for 40 min, and then 100% B for 55 min was used at a flow rate of 8 mL min−1 (solvent A: H2O; B: ACN). After purification, a purple solid was obtained (33.5 mg, 22%). 1H NMR (850 MHz, chloroform-d) δ 7.71 (s, 1H), 7.63 (d, J = 16.3 Hz, 1H), 7.35 (d, J = 7.74 Hz, 1H), 7.32 (t, J = 7.91 Hz, 1H), 7.28 (s, 1H), 7.22 (d, J = 7.91 Hz), 7.18 (s, 1H), 7.12 (t, J = 1.7 Hz, 1H), 6.95 (d, J = 3.74 Hz, 1H), 6.94(dd, J = 8.14 Hz, 1H), 6.75 (s, 1H), 6.47 (q, J = 3.74 Hz, 3H), 3.88(s, 3H), 2.32 (s, 3H). 13C NMR (214 MHz, chloroform-d) δ 159.97, 158.61, 144.57, 139.88, 139.14, 137.83, 137.17, 133.21, 129.88, 126.20, 123.33, 120.73, 118.81, 117.24, 116.54, 115.92, 112.68, 55.37, 11.49. LC-MS (ESI†) [M + H]+, m/z calcd for C19H17BF2N2O 338.1, found: 339.0.2.11. Synthesis of compound G
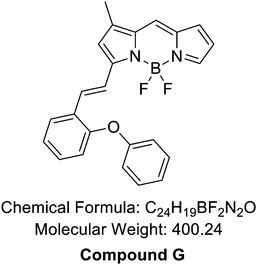
The reaction followed the below procedure. BD (30 mg, 136 μmole), 2-phenoxybenzaldehyde (32.3 mg, 164 μmole), acetic acid (47 μL, 818 μmole), and pyrrolidine (69 μL, 818 μmole) were dissolved in ACN (5 mL) and stirred at 70 °C for 30 min. The collected mixture was concentrated in vacuo. The residue was purified via high-performance liquid chromatography (HPLC) carried out using a Prep. HPLC (Shimadzu) with a PDA detector with a C18(2) Luna column (5 μm, 250 mm × 21.2 mm, 100 Å). A gradient elution of 60% B for 5 min, then 60% B to 100% B for 40 min, and then 100% B for 55 min was performed at a flow rate of 8 mL min−1 (solvent A: H2O; B: ACN). After purification, a purple solid was obtained (11.1 mg, 20%). LC-MS (ESI†) [M + H]+, m/z calcd for C24H19BF2N2O 400.1, found: 401.3. Results and discussion
3.1. Development of an AGE-selective probe
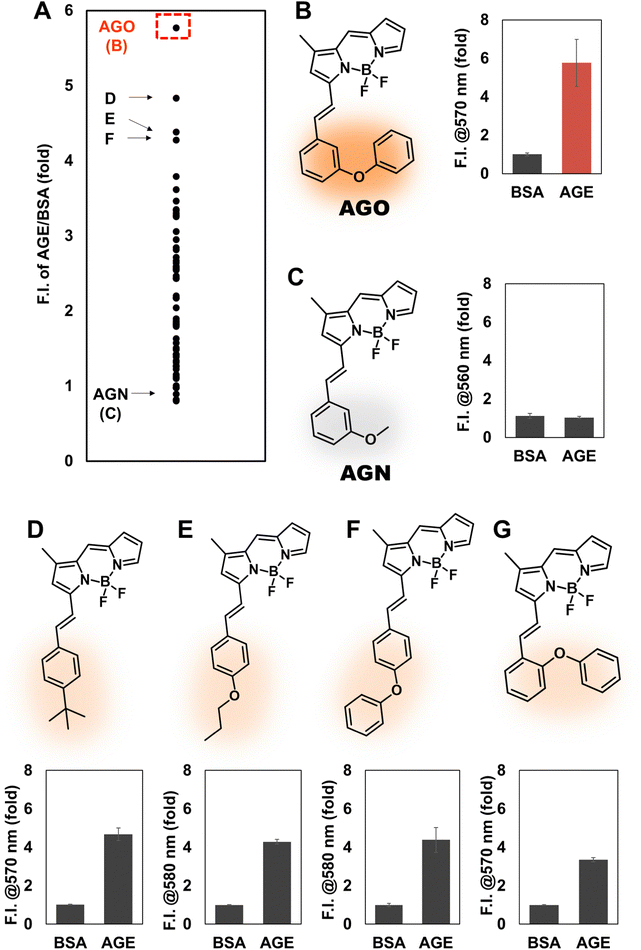
To develop an AGE-selective probe, we began by generating glycation samples in vitro using BSA, a standard protein for inducing glycation, and ribose, known for rapidly forming AGEs compared to other monosaccharides.13 We prepared a solution of BSA suspended with D-ribose to achieve final concentrations of 1.5 mM BSA and 1 M ribose, followed by incubation at 37 °C (Fig. 1B). Subsequently, we established a systematic screening system (Fig. 1C). Initially, we carefully selected potential fluorescence libraries from the DOFL with a higher likelihood of targeting AGEs. We opted for boron dipyrromethene (BODIPY)-based libraries, comprising 2080 compounds, given BODIPY's highly lipophilic neutral fluorophore characteristics,14 suggesting compatibility with AGEs. BSA and AGE samples were then added to 384-well plates, followed by incubation with BODIPY compounds (5 μM) for 30 minutes. Following plate reading, we filtered the results based on complete emission wavelength shapes, and subsequently analyzed AGE selectivity by calculating the fluorescence intensity ratio of AGEs to BSA. The resulting numerical values were plotted on a one-dimensional graph (Fig. 2A). Remarkably, one compound exhibited the highest selectivity for AGEs over BSA (approximately 6-fold), and we named it AGO (Advanced Glycation end products Orange) (Fig. 2B).AGO comprised a BODIPY fluorophore with a meta-positioned phenoxybenzyl ring. To explore structural preferences for AGEs, we identified three molecular compositions exhibiting over 4-fold selectivity to AGEs compared to BSA (Fig. 2E and F). Intriguingly, one compound featured a phenoxybenzyl derivative in a para position (Fig. 2F), also categorized as AGO. Given our hypothesis that this moiety may confer a specific ability to sense AGEs, we synthesized a BODIPY compound containing an ortho-positioned phenoxybenzyl ring not present in the DOFL (Fig. 2G). Although the molecule could discriminate AGEs from BSA, its capability was not superior to AGO. Furthermore, we verified that the methoxybenzyl moiety could not distinguish between AGEs and BSA, designating it as AGN (Advanced Glycation end products Negative) (Fig. 2C). Therefore, we concluded that a phenoxybenzyl derivative with an appropriate attachment (meta position) would confer a high capacity to sense AGEs.
3.2. Confirmation of the selectivity of AGO compared to autofluorescence
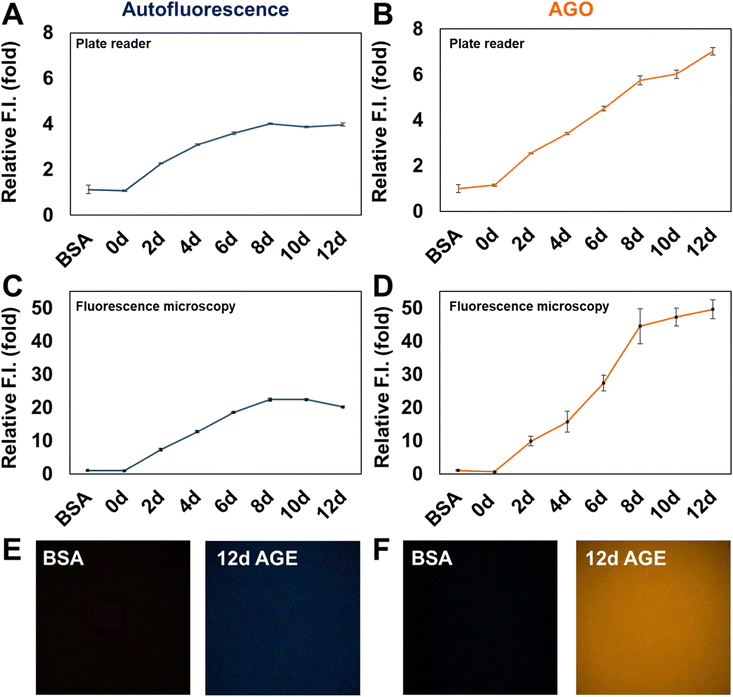
Inspired by the selectivity of AGO, we aimed to assess its ability to systematically detect AGEs. Initially, we prepared AGE samples derived from the incubation of BSA and ribose over various time intervals, ranging from 2 to 12 days. The formation of AGEs was monitored by measuring their autofluorescence using a plate reader (excitation: 370 nm; emission: 420 nm) (Fig. 3A). The signal exhibited a continuous increase and then a plateau could be observed after 8 days. In contrast, AGO consistently maintained its ability to detect AGEs up to 12 days (Fig. 3B). Furthermore, we evaluated its selectivity using fluorescence microscopy to facilitate wider application. Consistent with previous findings, while autofluorescence increased by day 8, reaching its limit (Fig. 3C), AGO continued to effectively detect AGEs (Fig. 3D). Additionally, the displayed images demonstrated that AGO offers greater sensitivity and selectivity in discriminating AGEs from BSA compared to autofluorescence measurements (Fig. 3E and F).3.3. AGO sensitivity to AGEs derived by different types of carbohydrates
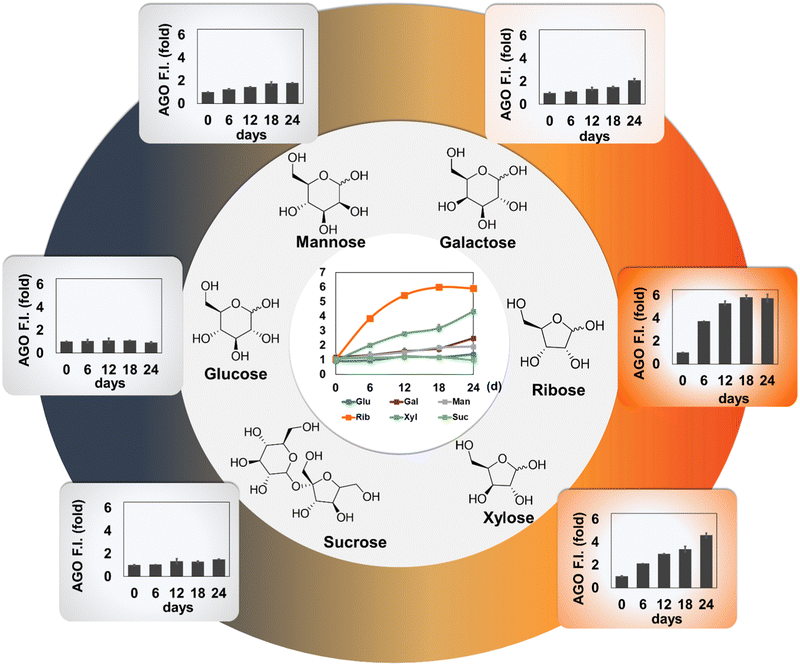
We then investigated how AGO behaves in AGEs formed by different types of carbohydrates. We prepared three pyranoses (glucose, galactose, and mannose), two furanoses (ribose and xylose), and sucrose, a disaccharide composed of fructose and glucose. Since it has been reported that each carbohydrate may require a different period to reach maximum yields of AGEs,15–17 we extended the incubation time to 24 days. Subsequently, we measured autofluorescence to confirm the proper formation of AGEs. Interestingly, the autofluorescence of ribose, when incubated with BSA, peaked rapidly at 6 days compared to other saccharides (Fig. S1, ESI†). This observation aligns with previous research indicating that ribose has a higher ability to generate AGEs than other carbohydrate types.13,16 Additionally, xylose and galactose demonstrated their capacity to form AGEs, although they seemed to lag behind ribose in terms of record timing. This observation led us to realize that pentose has quicker kinetics than hexose. This difference could be attributed to the ratio of the opened form (aldehyde), which is approximately 5-fold higher in pentose (around 0.1%) compared to that in hexose (∼0.02%).18 Furthermore, unequal competence may stem from structural stability, as a pentose ring can produce an unstable aldofuranose, described as “puckered”, which is susceptible to reaction with amino acids.19 This preference was reflected by AGO, which demonstrated a preference for detecting pentose-derived AGEs, particularly in ribose (Fig. 4). These results indicate that AGO exhibits sensitive detection ability for AGEs generated by furanoses, especially ribose, surpassing AGE autofluorescence.3.4. AGO selectivity in a glycation mimicking model
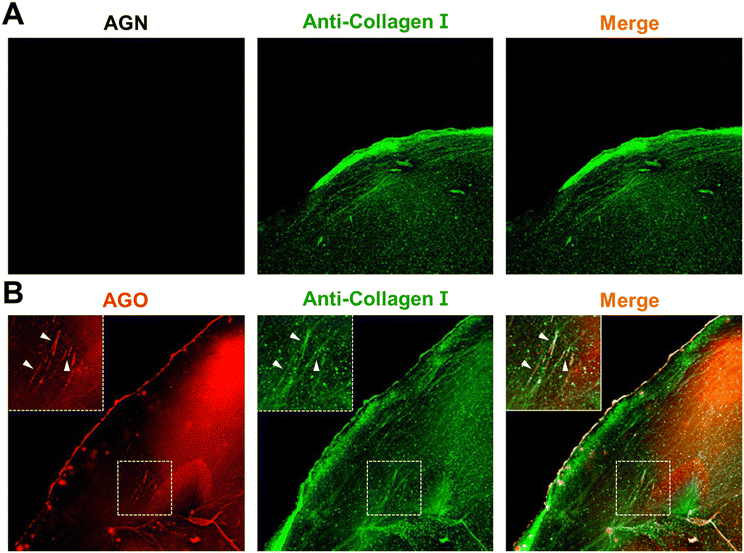
To delve deeper into the selectivity of AGO, we constructed a glycated collagen framework, mimicking the natural glycation of the matrix. Collagen serves as a susceptible target for glycation product formation due to several factors: (1) its prevalence as one of the most abundant proteins in the human body;20 (2) collagen's long half-life, increasing the likelihood of exposure to sugars;21 (3) its unique triple helical configuration consisting of lengthy amino acid chains, which facilitates multiple sugar-binding sites and makes it conducive to glycation;20 (4) the presence of lysine residues in collagen, which are particularly prone to glycation reactions.22 For these reasons, we hypothesized that collagen could serve as a representative model for glycation environments, demonstrating biochemically induced AGE formation.Initially, a collagen solution underwent pre-glycation with D-ribose. The alterations in the mechanical and structural characteristics of the glycated matrix were investigated in previous work. The study characterized a non-enzymatically glycated collagen framework, marked by elongated, reinforced, and extensively crosslinked collagen fibers.23 Following the glycation process, collagen microgels were treated with 10 μM AGO, and samples stained with AGN (negative control) were prepared together to validate the selectivity of AGO after staining with anti-collagen I. In comparison to AGN, which lacked the ability to detect AGEs (Fig. 5A), the images clearly demonstrated that AGO could stain AGEs (Fig. 5B), with fluorescence overlapping the signal of the collagen antibody. This result revealed the capability of AGO to selectively detect AGEs even in a glycation mimicking model.
4. Conclusions
The accumulation of AGEs has emerged as a significant contributor, among the numerous factors implicated in aging. Despite their importance, the relationship between aging and AGEs remains poorly understood due to the lack of reliable monitoring tools. To address this need, we developed a novel probe, AGO, showing high selectivity for AGEs over 2080 BODIPY-fluorescence compounds. Also, we found that phenoxybenzene enables sensitive detection of AGEs, along with its appropriate positioning, through structure–activity relationship studies. Moreover, AGO exhibited sensitive detection of AGEs over time, while autofluorescence reached its limit at a certain point. The potential of AGO to replace autofluorescence for measuring AGEs more accurately was revealed.Motivated by the sensitivity of AGO, we investigated its selectivity for AGEs derived from different types of carbohydrates. The results revealed that AGO can systematically measure the kinetics of AGE formation depending on glycation effects of carbohydrates.
Finally, we confirmed the selectivity of AGO in a glycated collagen model, mimicking natural phenomena.23 Given that AGEs can cross-link with collagen fibers, resulting in the formation of rigid and dysfunctional collagen structures,24,25 our results might make significant progress in understanding the role of AGEs in aging, further suggesting the potential of AGO as an effective diagnostic tool for detecting AGEs in vitro.
Author contributions
Conceptualization: H. Cho, N.-K. Hong, H.-Y. Kwon, N.-Y. Kang, L. M. C, and Y.-T. Chang; methodology: H. Cho, N.-K. Hong, I. Yong, L. M. C, P. Kim, and Y.-T. Chang; software: H. Cho and N.-K. Hong; validation: H. Cho, N.-K. Hong, and I. Yong; investigation: H. Cho, N.-K. Hong, I. Yong, H.-Y. Kwon, N.-Y. Kang, L. M. C, P. Kim, and Y.-T. Chang; data curation: H. Cho and N.-K. Hong; writing – original draft preparation: H. Cho.; writing – review and editing: Y.-T. Chang.; visualization: H. Cho, N-K. Hong, and I. Yong.; supervision: Y.-T. Chang.; project administration: Y.-T. Chang.; funding acquisition: H.-Y. Kwon, N.-Y. Kang, and Y.-T. Chang. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declareAcknowledgements
This research was supported by the Basic Science Research Institute Fund (2021R1A6A1A10042944 to Y.-T. C), the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2023R1A2C300453411 to Y.-T. C), the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2020R1A2C2009776 to N.-Y. K; RS-2023-00210972 to H.-Y. K), and the Ministry of Education (2020R1A6A1A03047902 to N.-Y. K.). H. C. is grateful for financial support from Hyundai Motor Chung Mong-Koo foundation.Notes and references
- R. Singh, A. Barden, T. Mori and L. Beilin, Diabetologia, 2001, 44, 129–146 CrossRef CAS PubMed.
- P. Gkogkolou and M. Bohm, Derm.-Endocrinol., 2012, 4, 259–270 CrossRef CAS PubMed.
- R. G. Paul and A. J. Bailey, Int. J. Biochem. Cell Biol., 1996, 28, 1297–1310 CrossRef CAS PubMed.
- J. Chaudhuri, Y. Bains, S. Guha, A. Kahn, D. Hall, N. Bose, A. Gugliucci and P. Kapahi, Cell Metab., 2018, 28, 337–352 CrossRef CAS PubMed.
- A. Perrone, A. Giovino, J. Benny and F. Martinelli, Oxid. Med. Cell. Longevity, 2020, 2020, 3818196 Search PubMed.
- J. S. Lee, Y. K. Kim, M. Vendrell and Y. T. Chang, Mol. BioSyst., 2009, 5, 411–421 RSC.
- S. W. Yun, N. Y. Kang, S. J. Park, H. H. Ha, Y. K. Kim, J. S. Lee and Y. T. Chang, Acc. Chem. Res., 2014, 47, 1277–1286 CrossRef CAS PubMed.
- Y. K. Choi, J. J. Kim and Y. T. Chang, Acc. Chem. Res., 2019, 52, 3097–3107 CrossRef CAS PubMed.
- X. Liu and Y. T. Chang, Chem. Soc. Rev., 2022, 51, 1573–1591 RSC.
- H. G. Lee, N. K. Hong and Y. T. Chang, Chem. Commun., 2023, 59, 9372–9375 RSC.
- W. Xu, C. Ren, C. L. Teoh, J. Peng, S. H. Gadre, H. W. Rhee, C. L. Lee and Y. T. Chang, Anal. Chem., 2014, 86, 8763–8769 CrossRef CAS PubMed.
- W. Xu, J. Bai, J. Peng, A. Samanta and Y. T. Chang, Chem. Commun., 2014, 50, 10398–10401 RSC.
- Y. Wei, L. Chen, J. Chen, L. Ge and R. Q. He, BMC Cell Biol., 2009, 10, 10 CrossRef PubMed.
- J. Wang, X. Guo, L. Li, H. Qiu, Z. Zhang, Y. Wang and G. Sun, Molecules, 2018, 23 Search PubMed.
- C. Y. Shen, C. H. Wu, C. H. Lu, Y. M. Kuo, K. J. Li, S. C. Hsieh and C. L. Yu, Molecules, 2019, 24 Search PubMed.
- I. Syrovy, J. Biochem. Biophys. Methods, 1994, 28, 115–121 CrossRef CAS PubMed.
- J. M. Lee and S. P. Veres, J. Appl. Physiol., 1985, 2019(126), 832–841 Search PubMed.
- Y. Zhu, J. Zajicek and A. S. Serianni, J. Org. Chem., 2001, 66, 6244–6251 CrossRef CAS PubMed.
- Y. Wei, C. S. Han, J. Zhou, Y. Liu, L. Chen and R. Q. He, Biochim. Biophys. Acta, 2012, 1820, 488–494 CrossRef CAS PubMed.
- S. Ricard-Blum, Cold Spring Harbor Perspect. Biol., 2011, 3, a004978 Search PubMed.
- N. Verzijl, J. DeGroot, S. R. Thorpe, R. A. Bank, J. N. Shaw, T. J. Lyons, J. W. Bijlsma, F. P. Lafeber, J. W. Baynes and J. M. TeKoppele, J. Biol. Chem., 2000, 275, 39027–39031 CrossRef CAS PubMed.
- M. Yamauchi and M. Sricholpech, Essays Biochem., 2012, 52, 113–133 CrossRef CAS PubMed.
- M. Jang, S. W. Oh, Y. Lee, J. Y. Kim, E. S. Ji and P. Kim, Acta Biomater., 2022, 141, 255–263 CrossRef CAS PubMed.
- H. Al-Atif, Dermatol. Pract. Concept., 2022, 12, e2022018 CrossRef PubMed.
- R. C. Hoy, D. N. D'Erminio, D. Krishnamoorthy, D. M. Natelson, D. M. Laudier, S. Illien-Junger and J. C. Iatridis, JOR Spine, 2020, 3, e1126 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00590b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |