Open Access Article
Open Access ArticleA matrix metalloproteinase activation probe for painting human tumours†
Bethany
Mills
 a,
Dominic
Norberg
a,
Kevin
Dhaliwal
a,
Ahsan R
Akram
a,
Mark
Bradley
a,
Dominic
Norberg
a,
Kevin
Dhaliwal
a,
Ahsan R
Akram
a,
Mark
Bradley
 b and
Alicia
Megia-Fernandez
b and
Alicia
Megia-Fernandez
 *b
*b
aCentre for Inflammation Research, Queen's Medical Research Institute, University of Edinburgh, 147 Little France Crescent, EH16 4TJ Edinburgh, UK
bEaStCHEM School of Chemistry, University of Edinburgh, David Brewster Road, EH9 3FJ Edinburgh, UK. E-mail: v1amegia@exseed.ed.ac.uk
First published on 16th July 2020
Abstract
A probe that allows specific ‘painting’ of human tumours is described. Probe activation was mediated by specific matrix metalloproteinases, resulting not only in disruption of a FRET pair, but in the generation of a fragment that “fluorescently paints” human tumours. This probe demonstrated rapid and effective human tumour labelling with the potential to allow margin detection during surgical resection.
The concept of tissue-based fluorescent labelling has gained attention as a method for identification of diseased tissue margins during intra-operative cancer surgery,1–6 made possible with key advances in imaging instrumentation.7,8 Pivotal to this has been the development of fluorescent probes which provide disease-mediated contrast.9 Approved fluorescent contrast agents in this area include compounds such as 5-aminolevulinic acid which is selectively up-taken and metabolized by cancerous tissues to generate protoporphyrin IX.10 Other optical agents in clinical studies11 include labelled nanobodies,12 peptides, such as chlorotoxin (tozuleristide)13 which is currently in phase II/III studies for pediatric CNS tumours, and the bis-cyclic peptide GE-137, which targets the human hepatocyte growth factor receptor (c-MET).14 There are also a wide variety of protease based probes,15–21 with cathepsins being a key target where signals are generated by either covalent modification of the enzyme22 or by FRET dequenching.23,24 Such probes have been able to detect margins in nonmelanoma skin cancer.25 Other examples of probes explored for cancer imaging include those targeting DPP-IV for esophageal cancer,26,27 folate receptor targeted probes,28 PSMA for prostate cancer29 or those developed by Tsien,30 whereby MMP-2 activation liberated a cell penetrating peptide that locally tagged proximal tissue. Matrix metalloproteinase 13 (MMP-13 or collagenase 3) is an endopeptidase overexpressed in the microenvironment around both lung tumours and fibrotic tissue, and has been shown to play a role in early invasive pulmonary adenocarcinoma.31 MMP-2 and -9 (gelatinase A and B) are cancer-associated endopeptidases overexpressed in a variety of malignant tumors and often associated with aggressiveness and poor prognosis.32,33 Here we report on the rational design, synthesis and evaluation on human cancerous tissue of a novel MMP-imaging agent (3) which allowed MMP-mediated ‘painting’ of resected human tumour tissue.
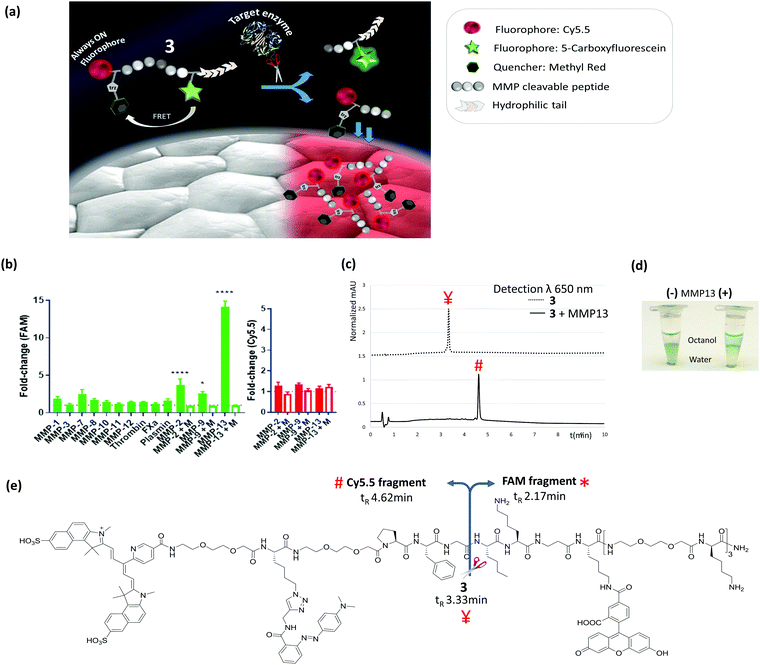
The designed probe (3) contained both a FRET pair between 5-carboxyfluorescein (FAM) and the quencher methyl red reporting on enzyme activity, as well as the incorporation of an ‘always-on’ far-red fluorophore (an in-house synthesized Cy5.5, ex/em 670/693 nm) with a spectral window distinct from FAM and away from tissue autofluorescence. The Cy5.5-fragment released enzymatically was responsible for the “tissue-painting” ability of the activated probe, which is attributable to the hydrophobicity of this fragment compared to the parent compound (Fig. 1a).
The peptide sequence (Pro-Phe-Gly-Nle-Lys-βAla, previously reported as MMP-2,9,13 substrate34,35) was synthesized by Fmoc solid-phase peptide synthesis on ChemMatrix resin using Oxyma/DIC as the coupling combination (Scheme 1). At the carboxy-terminus of the peptide, three replicates of bis-ethyleneglycol and D-lysine were added to ensure both solubility and stability against proteases.34 As part of this strategy we developed a novel Cy5.5 red dye, that was readily prepared on large scale in 4 steps, and contained a (5-carboxypyridin-2-yl) group to allow ready incorporation via an amide bond to the peptide by solid phase methods (ESI† for details). The Cy5.5, 5-carboxyfluorescein and methyl red were sequentially incorporated at the amino terminus of the peptide, on the Lys side chain (after Dde deprotection) and conjugated to the Lys(N3) residue via azide/alkyne cycloaddition respectively (Scheme 1 and ESI†). Probe 3 was purified and characterized by RP-HPLC and MALDI TOF MS and was fully aqueous soluble (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P −1.5) (ESI† for details).
P −1.5) (ESI† for details).
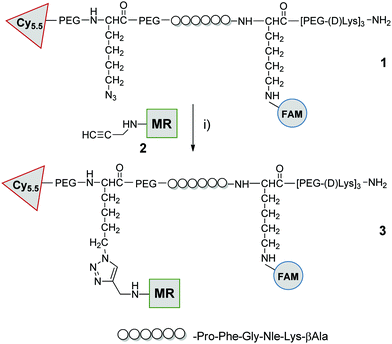 | ||
| Scheme 1 Probe synthesis: compound 1 was prepared by SPPS on chemmatrix resin as detailed in ESI,† after cleavage from the resin it was reacted with methylred-alkyne (2) by CuAAC click chemistry: (i) 2, CuSO4/ascorbate, tris(3-hydroxypropyltriazolylmethyl)amine, aminoguanidine, H2O, 30 °C, 5 h. FAM = 5-carboxyfluorescein, MR = methyl red (see ESI† for details and characterization). | ||
Target and off-target proteases were incubated with 3. Within 15 min, as anticipated, an increase in green fluorescence (FAM signal) was measurable from the probe incubated with the active domains of MMP-2, -9 and -13 (Fig. 1b and Fig. S1, ESI†), which was blockable in the presence of pan-MMP inhibitor marimastat.
Probe 3 was stable against the off-target proteases (Fig. 1b), with cleavage by MMPs specifically at Gly↑Nle, as confirmed by MALDI TOF MS (Fig. S2, ESI†). Importantly the Cy5.5 residue did not interfere with fluorescence in the green channel following MMP activation, with data similar to the control compound 4 that contained FAM & methyl red only (see Fig. S3 and S4, ESI†). In solution the Cy5.5 fluorescence intensity remained constant before and after cleavage (Fig. 1b), demonstrating that its intensity was independent of the FRET pairing. HPLC analysis confirmed that the Cy5.5-fragment obtained after cleavage was much more hydrophobic than the other two components, with its retention factor shifting from k = 4.2 for parent compound 3, to k = 8.0 for the Cy5.5 fragment (Fig. 1c and e), while MMP-13 treatment of the peptide 3 in biphasic buffer/octanol led to migration of the cleaved Cy5.5-labelled peptide into the octanol phase (Fig. 1d and Fig. S5, ESI†) demonstrating the hydrophobicity “switch-on” upon cleavage.
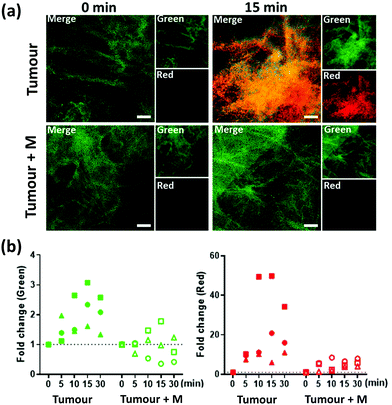
3 (5 μM) was applied onto human lung tumour tissue, from three individual patients: two with squamous cell carcinoma and one with adenocarcinoma (MMPs presence within these tissues was confirmed by gelatin zymography,36 Fig. S6, ESI†) and imaged over 30 min (Fig. 2 and ESI† videos) with a fibre-based imaging system,37 at each time point 300 frames were collected over 10 s. Marimastat (50 μM) was pre-incubated with a separate slice of the same tissue sample to serve as a control. An increase in FAM fluorescence (green) was observed over time, which was reduced in the presence of marimastat. Importantly, whilst the green fluorescence increased by some ∼2-fold above tissue autofluorescence during the imaging procedure, the Cy5.5 (red) fluorescence on the tissue increased by more than 20-fold, differing significantly from the results seen in vitro, and attributable to the hydrophobic nature of the cleaved Cy5.5 fragment. Thus, during enzymatic cleavage, the released Cy5.5 component immediately ‘sticks’ to the tissue in the proximity of the MMP, thus acting as a ‘tumour paint’. By contrast, healthy tissue was interrogated by the same procedure and activation of probe 3 happened only after spiking the tissue with MMP-2 or MMP-13 (Fig. S7, ESI†).
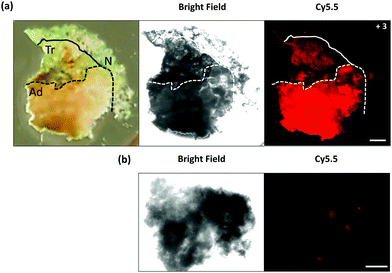
The tumour painting ability was further demonstrated by inspection of a freshly excised lung tissue sample (Fig. 3a), which contained tumour (adenocarcinoma), macroscopically normal tissue and a transitional zone in-between within the single slice. Compound 3 (50 μM) was added to the slice of lung tissue and imaged after 40 min with a wide-field fluorescence microscope. This procedure enabled the full tissue to be examined and fluorescence to be collected at a higher resolution and from a larger field of view compared to the fibre based imaging. A clear increase in brightness was demonstrated in the Cy5.5 fluorescence channel (red) within the adenocarcinoma region (4.1 fold increase), compared to the adjacent normal tissue, with a moderate level of fluorescence (2.1 fold increase) detectable in the transition zone, thus demonstrating that 3 can be utilized to ‘paint’ tumour margins on freshly resected tissue. A tissue slice from the same sample was imaged without the addition of 3 (Fig. 3b). Autofluorescence in the Cy5.5 channel was akin to that seen within the macroscopically normal zone of the tissue treated with compound 3, demonstrating that the adenocarcinoma did not strongly autofluorescence within this spectral region, and that the cleaved Cy5.5 fragment enabled tumour margin delineation, with the localized activated probe adhering onto the tissue, circumventing diffusion and loss of the signal.
In conclusion, this highly water soluble MMP imaging probe allows ‘tumour painting’ and provides a quick, simple and robust method of identifying areas of high MMP activity, with utility both in vitro, in situ (real-time fibre based imaging) and ex vivo (microscopic imaging) scenarios. The method has the potential to guide intraoperative surgery via fluorescent ‘painting’ of tumour margins. Its highly-specific activation (inhibited by marimastat) leads to novel and highly desirable tissue staining characteristics and adherence to cancerous human tumour tissue. Our approach offers advantages over other protease-activatable probes which may diffuse away from the target site.17 Other benefits include the speed of probe activation,38 demonstrated with the fibre-based imaging, and the ability to topically apply the imaging agent.
We would like to thank Engineering and Physical Sciences Research Council (EPSRC, UK) Interdisciplinary Research Collaboration grant EP/K03197X/1 and EP/R005257/1. AA is supported by a Cancer Research UK Clinician Scientist Fellowship (A24867). The research leading to these results has received funding from the EU Seventh Framework Program FP7 (No. 326465, AMF). Data used within this publication can be accessed at https://doi.org/10.7488/ds/2792.
Conflicts of interest
There are no conflicts to declare.References
- Q. T. Nguyen and R. Y. Tsien, Nat. Rev. Cancer, 2013, 13, 653–662 CrossRef CAS PubMed.
- Y. Zheng, H. Yang, H. Wang, K. Kang, W. Zhang, G. Ma and S. Du, Ann. Transl. Med., 2019, 7(Suppl 1), S6 CrossRef PubMed.
- M. J. Landau, D. J. Gould and K. M. Patel, Ann. Transl. Med., 2016, 4, 392 CrossRef PubMed.
- H. Orbay, J. Bean, Y. Zhang and W. Cai, Curr. Pharm. Biotechnol., 2014, 14, 733–742 Search PubMed.
- T. Nagaya, Y. A. Nakamura, P. L. Choyke and H. Kobayashi, Front. Oncol., 2017, 7, 314 CrossRef PubMed.
- K. R. Tringale, J. Pang and Q. T. Nguyen, Wiley Interdiscip. Rev.: Syst. Biol. Med., 2018, 10, e1412 CrossRef.
- A. V. Dsouza, H. Lin, E. R. Henderson, K. S. Samkoe and B. W. Pogue, J. Biomed. Opt., 2016, 21, 80901 CrossRef PubMed.
- G. Rossi, A. Tarasconi, G. Baiocchi, G. L. De' Angelis, F. Gaiani, F. Di Mario, F. Catena and R. Dalla Valle, Acta Biomed., 2018, 89, 135–140 Search PubMed.
- M. Koch and V. Ntziachristos, Annu. Rev. Med., 2016, 67, 153–164 CrossRef CAS PubMed.
- M. O. Chohan and M. S. Berger, J. Neuro-Oncol., 2019, 141, 517–522 CrossRef CAS PubMed.
- M. Gao, F. Yu, C. Lv, J. Choo and L. Chen, Chem. Soc. Rev., 2017, 46, 2237–2271 RSC.
- P. Debie, N. Devoogdt and S. Hernot, Antibodies, 2019, 8, 12 CrossRef CAS PubMed.
- C. G. Patil, D. G. Walker, D. M. Miller, P. Butte, B. Morrison, D. S. Kittle, S. J. Hansen, K. L. Nufer, K. A. Byrnes-Blake, M. Yamada, L. L. Lin, K. Pham, J. Perry, J. Parrish-Novak, L. Ishak, T. Prow, K. Black and A. N. Mamelak, Neurosurgery, 2019, 85, 641–649 CrossRef PubMed.
- J. Burggraaf, I. M. C. Kamerling, P. B. Gordon, L. Schrier, M. L. de Kam, A. J. Kales, R. Bendiksen, B. Indrevoll, R. M. Bjerke, S. A. Moestue, S. Yazdanfar, A. M. J. Langers, M. Swaerd-Nordmo, G. Torheim, M. V. Warren, H. Morreau, P. W. Voorneveld, T. Buckle, F. W. B. van Leeuwen, L.-I. Odegardstuen, G. T. Dalsgaard, A. Healey and J. C. H. Hardwick, Nat. Med., 2015, 21, 955–961 CrossRef CAS PubMed.
- M. Garland, J. J. Yim and M. Bogyo, Cell Chem. Biol., 2016, 23, 122–136 CrossRef CAS PubMed.
- C. Wang, Z. Wang, T. Zhao, Y. Li, G. Huang, B. D. Sumer and J. Gao, Biomaterials, 2018, 157, 62–75 CrossRef CAS PubMed.
- H.-Y. Hu, S. Gehrig, G. Reither, D. Subramanian, M. A. Mall, O. Plettenburg and C. Schultz, Biotechnol. J., 2014, 9, 266–281 CrossRef CAS PubMed.
- E. A. Lemke and C. Schultz, Nat. Chem. Biol., 2011, 7, 480 CrossRef CAS PubMed.
- M. Staderini, A. Megia-Fernandez, K. Dhaliwal and M. Bradley, Bioorg. Med. Chem., 2018, 26, 2816–2826 CrossRef CAS PubMed.
- Y. Urano, in Development of Novel Fluorogenic Probes for Realizing Rapid Intraoperative Multi-color Imaging of Tiny Tumors, ed. Y. Toyama, A. Miyawaki, M. Nakamura and M. Jinzaki, Make Life Visible, Springer, Singapore, 2020 Search PubMed.
- A. Mochida, F. Ogata, T. Nagaya, P. L. Choyke and H. Kobayashi, Bioorg. Med. Chem., 2018, 26, 925–930 CrossRef CAS PubMed.
- E. Segal, T. R. Prestwood, W. A. van der Linden, Y. Carmi, N. Bhattacharya, N. Withana, M. Verdoes, A. Habtezion, E. G. Engleman and M. Bogyo, Chem. Biol., 2015, 22, 148–158 CrossRef CAS PubMed.
- M. J. Whitley, D. M. Cardona, A. L. Lazarides, I. Spasojevic, J. M. Ferrer, J. Cahill, C.-L. Lee, M. Snuderl, D. G. Blazer, III, E. S. Hwang, R. A. Greenup, P. J. Mosca, J. K. Mito, K. C. Cuneo, N. A. Larrier, E. K. O'Reilly, R. F. Riedel, W. C. Eward, D. B. Strasfeld, D. Fukumura, R. K. Jain, W. D. Lee, L. G. Griffith, M. G. Bawendi, D. G. Kirsch and B. E. Brigman, Sci. Transl. Med., 2016, 8, 320–324 Search PubMed.
- J. J. Yim, M. Tholen, A. Klaassen, J. Sorger and M. Bogyo, Mol. Pharmaceutics, 2018, 15, 750–758 CrossRef CAS PubMed.
- Y. Liu, E. Walker, S. R. Iyer, M. Biro, I. Kim, B. Zhou, B. Straight, M. Bogyo, J. P. Basilion, D. L. Popkin and D. L. Wilson, J. Med. Imaging, 2019, 6, 016001 Search PubMed.
- H. Onoyama, M. Kamiya, Y. Kuriki, T. Komatsu, H. Abe, Y. Tsuji, K. Yagi, Y. Yamagata, S. Aikou, M. Nishida, K. Mori, H. Yamashita, M. Fujishiro, S. Nomura, N. Shimizu, M. Fukayama, K. Koike, Y. Urano and Y. Seto, Sci. Rep., 2016, 6, 26399 CrossRef CAS PubMed.
- Y. Kitagawa, S. Tanaka, Y. Kuriki, K. Yamamoto, A. Ogasawara, T. Nejo, R. Matsuura, T. Koike, T. Hana, S. Takahashi, M. Nomura, S. Takayanagi, A. Mukasa, M. Kamiya, Y. Urano and N. Saito, Front. Oncol., 2019, 9, 727 CrossRef PubMed.
- S. M. Mahalingam, S. A. Kularatne, C. H. Myers, P. Gagare, M. Norshi, X. Liu, S. Singhal and P. S. Low, J. Med. Chem., 2018, 61, 9637–9646 CrossRef CAS PubMed.
- M. Kawatani, K. Yamamoto, D. Yamada, M. Kamiya, J. Miyakawa, Y. Miyama, R. Kojima, T. Morikawa, H. Kume and Y. Urano, J. Am. Chem. Soc., 2019, 141, 10409–10416 CrossRef CAS PubMed.
- Q. T. Nguyen, E. S. Olson, T. A. Aguilera, T. Jiang, M. Scadeng, L. G. Ellies and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4317–4322 CrossRef CAS PubMed.
- M. Salaün, J. Peng, H. H. Hensley, N. Roder, D. B. Flieder, S. Houlle-Crépin, O. Abramovici-Roels, J.-C. Sabourin, L. Thiberville and M. L. Clapper, PLoS One, 2015, 10, e0132960 CrossRef PubMed.
- B. Bauvois, Biochim. Biophys. Acta, Rev. Cancer, 2012, 1825, 29–36 CrossRef CAS PubMed.
- S. Chakrabarti and K. D. Patel, Exp. Lung Res., 2005, 31, 599–621 CrossRef CAS PubMed.
- A. Megia-Fernandez, B. Mills, C. Michels, S. V. Chankeshwara, N. Krstajić, C. Haslett, K. Dhaliwal and M. Bradley, Org. Biomol. Chem., 2018, 16, 8056–8063 RSC.
- M. Bradley, S. V. Chankeshwara and A. Megia-Fernandez. WO Pat. 2016151299A1, 2016 Search PubMed.
- M. Toth and R. Fridman, in Metastasis Research Protocols: Volume I: Analysis of Cells and Tissues, ed. S. A. Brooks and U. Schumacher, Humana Press, Totowa, New Jersey, 2001 Search PubMed.
- N. Krstajić, B. Mills, I. Murray, A. Marshall, D. Norberg, T. H. Craven, P. Emanuel, T. R. Choudhary, G. O. S. Williams, E. Scholefield, A. R. Akram, A. Davie, N. Hirani, A. Bruce, A. Moore, M. Bradley and K. Dhaliwal, J. Biomed. Opt., 2018, 23, 1–12 Search PubMed.
- Y. Urano, M. Sakabe, N. Kosaka, M. Ogawa, M. Mitsunaga, D. Asanuma, M. Kamiya, M. R. Young, T. Nagano, P. L. Choyke and H. Kobayashi, Sci. Transl. Med., 2011, 3, 110–119 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional results, videos and experimental procedures. See DOI: 10.1039/d0cc03886e |
| This journal is © The Royal Society of Chemistry 2020 |