Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Does deamidation affect inhibitory mechanisms towards amyloid protein aggregation?†
Yuko P. Y.
Lam
 a,
Cookson K. C.
Chiu
a,
Cookson K. C.
Chiu
 a,
Christopher A.
Wootton
a,
Christopher A.
Wootton
 a,
Ian
Hands-Portman
a,
Ian
Hands-Portman
 b,
Meng
Li
b,
Meng
Li
 a,
Mark P.
Barrow
a,
Mark P.
Barrow
 a and
Peter B.
O'Connor
a and
Peter B.
O'Connor
 *a
*a
aDepartment of Chemistry, University of Warwick, Coventry, UK. E-mail: p.oconnor@warwick.ac.uk
bDepartment of Life Sciences, University of Warwick, Coventry, UK
First published on 30th July 2020
Abstract
Deamidated amyloid proteins have been shown to accelerate fibril formation. Herein, the results show the inhibition performance and the interaction site between site-specific inhibitor and amyloid protein are significantly influenced by deamidation; while the inhibition mechanism of non-site specific inhibitor shows no significant disruption caused by amyloid protein deamidation.
Deamidation is a non-enzymatic post-translational modification associated with protein ageing.1 Deamidation at asparagine (Asn) and glutamine (Gln) residues contributes to the formation of aspartic acid or iso-aspartic acid and glutamic acid or γ-glutamic acid respectively.1 Deamidated amyloid proteins have been shown to accelerate amyloid fibril formation;2–5 and iso-aspartic acid, one of the deamidated products, was found to have a greater acceleration effect compared to the other deamidation isomeric product – aspartic acid.2,3 Even though deamidated amyloid proteins have been shown to have a strong correlation with the rate of fibril formation, the effect of deamidation on the inhibition of amyloid protein aggregation has not yet been fully addressed.
Human islet amyloid polypeptide (hIAPP) is a 37 amino acid hormone with an intramolecular disulfide bond between Cys-2 and Cys-7.6 hIAPP is stored in pancreatic β-cell secretory granules and co-secreted with insulin to regulate blood glucose levels.7 In general, monomeric hIAPP is water soluble and inherently disordered, conversely hIAPP amyloid fibrils are water insoluble and are observed in type II diabetes (T2D) patients.6 hIAPP contains 6 Asn and 1 Gln residues; deamidation is observed at Asn-21, Asn-22, and Asn-35 residues after 1 week incubation at 37 °C.3 Deamidated hIAPP has been shown to accelerate amyloid fibril formation via disruption of the β-sheet structure around the Leu-27 residue.4 Many inhibitors have been reported to effectively inhibit hIAPP amyloid fibril formation;8 the inhibition performance of these inhibitors against deamidated hIAPP aggregation, however, has not yet been studied.
Human insulin (hINS), one of the most common inhibitors against hIAPP aggregation, contains 51 amino acid residues over 2 chains linked by an intra-strand disulfide bond and 2 inter-strand disulfide bonds.9 hINS is also an amyloid protein which aggregates rapidly at low pH (∼pH 2–3);10 while hIAPP aggregates much faster than hINS at neutral pH (∼pH 7).11 Heterodimers, containing a hINS and a hIAPP molecules, have been observed in previous studies which are believed to be the key complex which prevents the formation of amyloid fibrils.12
(−)-Epigallocatechin 3-gallate (EGCG) is an extract from green tea and has shown to be an effective inhibitor against various types of amyloid protein aggregation, including amyloid β, α-synuclein, and hIAPP.13,14 EGCG molecules attach to amyloid proteins non-specifically which is believed to re-direct the aggregation pathway into the formation of an off-pathway oligomer in order to prevent the generation of amyloid fibrils.14
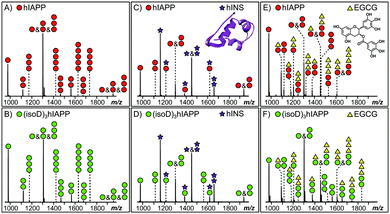
MS provides the high sensitivity and resolution required to study the interaction between early oligomers and inhibitors.15 Early hIAPP and (isoD)3hIAPP oligomers, from monomer up to pentamer, were observed in the MS spectra of the 10 μM solutions of hIAPP and (isoD)3hIAPP (Fig. 1A and B) respectively. These results are similar to the observations obtained in the previous studies,3,16 indicating both peptides are amyloidogenic and aggregate rapidly to form high order oligomers in solution. When 10 μM hIAPP and (isoD)3hIAPP were individually co-incubated with equimolar hINS solution, only the heterodimer complex (one hIAPP/(isoD)3hIAPP plus one hINS), as well as monomers and homodimers of hIAPP/(isoD)3hIAPP and hINS were observed in the MS spectra (Fig. 1C and D), suggesting the interactions between both the polypeptides and hINS are site-specific; which agrees with the previous results demonstrating the interaction between hIAPP and hINS.17 When 10 μM hIAPP and (isoD)3hIAPP solutions were instead mixed with equimolar of EGCG solution (Fig. 1E and F), early homo-oligomers of hIAPP/(isoD)3hIAPP, from monomer to trimer, as well as complexes composed of hIAPP/(isoD)3hIAPP and EGCG were observed in the MS spectra. The number of EGCG molecules attaching to the oligomers ranged from 0.5 (one EGCG molecule plus two polypeptides) to 3 (three EGCG molecules plus one polypeptide), indicating the interaction between the polypeptides and EGCG molecule is non-specific which is similar to the results observed by the Young et al. study.13 The results shown in the MS spectra of hIAPP and (isoD)3hIAPP mixed with hINS and EGCG demonstrate there is no significant difference between the early oligomers of wild-type and deamidated hIAPP attaching to either site specific (hINS) or non-site specific (EGCG) inhibitors.
The dissociation energy curve obtained from collisionally activated dissociation tandem MS (CAD MS/MS) shows the complexes of the polypeptides with hINS/EGCG involve fragile, non-covalent interactions (Fig. S1 and S2, ESI†). Electron capture dissociation tandem MS (ECD MS/MS) is a fast fragmentation process that can preserve the non-covalent interactions between molecules whilst fragmenting the protein backbone to generate sequence information.18 ECD MS/MS has been shown to successfully determine the aggregation sites between amyloid proteins as well as the interaction sites between ligands and amyloid proteins.16,19,20 The 7+ charge state heterodimer, [hIAPP + hINS], was isolated and fragmented by ECD MS/MS; monomer fragments of hIAPP and hINS as well as the heterodimer fragments were observed in the spectrum (Fig. S3, S4 and Table S1, ESI†). All heterodimer fragments were summarised (Fig. 2A) and used to determine the interaction regions between hIAPP and hINS units. Purely monomer fragments of hIAPP or hINS do no inform the interaction site explicitly,16 therefore, were not considered during structure elucidation. The ECD spectrum showed the c35 and c36 hIAPP fragments as well as a sequential line of hIAPP z-ion fragments (z3–34) attaching to an intact hINS species, indicating the primary interaction site exists at the residue Asn-35 on hIAPP which is towards the C-terminus of the polypeptide (Fig. 2F). In a previous study, we have demonstrated that Ser-28/29 and Asn-35 are two critical areas for hIAPP aggregation (Fig. 2E);16 the interaction observed above suggests that hINS interacts with hIAPP at its critical aggregation site (Asn-35) preventing the interaction with another hIAPP unit to form high-order oligomers and thereby inhibits the formation of amyloid fibrils in a single “capping” process to form a heterodimer of hIAPP and hINS.
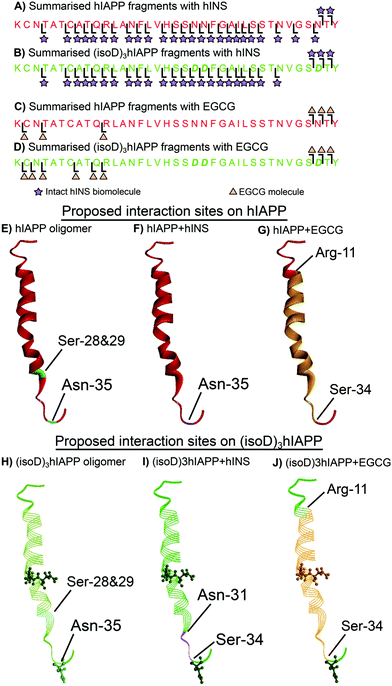 | ||
| Fig. 2 Summarised fragments of hINS attached to (A) hIAPP and (B) (isoD)3hIAPP. Summarised fragments of EGCG attached to (C) hIAPP and (D) (isoD)3hIAPP. The proposed interaction sites on (E) pure hIAPP (PDB 2KB8)21 with (F) hINS and (G) EGCG. The proposed interaction sites on (H) pure (isoD)3hIAPP with (I) hINS and (J) EGCG. | ||
ECD MS/MS was then applied to the heterodimer composed of an (isoD)3hIAPP and a hINS biomolecules (Fig. S5, S6 and Table S2, ESI†). In the ECD spectrum, (isoD)3hIAPP fragments of c34, c35, and c36 as well as a series of z-ions (z7–33) were shown to attach to an intact hINS molecule (Fig. 2B), indicating the interaction site between (isoD)3hIAPP and hINS locates between Asn-31 and Ser-33 residue (Fig. 2I), which is similar to binding with hIAPP, but shifts further away from the C-terminus of the polypeptide compared to the [hIAPP + hINS] complex (Fig. 2F). The ECD results of the two species binding with intact hINS demonstrates that deamidation influenced the interaction site between the polypeptides and the site-specific inhibitor – hINS, which may result in influencing the performance of the inhibitor.
ECD MS/MS was also used to determine the interaction site between EGCG and the polypeptides (Fig. S7, S8 and Tables S3, S4, ESI†). The c34–36 fragments and z27–35 fragments of hIAPP/(isoD)3hIAPP were shown to attach to an EGCG molecule in both ECD spectra (Fig. 2B and D), indicating the interaction site between hIAPP/(isoD)3hIAPP and EGCG are similar to each other but different from the interactions with hINS. The interaction region for both species was shown to be between Arg-11 and Ser-34 residues (Fig. 2G and J). This suggested that deamidation in hIAPP resulted in minimal disruption of the interaction site between the polypeptides and EGCG, suggesting EGCG may still be an effective inhibitor to prevent the aggregation of deamidated polypeptides. The large binding region also suggests that EGCG binds at several positions somewhat stochastically or that it engages in multiple hydrogen bonds within this region, distorting the peptide conformation accordingly.
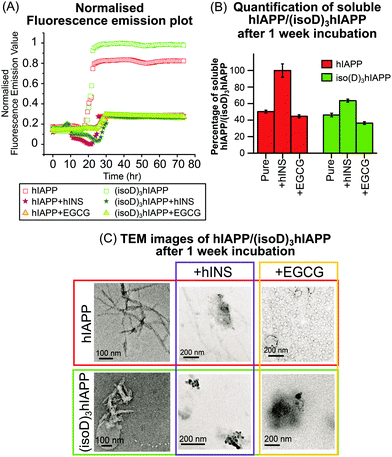
Fluorescence spectroscopy is useful to quantify amyloid fibrils in solution because of the formation of mature β-sheet fibrils which preferentially attach to thioflavin T (ThT) dye and produce a detectable fluorescence signal.22 In this experiment, pure hINS also generates amyloid fibrils which produce a fluorescence emission signal (Fig. S9C, ESI†). In order to focus on the fluorescence emission signal solely generated by the hIAPP and (isoD)3hIAPP fibrils (Fig. 3A); the fluorescence emission intensities of the ThT solution, hINS, and EGCG were baseline subtracted at each experimental time-point (Fig. S9, ESI†). The lag phases of the 50 μM hIAPP and (isoD)3hIAPP were both 21 hours; while the relative intensity of the amyloid fibrils formed in the (isoD)3hIAPP solution was 20% higher than that in the hIAPP solution (Fig. 3A), indicating deamidated hIAPP solution generates amyloid fibrils faster than wild-type solution, which is similar to previous observations.3 The lag phase of hIAPP and (isoD)3hIAPP incubated with equimolar hINS solutions were at 25 and 29 hours respectively; indicating hINS can effectively postpone the formation of amyloid fibrils in the non-deamidated and deamidated polypeptides solutions; meanwhile, a longer incubation time is required for (isoD)3hIAPP to form mature fibrils in the solution compared to hIAPP. The lag phase of hIAPP and (isoD)3hIAPP incubated with equimolar of EGCG were both at 26 hour, suggesting EGCG can also potentially delay the formation of amyloid fibrils for both deamidated and non-deamidated polypeptides to a similar degree. The relative intensities of the mature fibrils formed in the solutions of hIAPP and (isoD)3hIAPP incubated with either inhibitors (hINS or EGCG) were very similar. Both hINS and EGCG were shown to reduce the amount of hIAPP amyloid fibrils formed in the solutions by 66.5%; while the amount of (isoD)3hIAPP amyloid fibrils were reduced by 72.5%, suggesting both inhibitors are effective in reducing the formation of mature β-sheet amyloid fibrils consisting of either non-deamidated or deamidated polypeptides.
Transmission electron microscopy (TEM) is commonly used to observe the overall structures of mature aggregates and fibrils in solution. hIAPP and (isoD)3hIAPP samples, including solutions mixed with either insulin or EGCG, were incubated at 37 °C for 1 week and the structures of aggregates were observed using TEM. Elongated and branched fibrils were observed in the pure hIAPP incubated sample; while short and dense fibrils were found in the (isoD)3hIAPP incubated sample (Fig. 3C, left column), in which similar observations were shown in previous studies.3 When equimolar amounts of hINS were mixed with hIAPP for 1 week incubation, fewer amounts of elongated fibrils were observed in the TEM image, while dense amorphous aggregates were found in the mixture of (isoD)3hIAPP and hINS solution (Fig. 3C, middle column). The results imply deamidation did affect the inhibitory mechanism of hINS towards the polypeptide aggregation which results in the formation of amorphous aggregates. When equimolar amounts of EGCG and hIAPP were mixed and incubated for 1 week, globular vesicles were clearly observed in the TEM image with some dense amorphous aggregates attaching to them (Fig. 3C, top right column). Compared to the TEM images of incubated EGCG solution (Fig. S10, ESI†), the increased in size of the globular vesicles from 12 nm to 48 nm are believed to be a mixture of EGCG and hIAPP molecules. When equimolar amounts of EGCG were incubated with (isoD)3hIAPP, globular vesicles appeared to collapse and only dense amorphous aggregates were observed in the solutions. The TEM results suggest deamidated hIAPP not only affects the aggregate structures, but also the inhibitory mechanism of potential inhibitors toward its aggregation.
TEM images are used to observe the overall structure of aggregates, however, it is difficult to provide quantitative information of the aggregates or their composition, and thus it is difficult to quantitatively compare the inhibition performance between potential inhibitors. Herein, the performances of the potential inhibitors were justified by quantifying the amount of remaining soluble hIAPP or (isoD)3hIAPP in the fresh and 1 week incubated solutions (Fig. 3B). A 2-tailed t-test was performed to determine whether the differences measured between the fresh and incubated solutions were significant. The incubated solutions contained only 50% soluble hIAPP and 46% (isoD)3hIAPP quantifiable by MS and the t-test p values were less than 0.05, indicating both polypeptides aggregate rapidly in a solution and (isoD)3hIAPP aggregates faster than hIAPP in solution which is similar to the previous observation.3 The remaining soluble hIAPP in the [hIAPP + hINS] 1 week incubated solution was ∼100% and the t-test p value of the fresh and incubated solutions was greater than 0.05, suggesting hINS appears to be a good inhibitor for hIAPP aggregation as nearly all hIAPP can be recovered after the 1 week incubation period. In contrast, only 64% of soluble (isoD)3hIAPP was detected after 1 week incubation with a t-test value less than 0.05, indicating hINS has much less effect in preventing deamidated polypeptide aggregation. Interpretation of the results obtained in the MS spectrum (Fig. 1C and D), together with those from fluorescence spectroscopy (Fig. 3A), and TEM (Fig. 3C) experiments, we hypothesise that hINS can effectively cap each (isoD)3hIAPP monomer producing heterodimers in a similar manner to [hIAPP + hINS] (supported by MS spectra), but will eventually form amorphous aggregates (supported by fluorescence spectroscopy and TEM images) which is not a reversible process (supported by MS quantification). The levels of soluble hIAPP and (isoD)3hIAPP were significantly reduced by 55% and 63% in the 1 week EGCG incubated solution respectively, suggesting aggregates are formed in the incubated solutions and the rate of aggregate formation in (isoD)3hIAPP solution is higher than hIAPP solution. With the data observed via fluorescence spectroscopy (Fig. 3A) and TEM (Fig. 3C) experiments, we understand that EGCG triggers the formation of amorphous aggregates in a similar manner for both non-deamidated and deamidated polypeptides.
To conclude, our results demonstrate deamidation does not affect the binding stoichiometry between the early oligomers of amyloidogenic polypeptides and potential inhibitors. However, deamidation can potentially affect the interaction site of a site-specific inhibitor (hINS) which significantly alters the inhibitory pathway to generate irreversible amorphous aggregates. In contrast, the interaction site and inhibition performance of non-site specific inhibitor are not obviously influenced by deamidation. With the increase in focus of deamidation-induced changes in amyloid fibril formation and inhibitor efficacy, the data shown herein could inform future therapeutic development for some amyloid diseases In the future, it would be of interest and importance to compare the cytotoxic effects between wild-type and deamidated hIAPPs with various potential inhibitors to evaluate the dependency of these interactions to the cytotoxic effects.
Conflicts of interest
There are no conflicts to declare.References
- N. E. Robinson and A. Robinson, Molecular clocks: deamidation of asparaginyl and glutaminyl residues in peptides and proteins, Althouse Press, Oregon, 2004 Search PubMed.
- T. Shimizu, H. Fukuda, S. Murayama, N. Izumiyama and T. Shirasawa, J. Neurosci. Res., 2002, 70, 451–461 CrossRef CAS PubMed.
- Y. P. Y. Lam, C. A. Wootton, I. Hands-Portman, J. Wei, C. K. C. Chiu, I. Romero-Canelon, F. Lermyte, M. P. Barrow and P. B. O’Connor, Chem. Commun., 2018, 54, 13853–13856 RSC.
- E. B. Dunkelberger, L. E. Buchanan, P. Marek, P. Cao, D. P. Raleigh and M. T. Zanni, J. Am. Chem. Soc., 2012, 134, 12658–12667 CrossRef CAS PubMed.
- N. M. Kad, N. H. Thomson, D. P. Smith, D. A. Smith and S. E. Radford, J. Mol. Biol., 2001, 313, 559–571 CrossRef CAS PubMed.
- P. Westermark, C. Wernstedt, E. Wilander, D. W. Hayden, T. D. O'Brien and K. H. Johnson, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 3881–3885 CrossRef CAS PubMed.
- A. Lukinius, E. Wilander, G. Westermark, U. Engström and P. Westermark, Diabetologia, 1989, 32, 240–244 CrossRef CAS PubMed.
- P. Cao, P. Marek, H. Noor, V. Patsalo, L.-H. Tu, H. Wang, A. Abedini and D. P. Raleigh, FEBS Lett., 2013, 587, 1106–1118 CrossRef CAS.
- J. Brange and L. Langkjœr, Stability and Characterization of Protein and Peptide Drugs, Springer, 1993, pp. 315–350 Search PubMed.
- J. Brange, L. Andersen, E. D. Laursen, G. Meyn and E. Rasmussen, J. Pharm. Sci., 1997, 86, 517–525 CrossRef CAS PubMed.
- J. Haas, E. Vöhringer-Martinez, A. Bögehold, D. Matthes, U. Hensen, A. Pelah, B. Abel and H. Grubmüller, ChemBioChem, 2009, 10, 1816–1822 CrossRef CAS PubMed.
- M. Baram, S. Gilead, E. Gazit and Y. Miller, Chem. Sci., 2018, 9, 4244–4252 RSC.
- L. M. Young, J. C. Saunders, R. A. Mahood, C. H. Revill, R. J. Foster, L.-H. Tu, D. P. Raleigh, S. E. Radford and A. E. Ashcroft, Nat. Chem., 2015, 7, 73–81 CrossRef CAS PubMed.
- D. E. Ehrnhoefer, J. Bieschke, A. Boeddrich, M. Herbst, L. Masino, R. Lurz, S. Engemann, A. Pastore and E. E. Wanker, Nat. Struct. Mol. Biol., 2008, 15, 558–566 CrossRef CAS PubMed.
- A. E. Ashcroft, J. Am. Soc. Mass Spectrom, 2010, 21, 1087–1096 CrossRef CAS PubMed.
- Y. P. Y. Lam, C. A. Wootton, I. Hands-Portman, J. Wei, C. K. C. Chiu, I. Romero-Canelon, F. Lermyte, M. P. Barrow and P. B. O’Connor, J. Am. Soc. Mass Spectrom, 2020, 31, 267–276 CrossRef CAS PubMed.
- A. C. Susa, C. Wu, S. L. Bernstein, N. F. Dupuis, H. Wang, D. P. Raleigh, J. E. Shea and M. T. Bowers, J. Am. Chem. Soc., 2014, 136, 12912–12919 CrossRef CAS PubMed.
- R. A. Zubarev and Curr. Opin, Biotechnology, 2004, 15, 12–16 CAS.
- F. Lermyte, J. Everett, Y. P. Y. Lam, C. A. Wootton, J. Brooks, M. P. Barrow, N. D. Telling, P. J. Sadler, P. B. O’Connor and J. F. Collingwood, J. Am. Soc. Mass Spectrom., 2019, 30, 2123–2134 CrossRef CAS.
- Y. Xie, J. Zhang, S. Yin and J. A. Loo, J. Am. Chem. Soc., 2006, 128, 14432–14433 CrossRef CAS.
- S. M. Patil, S. H. Xu, S. R. Sheftic and A. T. Alexandrescu, J. Biol. Chem., 2009, 284, 11982–11991 CrossRef CAS.
- P. Arosio, T. P. Knowles and S. Linse, Phys. Chem. Chem. Phys., 2015, 17, 7606–7618 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc03548c |
| This journal is © The Royal Society of Chemistry 2020 |