Open Access Article
Open Access ArticleRevealing the transient conformations of a single flavin adenine dinucleotide using an aerolysin nanopore†
Meng-Yin
Li
ab,
Ya-Qian
Wang
b,
Yi-Lun
Ying
 a and
Yi-Tao
Long
a and
Yi-Tao
Long
 *a
*a
aState Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, 210023, Nanjing, P. R. China. E-mail: yitaolong@nju.edu.cn
bSchool of Chemistry and Molecule Engineering, East China University of Science and Technology, 200237, Shanghai, P. R. China
First published on 23rd September 2019
Abstract
Flavin adenine dinucleotide (FAD) as a cofactor is involved in numerous important metabolic pathways where the biological function is intrinsically related to its transient conformations. The confined space of enzymes requires FAD set in its specific intermediate conformation. However, conventional methods only detect stable conformations of FAD molecules, while transient intermediates are hidden in ensemble measurements. There still exists a challenge to uncover the transient conformation of each FAD molecule, which hinders the understanding of the structure–activity relationship of the FAD mechanism. Here, we employ the electrochemically confined space of an aerolysin nanopore to directly characterize a series of transient conformations of every individual FAD. Based on distinguishable current blockages, the “stack”, “open”, and four quasi-stacked FADs are clearly determined in solution, which is further confirmed by temperature-dependent experiments and mutant aerolysin assay. Combined with molecular dynamics simulations, we achieved a direct correlation between the residual current ratio (I/I0) and FAD backbone angle. These results would facilitate further understanding of the structure–activity relationship in the flavoprotein.
Introduction
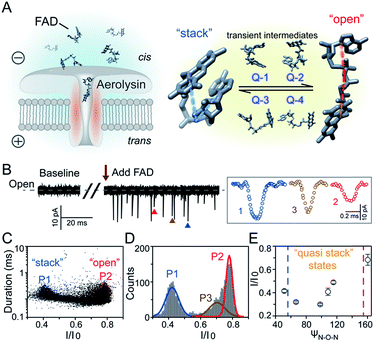
Flavoproteins are of great importance as they are involved in a wide range of biological processes, including photosynthesis, DNA repair and anti-oxidative stress.1 In particular, flavin adenine dinucleotide (FAD) is one of the most common flavin cofactors for electron transportation and photoreceptors in living systems.2–6 According to previous studies, FAD exhibits conformational dynamics in performing its biological functions, which is due to the local orientation of isoalloxazine and adenine.7,8 Conventional time-resolved fluorescence spectroscopy showed that FAD could be stabilized in two main shaped structures in aqueous solution.8,9 One is the “stack” form in which isoalloxazine and adenosine moieties face each other, while another is the elongated “open” form. However, X-ray crystal structures in the Protein Data Bank show that FAD adopts its intermediate conformations rather than the “stack” or “open” form within many local host enzyme environments.10 Therefore, the various binding conformations of FAD affect the diversity of the functions of FAD-based analogues, leading to a huge influence on the activity of related enzymes that are implicated in numerous disease pathways.11 Although several “partially stacked” conformations of FAD were suggested by computer modelling-combined with ion mobility spectrometry (IMS), many transient intermediates and conformation distribution are still hidden in these ensemble measurements. Therefore, there is an urgent need to uncover the transient conformations of FADs by using single molecule techniques.Nanopore technology as a powerful single molecule analysis has been used in nucleic acid sequencing, protein/peptide detection, enzyme activity measurements and detection of different conformations.12–27 A single biological membrane protein molecule provides a single-biomolecule interface for analysis of single molecules. The detection capability of biological nanopores is largely determined by the size and charge distribution of the sensing interface and interactions between the sensing interface and the analyte.28 Aerolysin, the founding member of a major pore-forming toxin family, has a long and narrow β-barrel with a high concentration of charged amino acid residues inside.29,30 A single aerolysin nanopore restricts the target molecule in this confined space, leading to the discrimination of oligonucleotides and peptides.31–37 Herein, we use the highly confined space inside the aerolysin nanopore to reveal the conformations for every individual FAD molecule by direct reading its nanopore blockages (Fig. 1A). The various conformations of FAD could be directly observed in the current recording at the single molecule level (Fig. 1B). Based on statistical analysis, the two main current populations were assigned to stable “stack” and “open” conformations (Fig. 1C and D), which was further confirmed by a series of temperature-dependent experiments, mutant aerolysin assay, and molecular dynamics (MD) simulations. More importantly, we determined four quasi-stacked conformations of FAD which facilitate dynamic transformation of the stack form FAD into the open form FAD in various polarity solvents. Our results provide new insights for studying the transient conformations of a single FAD molecule, which may enhance the understanding of the structure–activity relationship in the flavoprotein.
Results and discussion
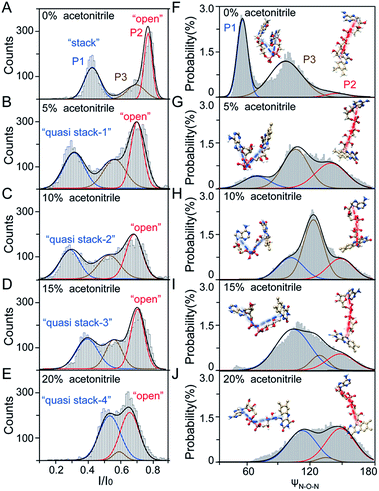
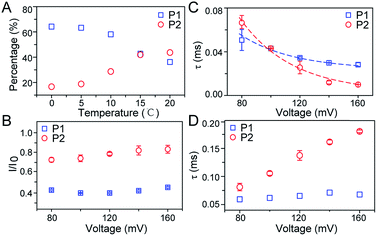
As illustrated in Fig. 1A, a FAD molecule is composed of a flavin mononucleotide and an adenosine monophosphate. Under the condition of pH = 8.0, the negatively charged FAD could be driven from the cis to trans side of aerolysin at positive applied voltage. The narrow lumen of aerolysin provides the electrochemical confinement of the single FAD molecule, which induces the substantial deep blockages (Fig. 1B). The statistical analysis of every single event shows that the FAD exhibits three current populations within the aerolysin nanopore. As shown in Fig. 1D, the I/I0 histogram could be fitted to three Gaussian distributions with a peak value of I/I0 centered at P1 of 0.42 ± 0.01, P2 of 0.78 ± 0.02 and P3 of 0.71 ± 0.02, respectively. Here, I represents the value of residual current and I0 is the open pore current, leading to a blockade current ratio of I/I0. The well-separated peaks of P1 and P2 occupy the majority of the events. However, less than 20% events fall into P3 which exhibits a large overlap with P2. According to previous time resolved fluorescence studies and computer modeling,8,38 the FAD molecule has unique structure dynamics in water, which is mainly stabilized in two distinct conformations as “open” and “stacked” (Fig. 1A). We suggest that the stabilized “open” and “stack” forms induce the events in P1 and P2. Since the “stack” conformation of FAD has the larger volume and larger cross-section (ESI Fig. S1†), which might also introduce stronger interaction between the FAD molecule and amino acid residues in the aerolysin lumen, leading to a deeper current blockage than the “open” form of FAD. Therefore, we propose that the “stack” FAD induces larger blockages in P1 while the “open” FAD generates the smaller blockages in P2. Since previous studies of the MD simulation reveal that FAD could adopt “partially stacked” as its metastable state,8 it is reasonable to speculate that the “partially stacked” conformation of FAD gives the current signal in P3. Note that the control experiments on K238Q, R220Q and R220A mutant aerolysins further support that the distinct current levels are caused by the FAD conformations rather than multiple bindings in the aerolysin lumen (ESI Fig. S2–S4†).Previous studies demonstrate that the π–π interaction between adenine and flavin could stabilize the stacked conformation of FAD. The decrease of solvent polarity would weaken their π–π interaction, leading to the transformation of the FAD molecule from “stack” into “open” form. To confirm the assignment of three populations to the FAD conformations, we carried out a series of solvent dependent experiments. Here, the acetonitrile was chosen to modulate the solvent polarity in 1.0 M KCl solution. Due to low destructivity of acetonitrile to the lipid membrane, the I–V curves of the aerolysin-membrane system remain constant in acetonitrile–KCl (ACN–KCl) solution with the acetonitrile ratio (RACN) ranging from 5–20% (ESI Fig. S5†). The stability of the aerolysin in ACN–KCl solution is further confirmed by the control experiments of the module molecule (ESI Fig. S6†). As shown in Fig. 2A–E, with the increase of RACN in KCl solution, P2 has a relatively consistent I/I0 value of 0.67 ± 0.04. Since the acetonitrile facilities the presence of the stable “open” structure, P2 is assigned to the “open” FAD. However, P1 shows remarkable shift in the I/I0 value from 0.42 to 0.50 with the increase of RACN. In 20% ACN–KCl solution, P3 merges into P2 with a serious overlap. This result confirms that P1 could be assigned to the “stack” FAD. Note that P1 undergoes the nonmonotonic changes of the I/I0 value. At lower RACN (<10%), the I/I0 of P1 decreases with the increase of RACN, while that increases with RACN increasing to 10%. Therefore, the conformational change of the stacked FAD to the open form may undergo several metastable states with considerable lifetime in the solution. These results indicate that the introduction of acetonitrile would regulate the stacked conformation of FAD to the open form. Note that this conformational change is confirmed by NMR spectra (ESI Fig. S17–19†). Moreover, the probability of P1 decreases with the increasing temperature, while that of P2 shows a continuous increase. These results further support the assignment of these populations since stacked FAD has higher stability at lower temperature.39 (Fig. 3A and ESI Fig. S7†).
To further reveal the mechanism of conformational change of FAD, we constructed the MD simulations on conformation of FAD in KCl solution with RACN = 0–20%. The FAD molecule was solvated in the series of ACN–KCl mixed solution and equilibration was performed (see details in the Methods of the ESI, ESI Fig. S8–S11 and Table S2†). Based on the trajectories calculated by above simulations, the angle of the atom N9A in adenine, O3P between two phosphorus atoms and N10 in flavin (ψN–O–N) was used to characterize the conformation of the FAD molecule (Fig. 1A). As shown in Fig. 2F–J, the statistical ψN–O–N distributions for each frame also show three Gaussian peaks (P1, P2 and P3) in ACN–KCl solution with RACN = 0–20%. This simulation reveals that there are three dominant conformations existing in the ACN–KCl solution, which is similar to the nanopore experimental findings. Similar to the P2 behavior in the current histogram, P2 in ψN–O–N distributions shows a constant value of ∼160° which is independent of RACN. However, ψN–O–N of P1 and P3 gradually increases from 55° to 116° and 102° to 148° with RACN increasing to 20%, respectively. Eventually, the population of P1 and P3 merged together and their ψN–O–N reached that of P3. The gradual expansion of the backbone shown in P1 of the simulation results reveals that the acetonitrile facilitates the transformation of the “stack” into the partially unstacked, relatively open, or even to the open state. Therefore, there are consistent trends between the results obtained in nanopore experiments and the simulations, which further confirms the assignment of three populations in nanopore experiments. More importantly, both the experimental and simulation results illustrate that the introduction of acetonitrile could stabilize the intermediate conformations with a prolonged lifetime, which produces the clearly current peak and ψN–O–N peak shift of P1. Note that the difference of probability for three peaks on current blockage and ψN–O–N might be induced by the occupancy, lifetime and capture rate of aerolysin for different FAD conformations. Taking together, we further re-define the assignment of P1 in the RACN ranging from 5%, 10%, 15% and 20% from the “stack” state to the “quasi stack-1” (Q-1), “quasi stack-2” (Q-2), “quasi stack-3” (Q-3) and “quasi stack-4” (Q-4), respectively (Fig. 2G–F).
The MD results describe that these quasi stacked FADs are stabilized by the insertion of the acetonitrile molecule into the space between the isoalloxazine ring and the adenine (ESI Fig. S12†). This metastable structure is attributed to the acetonitrile–π interactions between the acetonitrile and aromatic nucleus as suggested in a previous study.40 Furthermore, the relationship between I/I0 of P1 and RACN (Fig. 1E) in solution reveals that the initial expansion of the FAD backbone (quasi stack-1) increases the cross-section and enhances the interaction between the aerolysin lumen, leading to a large blockage of I/I0 ∼ 0.42. Then, the continuous addition of acetonitrile makes the quasi stacked FAD molecule adopt a wider ψN–O–N (Fig. 2F–J). These more expanding structures of “Q-2”, “Q-3” and “Q-4” lead to an increase of I/I0 of P1. Also, the increase of the occupancy for P3 in the current histogram with the RACN (5% to 15%) shows that a certain “partially stacked” form of FAD exists and is stabilized in ACN–KCl mixed solution. Moreover, as RACN increases from 5% to 15%, the “partially stacked” FAD tends to be stabilized in wider backbone angles, while the initial introduction of acetonitrile (5%) would stabilize a narrower backbone angle for the “partially stacked” FAD compared to pure KCl solution. These results together suggest a direct correlation between the current blockage (I/I0) and intramolecular angle of the backbone for a single FAD molecule. Note that the conformation of FAD inside aerolysin is largely determined by its initial conformation entering the pore according to the simulations of the FAD in the confined space (ESI Fig. S13†). Therefore, the conformation of each FAD in solution could be qualitatively determined to its corresponding current blockage. In particular, P3 in KCl solution strongly indicates that FADs adopt a more open conformation with relatively long lifetime as “partially stacked” based on the I/I0 peak position.
To further investigate the conformation details of FAD inside the aerolysin nanopore, the voltage dependent experiments were further performed. We focus on two stable conformations as “stack” and “open” which correspond to P1 and P2, respectively. As shown in Fig. 3B, the I/I0 of P1 almost remains at a constant value of ∼0.40 with increased voltages from +80 mV to +160 mV (ESI Fig. S14†). These results reveal that the stacked FAD in P1 current population is more stable at the applied voltage, which could be ascribed to the strong π–π interaction between the adenine and flavin. In contrast, the I/I0 of P2 slightly increases with the voltage, suggesting the stretching of the open conformation due to the stronger electric field force exerted on FAD. As for duration, the events caused by two conformations both experience exponentially decreased statistical duration (τ) with the applied voltage (Fig. 3C, ESI S15 and S16†), which represents the translocation of the stacked and open FAD through the pore according to previous studies.19 Interestingly, the duration time of P1 is shorter than that of P2 at +80 mV, but larger at the applied voltages higher than +120 mV. This result suggests that the “stack” and “open” conformation may undergo different interactions with the aerolysin under an electric field. By fitting the voltage-dependent durations, we further estimated the effective charge (zinside) of FAD inside the aerolysin which is 0.73 for the “stack” and 0.76 for “open” form, respectively (see in the ESI†). The stacked conformation of FAD has a relatively stronger binding to the counterions than the open form, leading to a screening of its negative charge with a lower zinside value. Moreover, this strong binding with counterions for stacked FAD could stabilize its conformation in a confined space, which is consistent with the results that the more stabilized stacked form gives the voltage-independent current peak value at P1 distribution. Furthermore, the duration of P2 in K238Q mutant aerolysin shows an obvious increase with the applied voltage, suggesting a larger energy barrier for open FAD (Fig. 3D). Due to the unpaired negative charge in K238Q, this observation also agrees with the large counterions binding to the stacked conformation.
Conclusions
In summary, we achieved the direct monitoring of various FAD conformations at the single molecule level using an aerolysin nanopore, including the two stable conformations, “open” and “stack”, and its intermediate states. Combined with MD simulations, the correlation between the current ratio (I/I0) and FAD conformation provides the qualitative analysis for the transient “partially stacked” conformations of FAD, which cannot be achieved by other bulk measurements. The introduction of an acetonitrile molecule would stabilize the expanded intermediate conformation of quasi stacked FAD, which facilitates the conformational changes of FAD from the “stack” to “open” form. These results demonstrate the possibility for stabilization of the FAD molecule in its intermediate states with a certain organic environment. Moreover, our methods allow mapping conformation distribution of FAD under certain physiological conditions that are essential for deeply studying various metabolic enzymes.Furthermore, we demonstrated the stronger binding of counterions with the stacked FAD, leading to the stabilization of this conformation in a confined space. This counterion effect could be regarded as another factor that could affect the stability of FAD in its certain conformation. As a cofactor, the functions of FAD are restricted in a confined enzyme. While the interaction between FAD and the enzyme contributes greatly to the activity, the stability of certain conformations of FAD in this nano-confinement also influences the efficiency of related enzymes. By monitoring the ratio change of FAD conformation using the presented nanopore method, the preference of the FAD binding state towards the flavoprotein could be revealed in the future. Therefore, we anticipate that the new insight of conformation dynamics and the conformation stability of FAD will shed light on the function and effect mechanism in distinct flavoproteins. The single molecule method could be expanded to the detection and evaluation of the dynamic structures of other cofactors or signal molecules in living systems.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (21834001, 21922405 and 61871183), Shanghai Rising-Star Program (19QA1402300) and Excellent Research Program of Nanjing University (ZYJH004).References
- Z. C. Litman, Y. Wang, H. Zhao and J. F. Hartwig, Nature, 2018, 560, 355–359 CrossRef CAS PubMed.
- A. Orita, M. G. Verde, M. Sakai and Y. S. Meng, Nat. Commun., 2016, 7, 13230 CrossRef CAS PubMed.
- C. E. Lubner, D. P. Jennings, D. W. Mulder, G. J. Schut, O. A. Zadvornyy, J. P. Hoben, M. Tokmina-Lukaszewska, L. Berry, D. M. Nguyen, G. L. Lipscomb, B. Bothner, A. K. Jones, A.-F. Miller, P. W. King, M. W. W. Adams and J. W. Peters, Nat. Chem. Biol., 2017, 13, 655–659 CrossRef CAS PubMed.
- A. W. t. MacFarlane and R. J. Stanley, Biochemistry, 2003, 42, 8558–8568 CrossRef CAS PubMed.
- M. A. van der Horst and K. J. Hellingwerf, Acc. Chem. Res., 2004, 37, 13–20 CrossRef CAS PubMed.
- N. S. Scrutton, Science, 2017, 357, 872–873 CrossRef CAS PubMed.
- J. Hahn, M.-E. Michel-Beyerle and N. Rösch, J. Mol. Model., 1998, 4, 73–82 CrossRef CAS.
- P. A. W. van den Berg, K. A. Feenstra, A. E. Mark, H. J. C. Berendsen and A. J. W. G. Visser, J. Phys. Chem. B, 2002, 106, 8858–8869 CrossRef CAS.
- S. D. M. Islam, T. Susdorf, A. Penzkofer and P. Hegemann, Chem. Phys., 2003, 295, 137–149 CrossRef CAS.
- G. Kuppuraj, D. Kruise and K. Yura, J. Phys. Chem. B, 2014, 118, 13486–13497 CrossRef CAS PubMed.
- I. H. Jain, L. Zazzeron, R. Goli, K. Alexa, S. Schatzman-Bone, H. Dhillon, O. Goldberger, J. Peng, O. Shalem, N. E. Sanjana, F. Zhang, W. Goessling, W. M. Zapo and V. K. Mootha, Science, 2016, 352, 54–61 CrossRef CAS PubMed.
- J. J. Kasianowicz, E. Brandin, D. Branton and D. W. Deamer, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13770–13773 CrossRef CAS PubMed.
- G. H. Xie, P. Li, Z. L. Zhao, Z. P. Zhu, X. Y. Kong, Z. Zhang, K. Xiao, L. P. Wen and L. Jiang, J. Am. Chem. Soc., 2018, 140, 4552–4559 CrossRef CAS PubMed.
- P. C. Gao, Q. Ma, D. F. Ding, D. G. Wang, X. D. Lou, T. Y. Zhai and F. Xia, Nat. Commun., 2018, 9, 4557 CrossRef PubMed.
- M. J. Zhao, W. H. Wu and B. Su, ACS Appl. Mater. Interfaces, 2018, 10, 33986–33992 CrossRef CAS PubMed.
- D. Rotem, L. Jayasinghe, M. Salichou and H. Bayley, J. Am. Chem. Soc., 2012, 134, 2781–2787 CrossRef CAS PubMed.
- S. Wen, T. Zeng, L. Liu, K. Zhao, Y. Zhao, X. Liu and H.-C. Wu, J. Am. Chem. Soc., 2011, 133, 18312–18317 CrossRef CAS PubMed.
- W. Shi, A. K. Friedman and L. A. Baker, Anal. Chem., 2017, 89, 157–188 CrossRef CAS PubMed.
- A. Piruska, M. Gong, J. V. Sweedler and P. W. Bohn, Chem. Soc. Rev., 2010, 39, 1060–1072 RSC.
- C. Cao and Y.-T. Long, Acc. Chem. Res., 2018, 51, 331–341 CrossRef CAS PubMed.
- W. Si, Y. Zhang, J. J. Sha and Y. F. Chen, Nanoscale, 2018, 10, 19450–19458 RSC.
- S. Y. Chen, Y. L. Tang, K. Zhan, D. H. Sun and X. Hou, Nano Today, 2018, 20, 84–100 CrossRef CAS.
- H. C. Zhang, Y. Tian and L. Jiang, Nano Today, 2016, 11, 61–81 CrossRef CAS.
- M. Soskine, A. Biesemans, M. Maeyer and G. Maglia, J. Am. Chem. Soc., 2013, 135, 13456–13463 CrossRef CAS PubMed.
- M. Wanunu, T. Dadosh, V. Ray, J. Jin, L. McReynolds and M. Drndić, Nat. Nanotechnol., 2010, 5, 807–814 CrossRef CAS PubMed.
- W. Si and A. Aksimentiev, ACS Nano, 2017, 11, 7091–7100 CrossRef CAS PubMed.
- S. Loudwig and H. Bayley, J. Am. Chem. Soc., 2006, 128, 12404–12405 CrossRef CAS PubMed.
- Y.-L. Ying, C. Cao, Y. X. Hu and Y.-T. Long, Natl. Sci. Rev., 2018, 5, 450–452 CrossRef.
- M. T. Degiacomi, I. Iacovache, L. Pernot, M. Chami, M. Kudryashev, H. Stahlberg, F. G. van der Goot and M. Dal Peraro, Nat. Chem. Biol., 2013, 9, 623–629 CrossRef CAS PubMed.
- I. Iacovache, S. De Carlo, N. Cirauqui, M. Dal Peraro, F. G. van der Goot and B. Zuber, Nat. Commun., 2016, 7, 12062 CrossRef CAS PubMed.
- C. Cao, Y.-L. Ying, Z.-L. Hu, D.-F. Liao, H. Tian and Y.-T. Long, Nat. Nanotechnol., 2016, 11, 713–718 CrossRef CAS PubMed.
- C. Cao, J. Yu, M.-Y. Li, Y. Q. Wang, H. Tian and Y.-T. Long, Small, 2017, 13, 1702011 CrossRef PubMed.
- J. Yu, C. Cao and Y.-T. Long, Anal. Chem., 2017, 89, 11685–11689 CrossRef CAS PubMed.
- Y. Wang, K. Tian, X. Du, R.-C. Shi and L.-Q. Gu, Anal. Chem., 2017, 89, 13039–13043 CrossRef CAS PubMed.
- F. Piguet, H. Ouldali, M. Pastoriza- Gallego, P. Manivet, J. Pelta and A. Oukhaled, Nat. Commun., 2018, 9, 966 CrossRef PubMed.
- T. C. Sutherland, Y.-T. Long, R.-I. Stefureac, I. Bediako-Amoa, H.-B. Kraatz and J. S. Lee, Nano Lett., 2004, 4, 1273–1277 CrossRef CAS.
- S. Li, C. Cao, J. Yang and Y.-T. Long, ChemElectroChem, 2018, 6, 126–129 CrossRef.
- D. Voet and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1971, 68, 1151–1156 CrossRef CAS PubMed.
- M. Raszka and N. Kaplan, Proc. Natl. Acad. Sci. U. S. A., 1974, 81, 4546–4550 CrossRef PubMed.
- J. C. Molano-Arevalo, D. R. Hernandez, W. G. Gonzalez, J. Miksovska, M. E. Ridgeway, M. A. Park and F. Fernandez-Lima, Anal. Chem., 2014, 86, 10223–10230 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc03163d |
| This journal is © The Royal Society of Chemistry 2019 |