 Open Access Article
Open Access ArticleExploration of the polymorphic solid-state landscape of an amide-linked organic cage using computation and automation†
C. E.
Shields
 a,
T.
Fellowes
a,
T.
Fellowes
 a,
A. G.
Slater
a,
A. G.
Slater
 a,
A. I.
Cooper
a,
A. I.
Cooper
 a,
K. G.
Andrews
a,
K. G.
Andrews
 *b and
F. T.
Szczypiński
*b and
F. T.
Szczypiński
 *a
*a
aMaterials Innovation Factory and Department of Chemistry, University of Liverpool, 51 Oxford Street, Liverpool, L7 3NY, UK. E-mail: filip.szczypinski@liverpool.ac.uk
bDepartment of Chemistry, Durham University, Lower Mount Joy, South Rd, Durham, DH1 3LE, UK. E-mail: keith.g.andrews@durham.ac.uk
First published on 2nd May 2024
Abstract
Organic cages can possess complex, functionalised cavities that make them promising candidates for synthetic enzyme mimics. Conformationally flexible, chemically robust structures are needed for adaptable guest binding and catalysis, but rapidly exchanging systems are difficult to resolve in solution. Here, we use low-cost calculations and high-throughput crystallisation to identify accessible conformers of a recently reported organic cage by ‘locking’ them in the solid state. The conformers exhibit varying distances between the internal carboxylic acid groups, suggesting adaptability for binding a wide array of target guest molecules.
Molecules that possess permanent cavities are desirable for applications in both the solution and solid state, ranging from catalysis and sensing,1,2 to gas storage and separation,3,4 to permanently porous liquids.5 In particular, covalent organic cages have attracted attention due to their solution processability, chemical tunability, and unique host–guest chemistry.6–8 Like enzymatic receptors, organic cages can bind guests within their cavities,9 allowing the host to act as a supramolecular protecting group or template,10 or improving the rate or selectivity of a reaction.2,11 Most organic cages are synthesised through dynamic covalent chemistry and are often isolated as a high-symmetry species that precipitates from solution.12 When under thermodynamic control, such reactions must be carried out at high dilution since high concentrations can shift the equilibrium towards polymer or catenane formation. Hence, the solution-phase applications of cages formed through labile dynamic covalent chemistry are limited.6,13
To address the labile nature of bonding in organic cages, many groups have turned to post-synthetic modification strategies. Trapping of highly soluble symmetrical imine species by reduction to the corresponding amine prevents dynamic exchange in solution, and the resulting cages can easily be isolated by solvent removal.14–16 Another strategy is oxidation to the amide cage.17–19 Amide-linked organic cages are chemically stable relative to imine-linked cages, and retain their rigidity unlike amine-linked cages. Furthermore, amide moieties can act as additional interaction sites with molecules of varying polarities. Different relative orientations of the resulting amide groups lead to further stereoelectronic de-symmetrisation from a highly symmetric cage structure.20 Such de-symmetrisation is crucial for the strong binding of low symmetry guests, such as drug molecules and metabolites. Thus, amide cages provide a balance of flexibility and rigidity necessary for adaptive binding modes, akin to induced fit binding seen in enzymatic systems.9,21 The synthesis of low-symmetry structures is often hampered by poor reaction yields,22 or the need for careful precursor design and expensive high-level calculculations,23–26 although some discoveries can occur serendipitously.27 Recently, Andrews and Christensen reported synthesis of an amide-linked organic cage 1, using Pinnick oxidation to trap metastable imine cages (Fig. 1, left).19
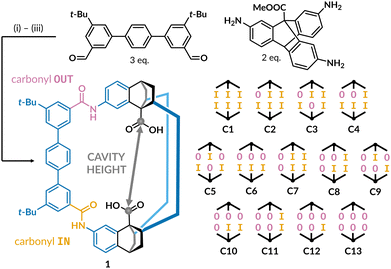 | ||
Fig. 1 Chemical structure of cage 1 and its thirteen possible amide configurations. The carbonyl group pointing outside the cavity is shown in purple and inside in orange. The cavity height is defined as the distance between the carbon atoms on the acid groups. Reaction conditions: (i) CF3COOH, toluene/THF (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); (ii) NaClO2, CH3COOH, tetramethylethylene; (iii) NaOH, dioxane/water (3 1); (ii) NaClO2, CH3COOH, tetramethylethylene; (iii) NaOH, dioxane/water (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).19 1).19 | ||
Cage 1 is a rare example of an endohedrally-functionalised cage with two carboxylic acid groups that point into the cavity and are accessible for guest binding.28–30 Furthermore, restricted rotation around the amide bonds results in 13 unique cage conformations where the six carbonyl groups point either into or out of the cavity (Fig. 1, right). Initial crystallisation by vapour diffusion yielded conformer C9, but low-temperature 1H NMR spectra of cage 1 in THF-d8 show only a symmetric set of peaks, suggesting either a single symmetrical conformer (corresponding to C1 or C13), or a structure highly fluxional on the NMR timescale.19,20 Understanding the conformational landscape of cage 1 is important because the properties of the cavity are expected to depend on the acid–acid distance (cavity height), which depends on the relative orientations of the amide groups. Here, we set out to isolate the different conformers of cage 1 in the solid state to aid the development of amide cages towards catalysis and sensing.
To identify which cage conformers can be accessed experimentally, we started with a computational analysis of the potential energy landscape. As cage 1 consists of 250 atoms connected by multiple rotatable bonds, we deemed it beyond the capabilities of the state-of-the-art crystal structure prediction methods.31 Furthermore, we anticipated that interactions with solvent molecules would greatly affect the actual crystal packing.32–34 Therefore, to inform our structural determination efforts, we limited our calculations to an extensive conformer search of an isolated cage structure in implicit THF solvent. The initial conformer scan using CREST with the GFN2-xTB method yielded twelve of the enumerated conformations of cage 1 within 50 kJ mol−1.35,36 Conformer C1 was too high in energy to be identified this way, which is unsurprising given the high strain caused by all the amide carbonyls pointing towards the centre of the cavity.20 Resulting conformers were further optimised with a low-cost composite B97-3c method designed by Brandenburg et al.37 (see ESI† for the resulting structures). We then calculated single point energies for all conformations with a number of dispersion-corrected DFT functionals and basis sets (Table S4, ESI†).38
Since our task was energy ranking of different conformers, it is perhaps unsurprising that double-zeta basis set results deviated significantly from larger basis sets (Table S4, ESI†) and completeness was assumed with def2-QZVP.39 Conformer C9 was identified as the lowest-energy conformer with PBE,40 PBE0,41 and B3LYP functionals,42 while conformer C13 was found to be preferred in the case of M06-2X,43 ωB97M-V,44 and ωB97X-D3.45 Given the observation of conformer C9 in the previous study,19 and substantially lower computation cost, we turned to PBE-D3(BJ)/def2-TZVP method for further analysis. Even though the energies of C5 and C9 were comparable at this level (within 1 kJ mol−1), we deemed it the best balance between chemical accuracy and computational cost. To test how shallow the potential energy landscape of cage 1 is, we performed a relaxed potential energy scan for all conformers at fixed acid-acid separations between 7 and 10 Å and identified five low-energy structures (C5, C9, C10, C12, and C13) within 10 kJ mol−1 (Fig. 2). Those conformers were also the five lowest-energy conformers for each tested functional. No interconversion between amide configurations was observed for those five structures during the potential energy scan.
 | ||
| Fig. 2 Five lowest-energy identified conformers of cage 1 (optimised with B97-3c, SMD = THF) and their relaxed potential energy scans (PBE-D3(BJ)/def2-TZVP, SMD = THF). | ||
Inspired by the conformationally-rich flat potential energy landscape suggested by these calculations, we designed a semi-automated crystallisation protocol to streamline our experimental search for different cage conformations. We first screened the solubility of cage 1 in 37 organic solvents using the commercially-available ChemSpeed SWING ISYNTH platform. Solvents and antisolvents were identified by solid dispensing of 1 (10 mg) and liquid dispensing of solvents (1 mL) into vials, followed by visual assessment of dissolution (for a full list of solvents, see Table S1, ESI†). We then developed an automated method for rapidly preparing solvent–antisolvent diffusion crystallisation experiments. Stocks of the cage were prepared in the ‘good’ solvents and then dispensed into vials. The antisolvent was carefully layered on top, and samples were left in a fume hood to crystallise. We adjusted the automatic liquid dispensing conditions to improve layer formation between the solvent and the antisolvent, to slow mixing and improve crystallinity of the resulting material (Section S1.2, ESI†). Around 90 crystallisation experiments were prepared over two separate screens using the ChemSpeed platform.
The initial strategy for the crystallisation experiments was to analyse samples by high-throughput powder X-ray diffraction (PXRD) and compare powder patterns to identify clusters of different polymorphs, which could indicate a different cage conformation, before attempting to obtain single crystals of any promising candidates. After two weeks, vials were visually assessed and any samples that appeared suitable for single-crystal analysis were set aside. The remaining samples containing precipitate were dried and analysed by PXRD (Table S2, ESI†). In principle, PXRD analysis is more amenable to high-throughput measurements in our setup due to the relative ease of sample preparation and analysis compared to single-crystal diffraction.34,46 However, the PXRD data obtained from the first crystallisation screen suggested the formation of solvates and also that structural changes occur upon desolvation of cage 1 (Table S3, ESI†). Hence, high-throughput assessment of whether samples contain different conformers was not ultimately possible from PXRD analyses alone, and we therefore focused on single-crystal diffraction for subsequent experiments.
All single crystals were examined by optical microscope to determine their morphology, both for samples obtained directly from the crystallisation screens or those grown manually using promising conditions identified by PXRD. Unit cell parameters were determined for suitable crystals. Crystals that possessed both a morphology and unit cell that closely matched a previously collected dataset were assumed to have the same crystal structure and were therefore not studied further. Several crystals were not stable to exposure of air. For some of these crystals it was possible to obtain the lattice parameters but not collect a full data set, while for others no data could be collected. The samples for which partial or complete single-crystal data could be collected are summarised in Table S3 (ESI†).
We identified all five of the predicted stable cage conformers (C5, C9, C10, C12, and C13) among the crystals where we could obtain a full dataset and structure solution. No other conformations were observed in any of the obtained crystal structures. Fig. 3 shows the isolated molecules of cage 1 in those conformations extracted from the crystal structures (see ESI† for crystallographic details). Furthermore, three new polymorphs of C9 were also identified.
The most commonly observed conformer was C9, which was present in the majority of the collected structures. Despite having the highest predicted energy of the five conformers, C10 was found in two crystal structures, as a co-crystal with either C9 or with C12. We hypothesise that C10 can substitute isomorphously for C9 and C12 in the extended structures, due to the small difference in cavity height between the three conformers, thus benefitting from the overall stability of the C9 and C12 structures. This suggests that crystallisation may be driven at least partially by the pore volume of the cage molecules. Additionally, cage conformations at either extreme of the predicted cavity height range (C5 and C13) are each found in only one crystal structure. The structure containing C5 has four cage molecules in the unit cell. Three of these molecules are in the C5 configuration, while the fourth molecule is disordered between C5 and C9. C5 and C9 have the largest and second-largest cavity heights, respectively, further suggesting that conformers with similar overall cavity heights may be able to interchange in the extended structures.
Interestingly, C5 and C13 were found to crystallise in separate experiments from the same solvent system, perhaps as a result of variations in stock concentration, which was not a factor we investigated during the initial automated screen. We also note that while our approach allowed the rapid identification of the target conformers, rotation of the amide bonds could result in very subtle changes to the unit cell, and hence some interesting polymorphs might have been missed. Clearly, the measurement of single-crystal data is a significant bottleneck to a thorough exploration of solid-state landscapes, which we will tackle in future studies.
Although we do not fully understand what drives the formation of the different conformers, crystallisation appears to be subtly influenced by a combination of factors, as reported previously for organic cages.32–34 Hirshfeld surface analysis shows that weak π–π and C–H–π interactions dominate the close contacts between molecules in the crystals (Fig. 4 and ESI,† Fig. S11). Intermolecular hydrogen bonding between amide groups did not appear to be a major driving force for the crystallisation of one conformer over another. Hydrogen bonding to solvent molecules was observed much more frequently. However, solvent molecules present substantial disorder in all observed diffraction experiments, suggesting that specific solvent interactions may perhaps not drive the preferential crystallisation in different cage conformers. A representative example of a Hirshfeld surface is shown in Fig. 4 (left), highlighting a single strong hydrogen bond between amide groups, and a large number of weaker interactions along the aromatic faces of the triptycene unit.
The C13 structure, which has no external H-bond donors, is the only crystal system observed here where no hydrogen bonding occurred between neighbouring cage molecules. In this case, crystal packing was only influenced by weaker van der Waals interactions. Unlike the close-packed arrangement of cage molecules in all other structures, the C13 molecules pack hexagonally to form 1-D solvent-filled pore channels along the crystallographic c axis (Fig. 4, right). Although porosity was not the focus of this work, this material could in principle exhibit permanent porosity, if the C13 crystals could be grown on a larger scale and rendered stable to desolvation. The polarity of the carboxylic acid groups in the cage cavity could make this or similar structures interesting from a CO2 capture perspective.
In conclusion, cage 1 exhibits different conformers in solution depending on the orientation of structural amide bonds. These cage conformers are in fast exchange and cannot be resolved on the NMR timescale. Typically, crystallisations of such dynamically evolving mixtures are also a difficult-to-reproduce stochastic process.47 Here, we demonstrate that it is possible to induce different conformations in the solid state via crystallisation. Low-cost computational modelling was used to identify five stable conformers of dynamic cage 1 (250 atoms). These discrete conformers were then realised in different polymorphs using a high-throughput crystallisation workflow using a commercial liquid handling robot that allows for standardisation, reproducibility46,48 and the generation of FAIR datasets over vast chemical space that facilitate future data science and machine learning efforts.49 Our setup can be transferred to molecular crystals where solvent composition may affect crystal formation or the complexity of the observed structures goes beyond state-of-the-art crystal structure prediction. A range of distances was observed between the two binding groups inside the cage cavity, validating our initial hypothesis that cage 1 can freely interchange between conformers in solution, thus underpinning future studies on binding and catalytic activity of dynamic organic cages.
This project received funding from the European Research Council under the European Union's Horizon 2020 research and innovation programme (grant agreement 856405), and the support of Diamond Light Source, instrument I19-1 (proposal CY30461). We thank the Royal Society for a University Research Fellowship (AGS, 201168) and for a Research Professorship (AIC, RSRP\S2\232003). KGA thanks the Royal Commission for the Exhibition of 1851 and Linacre College, Oxford.
Conflicts of interest
The authors declare no conflicts of interest.Notes and references
- S. La Cognata and V. Amendola, Chem. Commun., 2023, 59, 13668–13678 RSC.
- P. Bhandari and P. S. Mukherjee, ACS Catal., 2023, 13, 6126–6143 CrossRef CAS.
- G. Zhang, et al. , Acc. Chem. Res., 2021, 54, 155–168 CrossRef CAS PubMed.
- S. Yu, et al. , Mater. Chem. Front., 2023, 7, 3560–3575 RSC.
- K. Jie, et al. , Adv. Mater., 2021, 33, 2005745 CrossRef CAS PubMed.
- X. Yang, et al. , Chem. Rev., 2023, 123, 4602–4634 CrossRef CAS PubMed.
- D. Hu, et al. , Chem. Commun., 2022, 58, 11333–11346 RSC.
- T. Kunde, et al. , Eur. J. Org. Chem., 2021, 5844–5856 CrossRef CAS.
- G. Montà-González, et al. , Chem. Rev., 2022, 122, 13636–13708 CrossRef PubMed.
- V. Leonhardt, et al. , Chem. Sci., 2020, 11, 8409–8415 RSC.
- E. Ubasart, et al. , Nat. Chem., 2021, 13, 420–427 CrossRef CAS PubMed.
- D. Chakraborty and P. Sarathi Mukherjee, Chem. Commun., 2022, 58, 5558–5573 RSC.
- T. Hasell and A. I. Cooper, Nat. Rev. Mater., 2016, 1, 1–14 Search PubMed.
- M. Liu, et al. , J. Am. Chem. Soc., 2014, 136, 7583–7586 CrossRef CAS PubMed.
- T. Kunde, et al. , Chem. Commun., 2020, 56, 4761–4764 RSC.
- P. Li, et al. , Angew. Chem., Int. Ed., 2020, 59, 7113–7121 CrossRef CAS PubMed.
- P. Bhandari and P. S. Mukherjee, Chem. – Eur. J., 2022, 28, e202201901 CrossRef CAS PubMed.
- A. S. Bhat, et al. , Angew. Chem., Int. Ed., 2019, 58, 8819–8823 CrossRef CAS PubMed.
- K. G. Andrews and K. E. Christensen, Chem. – Eur. J., 2023, 29, e202300063 CrossRef CAS PubMed.
- K. G. Andrews, et al. , Chem. Sci., 2024, 15, 6536–6543 RSC.
- F. Jia, et al. , J. Am. Chem. Soc., 2020, 142, 3306–3310 CrossRef CAS PubMed.
- C. M. Thomas and N. G. White, Supramol. Chem., 2024 DOI:10.1080/10610278.2024.2324822.
- S. Klotzbach and F. Beuerle, Angew. Chem., Int. Ed., 2015, 54, 10356–10360 CrossRef CAS PubMed.
- L. Zhang, et al. , Angew. Chem., Int. Ed., 2020, 59, 20846–20851 CrossRef CAS PubMed.
- E. Berardo, et al. , Nanoscale, 2018, 10, 22381–22388 RSC.
- V. Abet, et al. , Angew. Chem., Int. Ed., 2020, 59, 16755–16763 CrossRef CAS PubMed.
- A. G. Slater, et al. , Mol. Syst. Eng., 2018, 3, 223–227 RSC.
- B. Chatelet, et al. , Chem. – Eur. J., 2014, 20, 8571–8574 CrossRef CAS PubMed.
- M. W. Schneider, et al. , Angew. Chem., Int. Ed., 2013, 52, 3611–3615 CrossRef CAS PubMed.
- H.-Y. Chen, et al. , Chem. Commun., 2017, 53, 3524–3526 RSC.
- J. D. Evans, et al. , Chem. Soc. Rev., 2017, 46, 3286–3301 RSC.
- W. Wang, et al. , ACS Appl. Mater. Interfaces, 2021, 13, 24042–24050 CrossRef CAS PubMed.
- D. P. McMahon, et al. , Faraday Discuss., 2018, 211, 383–399 RSC.
- P. Cui, et al. , Chem. Sci., 2019, 10, 9988–9997 RSC.
- P. Pracht, et al. , Phys. Chem. Chem. Phys., 2020, 22, 7169–7192 RSC.
- C. Bannwarth, et al. , J. Chem. Theory Comput., 2019, 15, 1652–1671 CrossRef CAS PubMed.
- J. G. Brandenburg, et al. , J. Chem. Phys., 2018, 148, 064104 CrossRef PubMed.
- S. Grimme, et al. , J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- J. P. Perdew, et al. , Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- J. P. Perdew, et al. , J. Chem. Phys., 1996, 105, 9982–9985 CrossRef CAS.
- P. J. Stephens, et al. , J. Phys. Chem., 1994, 98, 11623–11627 CrossRef CAS.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 Search PubMed.
- N. Mardirossian and M. Head-Gordon, J. Chem. Phys., 2016, 144, 214110 CrossRef PubMed.
- J.-D. Chai and M. Head-Gordon, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- A. Lunt, et al. , Chem. Sci., 2024, 15, 2456–2463 RSC.
- E. C. Lee, et al. , Matter, 2020, 2, 649–657 CrossRef.
- R. J. Arruda, et al. , Cryst. Growth Des., 2023, 23, 3845–3861 CrossRef CAS.
- M. D. Wilkinson, et al. , Sci. Data, 2016, 3, 160018 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: instrument setup, crystallographic data, and computational details. CCDC 2343720, 2343734, 2343766, 2343777, 2343780, 2343784, 2343788 and 2343792. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4cc01407c |
| This journal is © The Royal Society of Chemistry 2024 |


