Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Modulating halide leaving-group trends through recognition by bisboranes
Tong-Tong Liu†
 a,
Xiao-Wen Li†a,
Yun-Shu Cuia,
Zi-Hao Denga,
Feng Liu
a,
Xiao-Wen Li†a,
Yun-Shu Cuia,
Zi-Hao Denga,
Feng Liu *a,
Dan-Dan Zhai
*a,
Dan-Dan Zhai *a and
Zhang-Jie Shi
*a and
Zhang-Jie Shi *abc
*abc
aDepartment of Chemistry, Fudan University, Shanghai, 200438, P. R. China. E-mail: zjshi@fudan.edu.cn; zhaidandan@fudan.edu.cn; liufeng@fudan.edu.cn
bState Key Laboratory of Organometallic Chemistry, Chinese Academy of Sciences, Shanghai 200032, P. R. China
cCollaborative Innovation Center of Chemistry for Energy Materials (2011-iChEM), Shanghai, 201418, China
First published on 6th February 2026
Abstract
Modulating the intrinsic leaving-group tendency of halides remains a long-standing challenge in synthetic chemistry. Herein, we reported dynamic anion recognition and demonstrated its application in nucleophilic substitution reactions, enabling an apparent reversal of the halide leaving-group tendency sequence. A bidentate Lewis acid-based platform was developed to selectively bind halide ions, forming host–guest complexes that were characterized by NMR spectroscopy and X-ray crystallography. Competitive experiments with the host molecule have revealed tunable binding affinities for the halides based on the cavity size of the bisboron center. Moreover, anion exchange experiments have demonstrated that the dynamic binding of halides is primarily influenced by their nucleophilicity and ion radius. The recognition of organohalides by diborane hosts induced a reversal of the leaving ability of Br− and Cl− in diborane-catalyzed transformations, which deviated from the conventional sequence.
Introduction
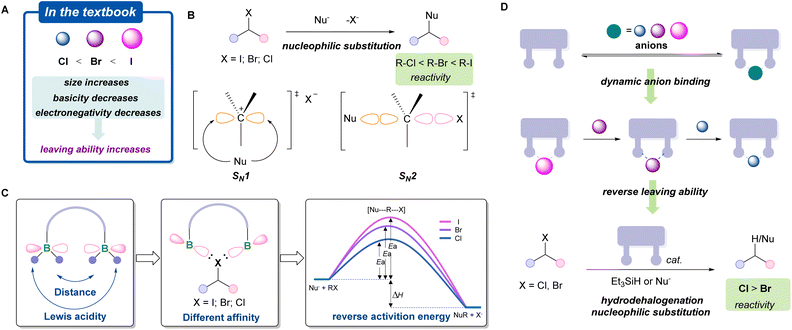
Group 17 halogen anions play a pivotal role in both physiological homeostasis and modern organic synthesis.1–4 In synthetic chemistry, organic halides represent one of the most widely utilized building blocks, with broad applications in medicinal chemistry and materials science.5–8 Owing to their high electronegativity and systematic variation in atomic size, halogens impart distinct reactivity to carbon–halogen bonds, enabling nucleophilic substitution, charge-transfer processes, and transition-metal-mediated cross-coupling reactions.9–15 In conventional reaction systems, the leaving-group ability of halides follows a well-established hierarchy (I− > Br− > Cl−), arising from systematic trends in electronegativity, atomic size, and basicity (Fig. 1A).16–18 This sequence has been validated across numerous classical organic reactions, such as nucleophilic substitution (SN1/SN2) (Fig. 1B), and is widely accepted as a guiding principle in modern organic synthesis.19Reversing intrinsic reactivity patterns represents a powerful strategy for expanding chemical reactivity space, as exemplified by umpolung chemistry.20–26 The ability to reverse the reactivity of organic halides has significant implications in a diversity of chemical transformations. To date, a general and well-developed strategy for reversing organohalide reactivity remains elusive. Boron-based poly-Lewis acids (PLAs) have recently emerged as effective platforms for anion binding via chelation effects.27–32 We envisioned that this feature may provide an alternative approach to modulate halide leaving behavior, as exemplified by SN2 reactions (Fig. 1C). Despite significant progress in anion binding chemistry, studies on dynamic halide binding and host–guest interactions involving diboranes remain limited, and structural evidence beyond fluoride complexes is scarce.33 Herein, we report a class of highly Lewis acidic diboranes capable of reversibly binding different halides through cavity-controlled recognition (Fig. 1D). Structural and spectroscopic studies reveal dynamic host–guest behavior, while catalytic investigations demonstrate that this diborane enables nucleophilic substitutions, Friedel–Crafts reactions, and reductions of alkyl chlorides and bromides with a distinct reversal of conventional halide leaving-group trends.
Results and discussion
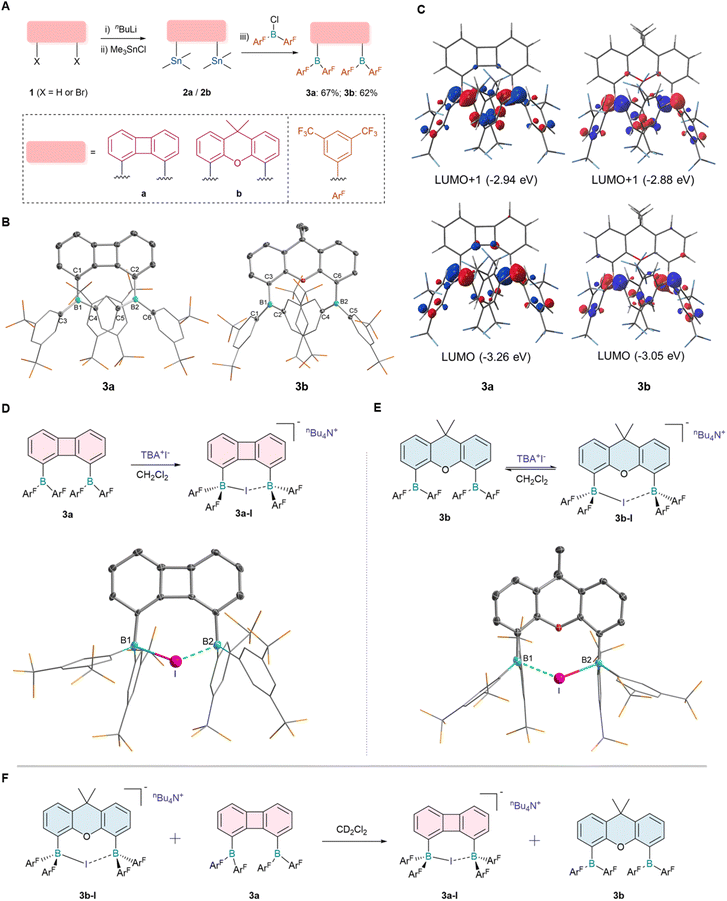
We postulate that the ideal bisborane systems should possess a spacious cavity to enable the reversible chelation of anions. Working from this assumption, electron-withdrawing groups were introduced to enhance the Lewis acidity of diborane centers, while biphenylene and 9,9-dimethylxanthene frameworks were selected to ensure differential binding affinities between two borane moieties and halides.34 Starting with compound 1, dilithium reagents were formed in situ with n-BuLi and TMEDA (where X = H). Subsequently, Me3SnCl was added, resulting in high-yield formation of di-stannyl compounds 2a and 2b (Fig. 2A). Treatment of 2a with bis(3,5-bis(trifluoromethyl)phenyl)chloroborane in hexane resulted in the precipitation of an orange-red solid, which was identified as the symmetric compound 3a by 1H NMR and 19F NMR analysis. Similarly, compound 3b was synthesized, which exhibited a broad resonance at δ = 68.11 ppm (11B NMR). This downfield signal was characteristic of a three-coordinate, electron-deficient boron center and was consistent with Lewis acidic behavior. Single crystal X-ray diffraction confirmed the structures of both compounds (Fig. 2B), where the boron centers adopted a trigonal planar geometry with a sum angle for C–B–C of 360.06° (3a) and 360.08° (3b). The parallel alignment of the two diaryl boron groups effectively minimized the steric hindrance associated with 3a and 3b. As expected, the boron–boron distance in 3a (4.048 Å) was significantly lower than that in 3b (4.620 Å). Density functional theory (DFT) calculations were performed to optimize the structures of 3a and 3b. The optimized geometries matched the experimental data (Fig. 2C). The lowest unoccupied molecular orbital (LUMO) and the second lowest unoccupied molecular orbital (LUMO+1) of 3a and 3b were dominated by the two boron pπ orbitals and partially delocalized into the π orbitals of the fluorinated aromatic rings.Having established the structure and electronic properties of the two diboranes, we further examined their binding affinity toward different anions. While diborane species with a large bite angle facilitated the chelation of polyatomic ions, studies addressing the synergistic chelation of monoatomic halide ions remained scarce.35,36 Synergistic chelation of monoatomic halide ions is geometrically disfavored by the relatively large boron–boron separation, however, this spatial constraint may be exploited to enable dynamic anion binding.37,38 The introduction of high Lewis acid-type boron substituents may compensate this spatial barrier. Unlike fluoride anions (F−), iodide anions (I−) exhibit considerably weaker binding affinity toward diboranes and have thus rarely been investigated. Therefore, we set out to investigate the interaction between iodide anions and diboranes 3a or 3b (Fig. 2). Upon adding TBAI to a dichloromethane solution of 3a, the color quickly faded and the 1H NMR spectrum revealed a set of new symmetric peaks, indicating that two sets of boryl substituents were equivalent in solution. Colorless single crystals suitable for X-ray crystallographic studies were obtained by recrystallization from a dichloromethane and n-hexane solution, confirming the structure of product 3a-I (Fig. 2D). In the solid state, iodide ions were chelated by both boron centers, where one boron center was slightly closer to the iodide anion. The B–I bond lengths were 2.407(6) Å(B1–I) and 2.469(6) Å(B2–I), respectively. This coordination mode with I− was also observed in diborane 3b, although a coordination dissociation equilibrium was observed in solution (Fig. 2E). The single crystal of complex 3b-I was obtained under low temperature conditions and the chelating coordination structure was determined by X-ray diffraction analysis. The boron–boron bond distance (B1–B2, 4.40 Å) was smaller than that in precursor 3b. The B–I bond lengths were determined to be 2.668(3) Å and 2.467(3) Å, respectively, with the resultant bond length discrepancy being more pronounced than that observed in 3a-I, which was indicative of a weaker chelation interaction. Subsequent to the synthesis and detailed structural characterization of these complexes, the anion-binding capabilities of the two diborane derivatives were evaluated via competitive binding experiments. Upon mixing 3a and 3b-I, 1H NMR spectroscopic monitoring revealed the quantitative transfer of I− from 3b-I to 3a, forming 3a-I and free 3b (Fig. 2F and S34), demonstrating that the cavity size of the boron moiety had a crucial influence on the binding affinity toward I−.
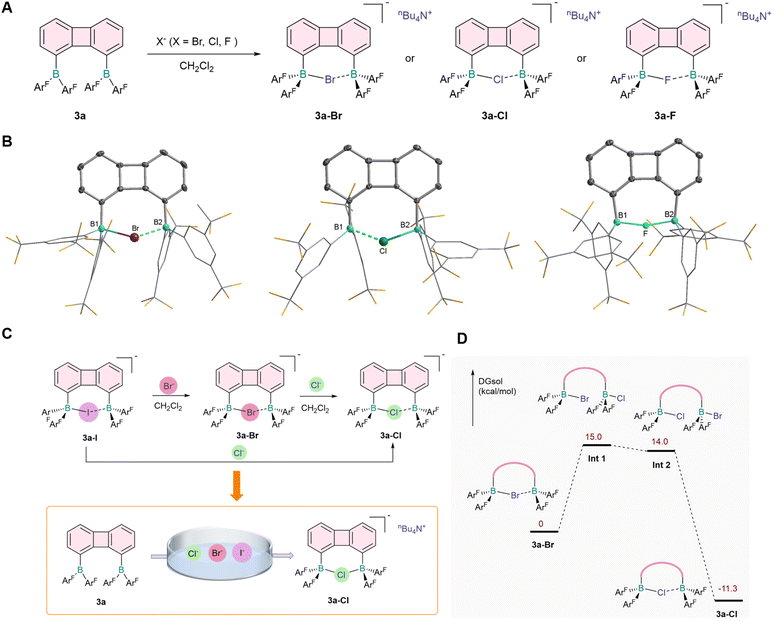
Given the higher affinity of 3a for I−, we chose 3a as a host in other halide binding studies. As expected, when stronger nucleophilic halide ions (Br−, Cl− and F−) reacted with 3a, the corresponding complexes (3a-Br, 3a-Cl and 3a-F) with anions chelated at bisboron centers were obtained (Fig. 3A). Based on periodic trends, an increase in atomic number within the same group gave rise to a larger ionic radius, which enabled halide ions to be more readily accommodated by diboranes with larger bite angles. The boron–boron bond distances exhibited a gradual decrease in the order of 3a-I < 3a-Br < 3a-Cl < 3a-F (Table 1), which was indicative of distinct interaction strengths between the bisboron moieties and halide ions. In addition, the B–X interactions were regulated by electrostatic effects, as evidenced by the single-crystal structural data (Fig. 3B), where the B–X bond lengths for Br−, Cl− and F− were determined to be 2.221(4), 2.056(2) and 1.914(4) Å, respectively. In comparison with the reported monomeric borane–halide adducts,39–41 the B–X bond lengths in these bisboron complexes were slightly elongated; this phenomenon was consistent with the chelation-assisted halide binding mode, in which the halide ions were coordinatively shared between two proximal boron centers.
Based on the experimental results, we confirmed that 3a was capable of synergistically chelating different halogen anions, providing a basis for dynamic anion binding studies. Host–guest complexation chemistry of 3a was then explored (Fig. 3C), with Br− employed to study competitive binding against I−. We found that 3a-I quantitatively converted to the 3a-Br complex within seconds upon the addition of Br− (see ESI, Fig. S35), demonstrating a higher affinity of the diborane 3a for Br−. Analogously, competitive binding experiments with Cl− and 3a-I also resulted in the formation of 3a-Cl (Fig. S37). Competitive binding experiments between Br− and Cl− further revealed the predominant generation of 3a-Cl (Fig. S36), showing the superior affinity of diborane 3a for Cl− relative to Br−.
To elucidate the anion exchange mechanism, we performed density functional theory (DFT) calculations and obtained the corresponding energy profile, using the Cl−/Br− exchange as a model system (Fig. 3D). An associative substitution pathway was proposed herein: Cl− first coordinated to one boron center of the 3a-Br adduct, leading to the formation of a dianionic intermediate; subsequent chelation of Cl− and terminal binding of Br− then occurred, a process facilitated by the rotation of the two Caryl–B single bonds. The exchange was completed followed by desorption of Br−. Notably, the addition of F− to a dichloromethane solution of 3a-Cl caused severe disruption of the 1H NMR signals (Fig. S38), indicating the formation of cross-multi-coordinated complexes. This reduced chelation selectivity of 3a toward Cl− and F− could be rationalized by combined comparable effects of anion ionic radius and charge density in their interactions with the boron centers of 3a. Furthermore, mixed competitive binding experiments in the presence of Cl−, Br− and I− confirmed that 3a exhibited prominent chelation selectivity for Cl− (Fig. 3C and S39), a preference attributable to the well-matched bisboron cavity and favorable electrostatic interactions between Cl− and the boron centers.
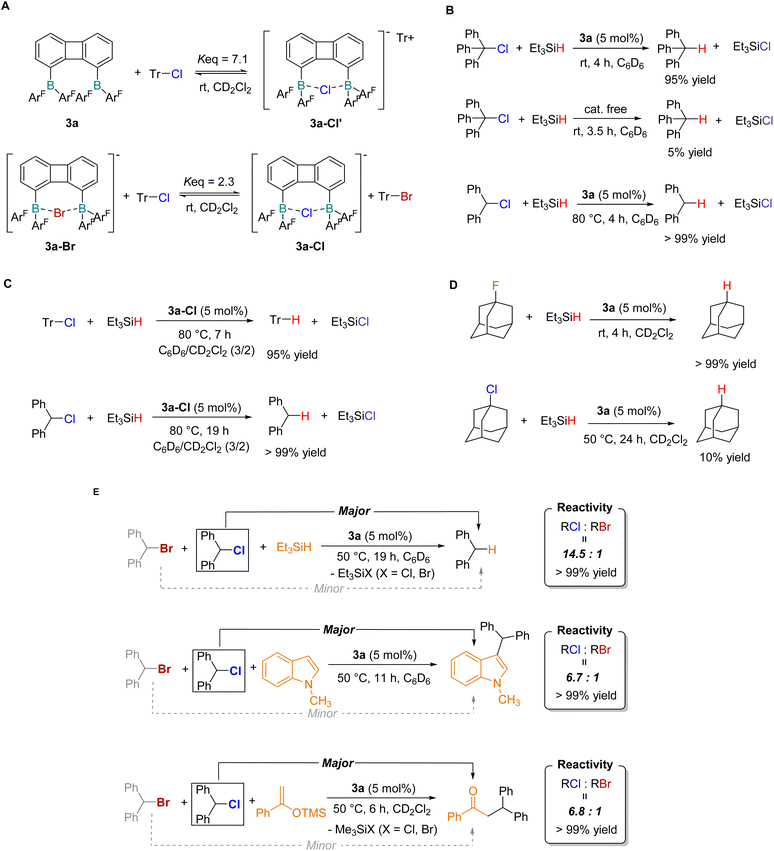
Furthermore, we investigated the reactivity of 3a toward covalent C-X bonds. Upon reaction of 3a with Ph3CCl in CD2Cl2 at room temperature, complex 3a-Cl′ was formed and the system reached an equilibrium state (Keq = 7.1), indicating the reversible activation of the C–Cl bond by 3a (Fig. 4A and S40). Analogously, the equilibrium reaction of 3a-Br with Ph3CCl yielded 3a-Cl as the anion exchange product (Fig. 4A and S42). This chelation equilibrium suggested a potential catalytic capability of 3a in C–X bond cleavage. We thus proposed a catalytic pathway where diborane 3a facilitated the reduction of C–X bonds by quenching of carbon cations with a weak hydride source. Experimental results confirmed this hypothesis: triphenylmethane was obtained in excellent yield when triethylsilane was employed as the reductant in the presence of a catalytic amount of 3a (Fig. 4B). When Ph2CHCl was used as the substrate, the reaction proceeded at a lower rate at room temperature, and complete conversion was achieved within 3 h upon heating to 80 °C. Control experiments demonstrated that these reactions did not occur in the absence of diborane catalysts. To further elucidate the role of the diborane 3a, we tested the catalytic performance of the 3a–Cl adduct in the reduction of C–Cl bonds with silanes, and the corresponding reduced products were obtained in high yields (Fig. 4C). These results suggested that diborane 3a may act as a catalyst rather than an initiator in the catalytic cycle. In addition, we investigated the reduction of the 1-fluoroadamantane using compound 3a as a catalyst at room temperature, and adamantane was quantitatively produced (Fig. 4D). In contrast, 1-chloroadamantane could not be effectively reduced, even under heating conditions.
Based on the results of halide affinity competition experiments and catalytic reactions described above, we proposed that diborane species could induce a reversal in the leaving group ability of halides in catalytic transformations. To demonstrate this proof of concept, three representative transformations of organohalides were evaluated to assess the selective activation of organic halides by bisboron catalysts (Fig. 4E). First, Ph2CHCl and Ph2CHBr were used to determine the selectivity of diborane-catalyzed reduction with silanes. The hydrogenated product Ph2CH2 was obtained in high efficiency in the presence of triethylsilane (1.0 equiv.) and catalyst 3a (5 mol%) at 50 °C, with the chloro-substituted substrate displaying significantly higher reactivity than its bromo congener (reactivity ratio = 14.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Under analogous reaction conditions with N-methylindole (1.0 equiv.) as the substrate, the Friedel–Crafts alkylation also exhibited a preference for Ph2CHCl over the bromo analogue (reactivity ratio = 6.7
1). Under analogous reaction conditions with N-methylindole (1.0 equiv.) as the substrate, the Friedel–Crafts alkylation also exhibited a preference for Ph2CHCl over the bromo analogue (reactivity ratio = 6.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). This reversed halide selectivity was further retained in bisboron-catalyzed nucleophilic substitution reactions with an enol silyl ether, albeit with a moderate reactivity ratio of 6.8
1). This reversed halide selectivity was further retained in bisboron-catalyzed nucleophilic substitution reactions with an enol silyl ether, albeit with a moderate reactivity ratio of 6.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The relatively modest selectivity observed in the nucleophilic substitution reaction was attributed to the presence of competing reaction pathways, including direct nucleophile participation and dynamic halide exchange at the bisboron center, which partially diminished the catalyst-controlled halide recognition process. Control experiments confirmed that a small amount of product formation occurred in the absence of catalyst 3a (Fig. S57 and S62). Additionally, a monoboron analogue (Fxyl)2BPh (3c) was synthesized, and under otherwise identical reaction conditions, it exhibited a substantial reduction in Cl/Br selectivity (Fig. S63–S66). Collectively, these findings demonstrated that the rational design of the bisboron framework enabled the reversal of halide leaving group reactivity, thus providing a versatile strategy for the precise control of reaction selectivity in organohalide transformations.
1. The relatively modest selectivity observed in the nucleophilic substitution reaction was attributed to the presence of competing reaction pathways, including direct nucleophile participation and dynamic halide exchange at the bisboron center, which partially diminished the catalyst-controlled halide recognition process. Control experiments confirmed that a small amount of product formation occurred in the absence of catalyst 3a (Fig. S57 and S62). Additionally, a monoboron analogue (Fxyl)2BPh (3c) was synthesized, and under otherwise identical reaction conditions, it exhibited a substantial reduction in Cl/Br selectivity (Fig. S63–S66). Collectively, these findings demonstrated that the rational design of the bisboron framework enabled the reversal of halide leaving group reactivity, thus providing a versatile strategy for the precise control of reaction selectivity in organohalide transformations.
Conclusions
In conclusion, we have developed a class of bidentate boron-based strong Lewis acids featuring a large bite angle, which enables chelation with monoatomic halide anions. These diboranes exhibit tunable and dynamic halide-binding behavior. Importantly, the prominent anion recognition ability of 3a has been successfully applied to the catalytic transformation of organohalides, achieving a reversal of the conventional reactivity of C–X bonds. These findings establish anion recognition as a promising strategy for modulating organohalide reactivity and provide valuable insights for the rational design of host-based catalytic systems for selective bond activation.Author contributions
L. T.-T. and S. Z.-J. directed the research and developed the concept of the reaction. L. T.-T. and L. X.-W. designed and carried out the experiments and characterized the products. C. Y. S. performed the computation. D. Z.-H. performed the data collection for some complexes. L. T.-T., L. X.-W., L. F., Z. D.-D., and S. Z.-J. analysed data and prepared the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
CCDC 2323469 (3a), 2323477 (3b), 2323472 (3a-I), 2323470 (3b-I), 2323475 (3a-F), 2323474 (3a-Cl), 2323473 (3a-Br) contains the supplementary crystallographic data for this paper.42a–gThe data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc10013e.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (22588201 for Z. S., 22201044 for D. Z., 22071029 for F. L.), and Fundamental Research Funds for the Central Universities (20720220010).References
- R. Dutzler, E. B. Campbell, M. Cadene, B. T. Chait and R. MacKinnon, Nature, 2002, 415, 287–294 CrossRef PubMed.
- T. A. Chew, B. J. Orlando, J. Zhang, N. R. Latorraca, A. Wang, S. A. Hollingsworth, D.-H. Chen, R. O. Dror, M. Liao and L. Feng, Nature, 2019, 572, 488–492 Search PubMed.
- L. Troian-Gautier, M. D. Turlington, S. A. M. Wehlin, A. B. Maurer, M. D. Brady, W. B. Swords and G. J. Meyer, Chem. Rev., 2019, 119, 4628–4683 Search PubMed.
- G. Cavallo, P. Metrangolo, T. Pilati, G. Resnati, M. Sansotera and G. Terraneo, Chem. Soc. Rev., 2010, 39, 3772–3783 RSC.
- S. P. Marsden, Contemp. Org. Synth., 1996, 3, 133–150 Search PubMed.
- V. Palani, M. A. Perea and R. Sarpong, Chem. Rev., 2022, 122, 10126–10169 Search PubMed.
- G. Cavallo, P. Metrangolo, R. Milani, T. Pilati, A. Priimagi, G. Resnati and G. Terraneo, Chem. Rev., 2016, 116, 2478–2601 Search PubMed.
- G. W. Gribble, ARKIVOC, 2018, 2018, 372–410 Search PubMed.
- W. P. Davey, Chem. Rev., 1925, 2, 349–367 CrossRef.
- B. M. Trost and I. Fleming, Comprehensive organic synthesis : selectivity, strategy, and efficiency in modern organic chemistry, Pergamon Press, Oxford, England, 1st edn, 1991 Search PubMed.
- M. B. Smith, March's Advanced Organic Chemistry: Reactions, Mechanisms, and Structure, John Wiley and Sons, 8th edn, 2019 Search PubMed.
- C.-L. Sun and Z.-J. Shi, Chem. Rev., 2014, 114, 9219–9280 CrossRef.
- P. Ruiz-Castillo and S. L. Buchwald, Chem. Rev., 2016, 116, 12564–12649 CrossRef PubMed.
- F. Alonso, I. P. Beletskaya and M. Yus, Chem. Rev., 2002, 102, 4009–4092 CrossRef.
- Q. Yang, Y. Zhao and D. Ma, Org. Process Res. Dev., 2022, 26, 1690–1750 CrossRef.
- R. Bruckner and M. Harmata, Organic Mechanisms: Reactions, Stereochemistry and Synthesis, Springer, Berlin, Heidelberg, 2010 Search PubMed.
- A. d. Meijere, S. Bräse and M. Oestreich, in Metal-Catalyzed Cross-Coupling Reactions and More, 2014, pp. 533–663 Search PubMed.
- R. B. Jordan, in Principles of Inorganic Chemistry: Basics and Applications, Springer International Publishing, Cham, 2024, pp. 777–804 Search PubMed.
- T. A. Hamlin, M. Swart and F. M. Bickelhaupt, ChemPhysChem, 2018, 19, 1315–1330 CrossRef.
- A. B. Smith and C. M. Adams, Acc. Chem. Res., 2004, 37, 365–377 CrossRef PubMed.
- K. Spielmann, G. Niel, R. M. de Figueiredo and J.-M. Campagne, Chem. Soc. Rev., 2018, 47, 1159–1173 RSC.
- S. Guin, D. Majee and S. Samanta, Chem. Commun., 2021, 57, 9010–9028 RSC.
- D. Seebach, Angew. Chem., Int. Ed., 1979, 18, 239–258 CrossRef.
- B. Shen, D. M. Makley and J. N. Johnston, Nature, 2010, 465, 1027–1032 CrossRef.
- Y. Wu, L. Hu, Z. Li and L. Deng, Nature, 2015, 523, 445–450 Search PubMed.
- X. Bugaut and F. Glorius, Chem. Soc. Rev., 2012, 41, 3511–3522 Search PubMed.
- H. E. Katz, J. Org. Chem., 1985, 50, 5027–5032 Search PubMed.
- V. C. Williams, W. E. Piers, W. Clegg, M. R. J. Elsegood, S. Collins and T. B. Marder, J. Am. Chem. Soc., 1999, 121, 3244–3245 Search PubMed.
- S. Solé and F. P. Gabbaï, Chem. Commun., 2004, 1284–1285, 10.1039/B403596H.
- M. Melaïmi, S. Solé, C.-W. Chiu, H. Wang and F. P. Gabbaï, Inorg. Chem., 2006, 45, 8136–8143 CrossRef.
- C.-H. Chen and F. P. Gabbaï, Chem. Sci., 2018, 9, 6210–6218 Search PubMed.
- C.-H. Chen and F. P. Gabbaï, Angew. Chem., Int. Ed., 2018, 57, 521–525 CrossRef PubMed.
- C. R. Wade, A. E. J. Broomsgrove, S. Aldridge and F. P. Gabbaï, Chem. Rev., 2010, 110, 3958–3984 CrossRef.
- W. E. Piers, G. J. Irvine and V. C. Williams, Eur. J. Inorg. Chem., 2000, 2000, 2131–2142 CrossRef.
- R. Teerasarunyanon, L. C. Wilkins, G. Park and F. P. Gabbaï, Dalton Trans., 2019, 48, 14777–14782 RSC.
- P. Niermeier, S. Blomeyer, Y. K. J. Bejaoui, J. L. Beckmann, B. Neumann, H.-G. Stammler and N. W. Mitzel, Angew. Chem., Int. Ed., 2019, 58, 1965–1969 CrossRef PubMed.
- W.-L. Jiang, B. Huang, X.-L. Zhao, X. Shi and H.-B. Yang, Chem, 2023, 9, 2655–2668 Search PubMed.
- R. Custelcean, Chem. Soc. Rev., 2014, 43, 1813–1824 RSC.
- A. Ben Saida, A. Chardon, A. Osi, N. Tumanov, J. Wouters, A. I. Adjieufack, B. Champagne and G. Berionni, Angew. Chem., Int. Ed., 2019, 58, 16889 CrossRef PubMed.
- V. Morozova, P. Mayer and G. Berionni, Angew. Chem., Int. Ed., 2015, 54, 14508 CrossRef PubMed.
- J. Zhou, S. J. Lancaster, D. A. Walker, S. Beck, M. Thornton-Pett and M. Bochmann, J. Am. Chem. Soc., 2001, 123, 223–237 CrossRef PubMed.
- (a) CCDC 2323469: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2hzrmw; (b) CCDC 2323477: Experimental Crystal Structure Determination, 2026, DOI:10.5517/ccdc.csd.cc2hzrw4; (c) CCDC 2323472: Experimental Crystal Structure Determination, 2026, DOI:10.5517/ccdc.csd.cc2hzrqz; (d) CCDC 2323470: Experimental Crystal Structure Determination, 2026, DOI:10.5517/ccdc.csd.cc2hzrnx; (e) CCDC 2323475: Experimental Crystal Structure Determination, 2026, DOI:10.5517/ccdc.csd.cc2hzrt2; (f) CCDC 2323474: Experimental Crystal Structure Determination, 2026, DOI:10.5517/ccdc.csd.cc2hzrs1; (g) CCDC 2323473: Experimental Crystal Structure Determination, 2026, DOI:10.5517/ccdc.csd.cc2hzrr0.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |