Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The time course of in vivo cellular responses to LNPs†
David
Loughrey‡
 *,
Kalina
Paunovska‡
,
Elisa Schrader
Echeverri
,
Karen E.
Tiegreen
and
James E.
Dahlman
*,
Kalina
Paunovska‡
,
Elisa Schrader
Echeverri
,
Karen E.
Tiegreen
and
James E.
Dahlman
 *
*
Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA. E-mail: david.loughrey@gmail.com; james.dahlman@bme.gatech.edu
First published on 11th February 2025
Abstract
Cells actively respond to drug delivery systems. However, the time course of cellular responses to lipid nanoparticles (LNPs) remains unclear. Here we characterized the transcriptomic response to LNPs carrying mRNA at different timepoints in vivo. Exposure to LNPs altered the expression of signaling pathways including endocytosis and lysosomal pathways as soon as one hour after administration. These pathways returned to their baseline state by 24 hours. Our data are consistent with the hypothesis that cells actively yet transiently respond to LNPs.
The FDA has approved three LNP-mRNA vaccines,1–3 one LNP-siRNA drug that targets hepatocytes, one GalNAc-ASO drug that targets hepatocytes, and four GalNAc-siRNA drugs that target hepatocytes.4 Notably, all six hepatocyte-targeting drugs enter cells through well-characterized mechanisms originally reported in mice,5 demonstrating the value of studying the biology of drug delivery. Given the success of these hepatocyte-targeting drugs, there is a growing interest in developing RNA therapies that target new cells and tissues.6 These therapies will also likely require scientists to understand the pathways that influence their delivery. One approach scientists have taken is to use RNA sequencing to study the transcriptomic response to LNPs in mice.7–9 However, these studies were not designed to reveal the dynamics of that response.
To test the hypothesis that LNPs lead to a dynamic and transient cellular response in vivo, we reasoned that it was best to use transcriptomics. Here we used low-input RNA bulk transcriptomic profiling to measure the time-dependent response to nanoparticles, focusing on Kupffer cells. We chose Kupffer cells for two reasons. First, they constitute the largest population of resident tissue macrophages in the body and play a key role in liver function. Second, they readily interact with nanomedicines10 and affect the efficiency of nanoparticle delivery.11 By measuring the transcriptomic response of Kupffer cells to LNPs one, three, six, and 24 hours after LNPs were administered, we identified time-dependent responses to LNPs. These time-course data also led us to the hypothesis that pre-dosing with the clinically relevant glucocorticoid dexamethasone may improve LNP delivery; subsequent experiments supported this hypothesis.
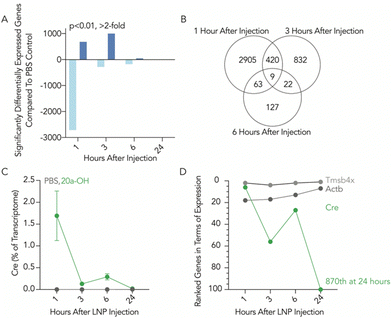
To assess the temporal response to LNP-mRNA exposure, we employed low-input RNA bulk sequencing technology. We treated mice with either a clinically relevant LNP composed to include the lipid cKK-E1212 and formulated to carry Cre mRNA,13 or a PBS control, and isolated Kupffer cells one, three, six, and 24 hours after injection using fluorescence-activated cell sorting (FACS) (Fig. S1a and b, ESI†). We performed low-input SMART-seq v4 using 1000 cells as an input with three replicates per condition (Table S1, ESI†). After 72 hours we used flow cytometry to show that the injected LNPs successfully delivered Cre mRNA, with 89% of Kupffer cells (CD45+CD68+) showing tdTomato expression above background (Fig. S1c, ESI†). We set a stringent threshold for significance (p < 0.01, fold change >2) for differentially expressed genes; relative to PBS-treated mice, we found 683 upregulated and 2714 downregulated genes one hour following LNP exposure. This total number of genes dropped after three hours, with 1000 up- and 283 downregulated genes after this timepoint, and then 46 up- and 175 downregulated genes after six hours. At the 24 hour timepoint, five genes were upregulated and zero downregulated (Fig. 1a and b). We learned two lessons from these data. First, the number of genes affected by LNPs was high, which is consistent with observations made in previous publications.14,15 Second, the maximum number of differentially expressed genes occurred after one hour. These data suggest that cells rapidly respond to LNPs.
We reasoned that RNA sequencing could also quantify the amount of exogenous mRNA we delivered into the cell relative to all the endogenous mRNA made by the cell as it functions. We therefore created a Cre pseudo-gene and quantified its reads at each timepoint. We found that 1.7% of the recorded transcriptome after one hour was exogenously delivered Cre mRNA (Fig. 1c). This meant that at one hour, Cre mRNA was the sixth most highly expressed gene in the cell (Fig. 1d). This amount of Cre relative to the entire transcriptome decreased to <0.3% at the three- and six-hour timepoints, and at 24 hours the Cre mRNA in LNP-treated mice was similar to the background reads from PBS-treated mice.
After quantifying the number of differentially expressed genes, we hypothesized that they could uncover pathways that were especially affected by LNP delivery. Using the Kyoto encyclopedia of genes and genomes (KEGG) analytical tool, we found that the endocytosis pathway in LNP-treated mice showed a large divergence relative to PBS-treated mice; at one hour there were 53 differentially regulated genes related to endocytosis, and at three hours there were 22 endocytosis-related genes. We noticed similar trends for the phagosome and lysosome; RNA transport, RNA metabolic phases, and RNA polymerase; and the TLR, MAPK, NF-κB, PI3K/Akt, and TNF signaling pathways (Fig. S2a, ESI†). Of these signaling pathways, TLRs are known to play a crucial role in the innate immune system, and are some of the first immune receptors to interact with LNPs. For example, we previously observed that TLR4 activation dramatically reduces LNP-mRNA delivery.16
The temporal response to LNP exposure can also be seen through gene localization along the TLR signaling pathway (Fig. 2a–c). After one hour, we saw gene colocalization at the surface of the cell (and within endosomes), and genes such as TLR1, TLR6, TLR7, and LBP were significantly differentially expressed (Fig. 2d). Within the cytoplasm, a number of genes linked to signaling pathways considered part of the cascade such as PI3K-Akt, JAK-STAT, and particularly MAPK were stimulated. After three hours none of the surface receptors associated with the TLR cascade were significantly dysregulated, yet 53% of these significantly differentially expressed genes are linked to immune response in the nucleus (Fig. S2b, ESI†). Similar temporal cascade effects can be seen in the PI3K-Akt signal transduction pathway, which has been previously linked to nanoparticle delivery17,18 (Fig. S2c–e, ESI†).
The endocytosis pathway had some of the most striking numbers of dysregulated genes (Fig. S3a and b, ESI†), but with a more complex spatio-temporal distribution of where the genes were colocalizing (Fig. S3c and d, ESI†). After one hour, we saw gene colocalization around growth factors related to both clathrin- and dynamin-dependent endocytosis (Fig. S3d, ESI†). At the endosomal level after one hour, we saw 21 genes downregulated related to early and late endosomes. After three hours, the total number of significantly regulated genes dropped, and where they colocalized on the endosomal pathway shifted.
The genes highlighted in these data should be further studied to understand their impact on drug delivery, with particular emphasis on LNP-specific ligand–receptor interactions that could be involved in LNP uptake and intracellular trafficking (Table S2, ESI†). Analysis of all the differentially expressed genes reveals that only one gene was significantly upregulated at all four timepoints (CCL7). This inflammatory cytokine has also been shown to be upregulated in the thigh muscle fibroblasts of BALB/c mice after intramuscular injection of an LNP with or without an mRNA cargo, indicating a cytokine response at the injection site which depends on one of the LNP lipid components.9
The transcriptomic data provided evidence suggesting that pre-treating mice with dexamethasone could improve mRNA delivery to Kupffer cells. Specifically, dexamethasone suppresses TLR1 and TLR6,19 and has also been shown to modulate TLR4 activation,20 which was previously linked to reduced LNP-mRNA delivery.16 The induction of CCL7 in human peripheral blood-derived mast cells that are stimulated with an anti-IgE Ab has been shown to be significantly inhibited by dexamethasone.21 These accumulated data led us to test our hypothesis by measuring mRNA Kupffer cell delivery using flow cytometry at four different LNP doses with a fixed 2.5 mg kg−1 dexamethasone pre-treatment one hour prior to LNP administration (Fig. 3a). mRNA delivery increased in Kupffer cells and some other liver cell types (Fig. S4a, ESI†). Notably, at 0.3 mg kg−1 LNP-mRNA, we observed a 124% increase in delivery to Kupffer cells in mice pre-treated with dexamethasone; we also observed an 82% increase in delivery at the 1.0 mg kg−1 dose (Fig. 3a). We then performed a dexamethasone dose response study, and found dose-dependent increases in delivery (Fig. 3b and Fig. S4b, ESI†). Cytokine concentrations in plasma three hours after dosing mice with low and high doses of LNP with and without dexamethasone showed only TNF-α, CCL2 and CXCL10 induced at the 3.0 mg kg−1 LNP dose (Fig. 3c). CCL2 and CXCL10 were inhibited by dexamethasone. These data supported the hypothesis from the transcriptomic profiles that dexamethasone pre-treatment could also improve delivery in vivo, rather than just improve the anti-inflammatory safety profile.22
In this work, we quantified the transcriptomic response to LNPs at different timepoints as well as the actual amount of exogenous mRNA present in the immune cells. We noted that Cre mRNA was the sixth most highly expressed gene in the cell one hour after LNP administration, constituting 1.7% of the transcriptome. Exposure to LNPs was also shown to significantly alter the expression of the endocytosis and lysosome pathways, as well as important signaling pathways such as TLR, MAPK, and TNF. The temporal differential expression can be seen to progress through the cell with large numbers of genes colocalizing at the membrane level one hour post injection, before the effects being seen in the nucleus three hours post injection. In contrast with other work,14 no significant pro-inflammatory response is witsnessed at any of the measured timepoints. Examining the transcriptomic profile led us to show that pre-dosing with the clinically relevant glucocorticoid dexamethasone could improve subsequent LNP-mRNA delivery in vivo.
It is important to acknowledge the limitations of this work. First, we did not establish how Kupffer cellular heterogeneity affects delivery; research suggests two distinct Kupffer cell populations.23,24 Second, we did not deconvolute the role played by the mRNA cargo and the LNP lipid components by characterizing the transcriptomic response to an LNP without a payload. Therefore, the genes highlighted in this work refer to the in vivo cellular response to LNPs encapsulating mRNA. Third, the exogenous mRNA counts represented the biodistribution of the mRNA rather than functional delivery or protein levels. An ideal readout would be able to show the biodistribution of many chemically distinct LNPs, the functional delivery measured as the encoded protein, and the transcriptome of transfected cells.25 Regardless, the list of genes provided in this study may inform future studies as scientists continue to better understand the dynamic responses of cells to LNP-mRNA drugs.
Data availability
The data supporting this article have been included in the main article and as part of the ESI.† All RNA sequencing data are available at SRA (PRJNA1190958). Figure images were created with BioRender.Conflicts of interest
J. E. D. advises Readout Capital, Nava Therapeutics, and Edge Animal Health. For all other authors, there are no conflicts to declare.Notes and references
- L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller and T. Zaks, N. Engl. J. Med., 2020, 384, 403–416 CrossRef PubMed.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck, L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2603–2615 CrossRef CAS PubMed.
- E. Wilson, J. Goswami, A. H. Baqui, P. A. Doreski, G. Perez-Marc, K. Zaman, J. Monroy, C. J. Duncan, M. Ujiie, M. Rämet, L. Pérez-Breva, A. R. Falsey, E. E. Walsh, R. Dhar, L. Wilson, J. Du, P. Ghaswalla, A. Kapoor, L. Lan, S. Mehta, R. Mithani, C. A. Panozzo, A. K. Simorellis, B. J. Kuter, F. Schödel, W. Huang, C. Reuter, K. Slobod, S. K. Stoszek, C. A. Shaw, J. M. Miller, R. Das and G. L. Chen, N. Engl. J. Med., 2023, 389, 2233–2244 CrossRef CAS PubMed.
- J. R. Androsavich, Nat. Rev. Drug Discovery, 2024, 23, 421–444 CrossRef CAS PubMed.
- A. Akinc, W. Querbes, S. De, J. Qin, M. Frank-Kamenetsky, K. N. Jayaprakash, M. Jayaraman, K. G. Rajeev, W. L. Cantley, J. R. Dorkin, J. S. Butler, L. Qin, T. Racie, A. Sprague, E. Fava, A. Zeigerer, M. J. Hope, M. Zerial, D. W. Sah, K. Fitzgerald, M. A. Tracy, M. Manoharan, V. Koteliansky, A. Fougerolles and M. A. Maier, Mol. Ther., 2010, 18, 1357–1364 CrossRef CAS PubMed.
- D. Loughrey and J. E. Dahlman, Acc. Chem. Res., 2022, 55, 13–23 CrossRef CAS PubMed.
- A. Radmand, M. P. Lokugamage, H. Kim, C. Dobrowolski, R. Zenhausern, D. Loughrey, S. G. Huayamares, M. Z. C. Hatit, H. Ni, A. Del Cid, A. J. Da Silva Sanchez, K. Paunovska, E. Schrader Echeverri, A. Shajii, H. Peck, P. J. Santangelo and J. E. Dahlman, Nano Lett., 2023, 23, 993–1002 CrossRef CAS PubMed.
- M. Z. C. Hatit, C. N. Dobrowolski, M. P. Lokugamage, D. Loughrey, H. Ni, C. Zurla, A. J. Da Silva Sanchez, A. Radmand, S. G. Huayamares, R. Zenhausern, K. Paunovska, H. E. Peck, J. Kim, M. Sato, J. I. Feldman, M.-A. Rivera, A. Cristian, Y. Kim, P. J. Santangelo and J. E. Dahlman, Nat. Chem., 2023, 15, 508–515 CrossRef CAS PubMed.
- S. Kim, J. H. Jeon, M. Kim, Y. Lee, Y. H. Hwang, M. Park, C. H. Li, T. Lee, J. A. Lee, Y. M. Kim, D. Kim, H. Lee, Y. J. Kim, V. N. Kim, J. E. Park and J. Yeo, Nat. Commun., 2024, 15, 7226 CrossRef CAS PubMed.
- K. M. Tsoi, S. A. MacParland, X. Z. Ma, V. N. Spetzler, J. Echeverri, B. Ouyang, S. M. Fadel, E. A. Sykes, N. Goldaracena, J. M. Kaths, J. B. Conneely, B. A. Alman, M. Selzner, M. A. Ostrowski, O. A. Adeyi, A. Zilman, I. D. McGilvray and W. C. Chan, Nat. Mater., 2016, 15, 1212–1221 CrossRef CAS PubMed.
- A. J. Tavares, W. Poon, Y. N. Zhang, Q. Dai, R. Besla, D. Ding, B. Ouyang, A. Li, J. Chen, G. Zheng, C. Robbins and W. C. W. Chan, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E10871–E10880 CAS.
- Y. Dong, K. T. Love, J. R. Dorkin, S. Sirirungruang, Y. Zhang, D. Chen, R. L. Bogorad, H. Yin, Y. Chen, A. J. Vegas, C. A. Alabi, G. Sahay, K. T. Olejnik, W. Wang, A. Schroeder, A. K. Lytton-Jean, D. J. Siegwart, A. Akinc, C. Barnes, S. A. Barros, M. Carioto, K. Fitzgerald, J. Hettinger, V. Kumar, T. I. Novobrantseva, J. Qin, W. Querbes, V. Koteliansky, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3955–3960 CrossRef CAS PubMed.
- C. D. Sago, M. P. Lokugamage, K. Paunovska, D. A. Vanover, C. M. Monaco, N. N. Shah, M. Gamboa Castro, S. E. Anderson, T. G. Rudoltz, G. N. Lando, P. Munnilal Tiwari, J. L. Kirschman, N. Willett, Y. C. Jang, P. J. Santangelo, A. V. Bryksin and J. E. Dahlman, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9944–E9952 CrossRef CAS PubMed.
- S. Ndeupen, Z. Qin, S. Jacobsen, A. Bouteau, H. Estanbouli and B. Z. Igyarto, iScience, 2021, 24, 103479 CrossRef CAS PubMed.
- B. R. Kingston, Z. P. Lin, B. Ouyang, P. MacMillan, J. Ngai, A. M. Syed, S. Sindhwani and W. C. W. Chan, ACS Nano, 2021, 15, 14080–14094 CrossRef CAS PubMed.
- M. P. Lokugamage, Z. Gan, C. Zurla, J. Levin, F. Z. Islam, S. Kalathoor, M. Sato, C. D. Sago, P. J. Santangelo and J. E. Dahlman, Adv. Mater., 2020, 32, e1904905 CrossRef PubMed.
- K. Paunovska, A. Da Silva Sanchez, M. T. Foster, D. Loughrey, E. L. Blanchard, F. Z. Islam, Z. Gan, A. Mantalaris, P. J. Santangelo and J. E. Dahlman, Sci. Adv., 2020, 6, eaba5672 CrossRef CAS PubMed.
- K. Unfried, U. Sydlik, K. Bierhals, A. Weissenberg and J. Abel, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2008, 294, L358–367 CrossRef CAS PubMed.
- R. Broering, M. Montag, M. Jiang, M. Lu, J. P. Sowa, K. Kleinehr, G. Gerken and J. F. Schlaak, Int. Immunol., 2011, 23, 537–544 CrossRef CAS PubMed.
- Y. Ge, Y. Xu, W. Sun, Z. Man, L. Zhu, X. Xia, L. Zhao, Y. Zhao and X. Wang, Gene, 2012, 508, 157–164 CrossRef CAS PubMed.
- A. Kato, R. T. Chustz, T. Ogasawara, M. Kulka, H. Saito, R. P. Schleimer and K. Matsumoto, J. Immunol., 2009, 182, 7233–7243 CrossRef CAS PubMed.
- M. T. Abrams, M. L. Koser, J. Seitzer, S. C. Williams, M. A. DiPietro, W. Wang, A. W. Shaw, X. Mao, V. Jadhav, J. P. Davide, P. A. Burke, A. B. Sachs, S. M. Stirdivant and L. Sepp-Lorenzino, Mol. Ther., 2010, 18, 171–180 CrossRef CAS PubMed.
- G. De Simone, F. Andreata, C. Bleriot, V. Fumagalli, C. Laura, J. M. Garcia-Manteiga, P. Di Lucia, S. Gilotto, X. Ficht, F. F. De Ponti, E. B. Bono, L. Giustini, G. Ambrosi, M. Mainetti, P. Zordan, A. P. Benechet, M. Rava, S. Chakarov, F. Moalli, M. Bajenoff, L. G. Guidotti, F. Ginhoux and M. Iannacone, Immunity, 2021, 54, 2089–2100 CrossRef CAS PubMed.
- C. Bleriot, E. Barreby, G. Dunsmore, R. Ballaire, S. Chakarov, X. Ficht, G. De Simone, F. Andreata, V. Fumagalli, W. Guo, G. Wan, G. Gessain, A. Khalilnezhad, X. M. Zhang, N. Ang, P. Chen, C. Morgantini, V. Azzimato, W. T. Kong, Z. Liu, R. Pai, J. Lum, F. Shihui, I. Low, C. Xu, B. Malleret, M. F. M. Kairi, A. Balachander, O. Cexus, A. Larbi, B. Lee, E. W. Newell, L. G. Ng, W. W. Phoo, R. M. Sobota, A. Sharma, S. W. Howland, J. Chen, M. Bajenoff, L. Yvan-Charvet, N. Venteclef, M. Iannacone, M. Aouadi and F. Ginhoux, Immunity, 2021, 54, 2101–2116 CrossRef CAS PubMed.
- C. Dobrowolski, K. Paunovska, E. Schrader Echeverri, D. Loughrey, A. J. Da Silva Sanchez, H. Ni, M. Z. C. Hatit, M. P. Lokugamage, Y. Kuzminich, H. E. Peck, P. J. Santangelo and J. E. Dahlman, Nat. Nano, 2022, 17, 871–879 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc06659f |
| ‡ David Loughrey and Kalina Paunovska contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |