Open Access Article
Open Access ArticleTwo-dimensional SERS encoding method for on-bead peptide sequencing in high-throughput bioanalysis†
Sinyoung
Jeong‡
a,
Homan
Kang§
b,
Myeong Geun
Cha
a,
Sung Gun
Lee
a,
Jaehi
Kim
b,
Hyejin
Chang
 c,
Yoon-Sik
Lee
b and
Dae Hong
Jeong
c,
Yoon-Sik
Lee
b and
Dae Hong
Jeong
 *ad
*ad
aDepartment of Chemistry Education, Seoul National University, Seoul, 08826, Republic of Korea. E-mail: jeongdh@snu.ac.kr
bSchool of Chemical & Biological Engineering, Seoul National University, Seoul, 08826, Republic of Korea
cDepartment of Science Education, Kangwon National University, Chuncheon, 24341, Republic of Korea
dCenter for Educational Research, Seoul National University, Seoul 08826, Republic of Korea
First published on 13th February 2019
Abstract
We developed a ready-to-read on-bead peptide encoding method for high-throughput screening bioassays. With two-dimensional surface-enhanced Raman scattering nano-identifiers (2D-SERS IDs) which are concurrently labelled with two SERS codes (coupling steps and kinds of amino acid), we could possibly generate more than 10 trillion codes with only 30 Raman label compounds.
High-throughput screening (HTS) for compound libraries is an essential element for drug discovery, disease biomarker identification, gene screening, and biomolecular profiling. One of the key challenges in HTS bioassays is to establish a reliable and efficient encoding technique for identifying a leading compound among a large compound library. Thus far, several molecular tagging approaches have been proposed to encode combinatorial peptide libraries on a solid polymer bead, such as attaching oligonucleotides,1 secondary amines,2 and haloaromatic molecules3 on solid support microbeads in parallel. Although these approaches can offer a high encoding capability, they require chemical reactions to attach molecular tags on the polymer beads that may cause undesirable side reactions against library synthesis. Furthermore, decoding processes in those approaches are complicated and labour-intensive for requiring sample preparation prior to identifying molecular tags on the beads.4 Alternatively, spectroscopically and optically pre-encoded particles such as barcoded polymer beads,5,6 fluorescently encoded silica microparticles,7–9 quantum dot embedded microparticles,10,11 and surface-enhanced Raman scattering (SERS) nanoparticles (NPs) on the solid support beads12 have been proposed to offer a simple and efficient encoding/decoding process as well as a high encoding capability. In the perspective of practical application, however, their ability to generate a sufficient number of unique codes for a large compound library are still limited due to the spectral overlap between fluorescence signals13 or the limited number of available Raman label compounds having unique bands without spectral overlapping.14 The SERS assisted label-free on-bead peptide sequencing methods have been also proposed.15,16 Such methods showed a great potential in one-bead-one-compound (OBOC) combinatorial library screening by identifying the peptide sequence on the bead based on intrinsic SERS signals from amino acids. However, the SERS spectra from multiple amino acids of a single peptide are complicated and are possibly contaminated by reacting with biomolecules during HTS bioassay. In this context, there are still some challenges to be addressed to establish a simple and efficient encoding method for HTS bioassays. First of all, a practically sufficient number of unique codes must be generated to encode a huge number of compound candidates. Moreover, the encoding and decoding process needs to be easy and straightforward. Lastly, the encoded codes should be stable and reliable under bioassay conditions.
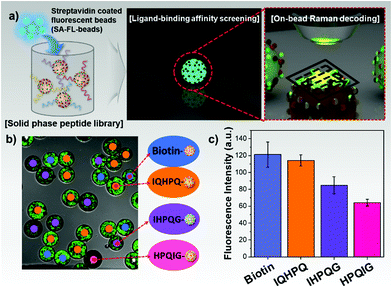
Herein, we proposed a practical and reliable encoding method for HTS bioassays using two-dimensional surface enhanced Raman scattering (SERS) nano-identifiers (2D-SERS IDs). As illustrated in Fig. 1a, the 2D-SERS IDs were concurrently labelled with two kinds of Raman label compounds (RLCs) having well separated spectral bands each other; one is for a coupling step code and another is for a code of amino acid (AA) at that coupling step. On the basis of this 2D-SERS encoding strategy, in theory, by using only 30 kinds of RLCs (20 AA codes and 10 coupling step codes), all possible combinations of decamer peptide (over 10 trillion peptides) could be completely and uniquely encoded. After synthesis of a compound library with this encoding process (Fig. 1b), conducting bioassays and decoding for screening leads an easy identification of each peptide sequence on the beads with an only single Raman scanning on the surface of 2D-SERS IDs encoded polymer micro-bead without a prior preparation (Fig. 1c). As a proof of concept, the HTS bioassay for a peptide library on solid supports which was encoded with 2D-SERS IDs was performed to identify on-bead ligand-binding affinities against streptavidin.
2D-SERS IDs comprise silver NPs embedded silica nanospheres (Ag-SiNPs) for a high signal sensitivity and are ∼0.5 μm in diameter as shown in Fig. S1 and S2a in ESI.† Ag-SiNPs showed a strong absorbance near 400 nm wavelength with a broad spectral feature, revealing an abundance of the silver NPs and their strong coupling of surface plasmons (Fig. S2b in ESI†). As an optical encoder, the 2D-SERS IDs showed a strong signal up to a single particle detectable level. It could be attributed to a large number of embedded silver NPs on a silica nanosphere that can generate highly localized electromagnetic fields and lead to significantly enhance the RLCs’ signals.17 Moreover, since those embedded silver NPs structure are randomly oriented, the 2D-SERS IDs do not rely on the polarizability of the incident photoexcitation source to generate SERS effects that offer reliably consistent SERS signals. Finally, as shown in Fig. S2a (ESI†), 2D-SERS IDs were coated with silica layer for both chemical and physical stability.
To demonstrate encoding capability of 2D-SERS codes, as shown in Fig. 2, the representative 25 kinds of 2D-SERS codes were successfully generated by combination of the 5 codes for coupling steps and the 5 codes for type of AAs using 10 kinds of RLCs. It implies that a penta-peptide library containing all possible combination of chosen 5 AAs can be uniquely encoded using these 2D-SERS codes. Thus far, we have found 44 kinds of commercially available Raman label compounds having simple aromatic structure and different functional groups, such as thiol, disulfide, amine, cyano, and isothiocyanate (Fig. S3 in ESI†), allowing of encoding micro-beads with peptide libraries of more than 10 sequences of 20 AAs. Since all RLCs could have different Raman cross-sections and surface affinities, the labelling concentrations of two RLCs need to be adjusted and optimized to generate two different unique SERS signals from a 2D-SERS ID.
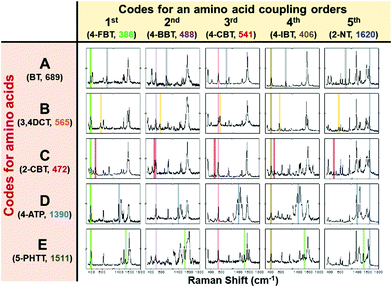 | ||
| Fig. 2 Representative 25 kinds of 2D-SERS codes using 10 kinds of Raman label compounds including 5 coupling step codes and 5 amino acid codes. | ||
The peptide libraries were prepared on commercially available TentaGel (TG) polymer beads (ca. 90 μm in diameter). As shown in Fig. 1b, in order to grant the SERS codes on TG beads after conducting peptide synthesis, 2D-SERS IDs corresponding to a peptide sequence were simply mixed with swollen TG beads in N-methylpyrrolidone (NMP) and then washed with ethanol (EtOH), as a shrinking agent, for several times. During this shrinking process, the polymer chains of TG bead collapsed, resulting that the added 2D-SERS IDs were physically buried halfway on the surface of TG bead (Fig. S4 in ESI†).12,18 Additionally, we found that the number density of the 2D-SERS IDs on the TG beads seemed to increase linearly as the concentration of 2D-SERS IDs increase (Fig. S5 in ESI†). From this result, the encoding concentration of 2D-SERS IDs for each step was decided to 0.1 wt% from the experimental result.
As a deconvolution process, with a single Raman scan on the surface of TG beads, we could readily identify the peptide sequence on the TG bead. As shown in Fig. 1c, firstly, each coupling step is identified in the result of SERS intensity maps from the 2D-SERS IDs encoded TG beads. And then, each kind of AA at corresponding coupling step can be identified by the AA code from the SERS spectrum at the location; thus, by analysing the SERS intensity maps and spectra concurrently, a peptide sequence on the bead can be efficiently and rapidly identified.
Next, in order to demonstrate the reliability of the 2D-SERS ID encoding strategy on TG microbeads, the stability of the encoded 2D-SERS IDs on TG beads was evaluated under various bioassay conditions including swelling solvent (dichloromethane, DCM), blocking solution (phosphate buffered saline [PBS] containing 3% bovine serum albumin [BSA], pH 7.0) for preventing non-specific binding, and washing solution (1% Tween 20 containing PBS). To grant the 2D-SERS codes on TG beads, the 2D-SERS ID labelled by 4-FBT and BT (2D-SERS ID[4FBT/BT]) was added to the suspension of TG beads in NMP. After incubating for 5 min, the encoded TG beads were washed with EtOH to anchor the 2D-SERS IDs onto the surface of shrunk TG beads and to remove out free 2D-SERS IDs. The 2D-SERS ID[4FBT/BT] encoded TG beads were then incubated in each condition for 1 h at room temperature. The number of encoded 2D-SERS IDs in a 900 μm2 surface area of TG bead was counted from scanning electron microscope (SEM) images of 2D-SERS IDs encoded TG bead after being incubated under bioassay conditions (Fig. S6 in ESI†). Compared to the initial number of 2D-SERS IDs on encoded microbeads in NMP without any other treatments, the number of encoded 2D-SERS IDs did not significantly decrease under any conditions. To evaluate the further stability of encoded 2D-SERS IDs, we investigated the cross contamination between 2D-SERS IDs[4FBT/BT] encoded TG bead and 2D-SERS IDs[4BBT/4ATP] encoded TG beads under bioassay condition. After mixing two differently encoded TG beads in PBS for 1 h, the SERS intensity maps at 386 cm−1 for 4-FBT, and 488 cm−1 for 4-BBT were randomly obtained from the mixture of encoded beads. As shown in Fig. S7 (ESI†), each encoded TG bead only showed their original SERS code without the other SERS code, indicating that the encoded 2D-SERS IDs were not dislodged or exchanged during the high-throughput bioassay (Fig. S7 in ESI†). Additionally, the signal stability of 2D-SERS ID was also evaluated under various conditions in peptide synthesis and bio-assay. The 2D-SERS IDs[4FBT/BT] were incubated for 1 h in NMP, DCM, EtOH, piperidine, coupling agents (containing BOP, HOBt and DIEA), trifluoroacetic acid, 3% BSA solution, and 1% Tween20 solution. After being treated with various solutions, the intensity at 386 cm−1 which is a Raman signature of 4-FBT on 2D-SERS ID[4FBT/BT] was not changed. The structure of 2D-SERS ID[4FBT/BT] was also maintained under all conditions (Fig. S8, ESI†). This feature could be attributed that silica shell of 2D-SERS ID protects the RLCs on the Ag-SiNPs and its plasmonic nanostructure from external medium and unexpected contamination.19 Collectively, these results indicated that the encoded 2D-SERS IDs on the TG beads are highly stable and reliable even after conducting the bioassay with solid phase peptide libraries.
As a proof of concept experiment, the on-bead peptide-protein binding assay was performed to demonstrate the feasibility of 2D-SERS IDs based encoding strategy for high-throughput screening with on-bead peptide libraries as illustrated in Fig. 3a. The three kinds of penta-peptides containing HPQ sequence (IQHPQ, IHPQG, and HPQIG) were synthesized on the TG beads via standard fluorenylmethyloxycarbonyl (Fmoc) chemistry. Since these HPQ containing penta-peptides have been known to have different affinities to streptavidin,18,20 the biotin was used as a positive control. All peptides were synthesized in parallel and 2D-SERS IDs corresponding to each peptide sequence were then encoded on the TG bead. To verify peptide synthesis, the synthesized penta-peptides were analyzed by mass spectroscopy (MS) and all peptides showed a clear peak corresponding to their masses (Fig. S9, ESI†). These 4 kinds bioligands-loaded TG beads were reacted with streptavidin coated fluorescent NPs (SA-F-NPs, Sphero™, ∼400 nm in diameter) for 30 min. After that, the fluorescence signals at ca. 550 nm were measured using confocal laser scanning microscope to quantitatively evaluate the affinities of bio-ligands toward streptavidin (Fig. 3b). The peptide sequences on the beads were then identified by decoding 2D-SERS IDs encoded on TG-beads with Raman scanning. The 2D-SERS ID codes corresponding to three penta-peptides were shown in Fig. S10 (ESI†). As shown in Fig. 3c, the fluorescence intensities of each bead revealed the different affinities against streptavidin according to their peptide sequences. Therefore, based on the results, we found that the IQHPQ has the highest affinity against streptavidin among three peptide candidates and that was similar to biotin's affinity. In contrast, IHPQG showed intermediate binding affinity and HPQIG showed lowest binding affinity against streptavidin. These binding affinities were consistent with previous reports.18,21,22 These results indicated that the 2D-SERS IDs based peptide encoding and bioassay method can be utilized to quantitatively screen biomolecules in a large bioligand library with high sensitivity, reliability, and efficiency.
In conclusion, we developed a simple and efficient 2D-SERS IDs based encoding strategy for high-throughput screening with on-bead compound library. We demonstrated that the 2D-SERS IDs could be easily and stably encoded on the surface of TG beads after synthesizing peptide libraries via solvent-driven swelling and shrinking process. By incorporating the two classifications of SERS codes into coupling steps and kinds of AA, a sufficiently large number of unique codes could be generated that could possibly encode every possible peptide combination of amino acid using a small number of RLCs. Additionally, we were able to quantitatively assess the affinities of on-bead bioligands against their target as well as to identify the on-bead peptide sequences by a single Raman scanning. Based on these results, we believe that the 2D-SERS IDs encoding system can offer a highly efficient and reliable on-bead peptide sequencing method as well as a high-throughput screening platform that can be used in a drug discovery and biomolecule profiling. It is also worth mentioning that this approach could be readily extended into monitoring and identifying OBOC in ‘split-and-mix’ combinatorial libraries since this method offers high stability and reliability of the unique codes on the polymer beads under peptide synthesis and bioassay conditions as well as a simple physical encoding mechanism on the polymer bead during peptide synthesis.
This work was supported by National Research Foundation (NRF) of Korea of the Ministry of Science, ICT & Future Planning, Nuclear R&D Program (NRF-2016M2A2A4A03913-619) and the Korean Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI17C1264).
Conflicts of interest
There are no conflicts of interest to declare.Notes and references
- M. C. Needels, D. G. Jones, E. H. Tate, G. L. Heinkel, L. M. Kochersperger, W. J. Dower, R. W. Barrett and M. A. Gallop, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 10700–10704 CrossRef CAS.
- Z. J. Ni, D. Maclean, C. P. Holmes, M. M. Murphy, B. Ruhland, J. W. Jacobs, E. M. Gordon and M. A. Gallop, J. Med. Chem., 1996, 39, 1601–1608 CrossRef CAS PubMed.
- M. H. J. Ohlmeyer, R. N. Swanson, L. W. Dillard, J. C. Reader, G. Asouline, R. Kobayashi, M. Wigler and W. C. Still, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 10922–10926 CrossRef CAS.
- R. Wilson, A. R. Cossins and D. G. Spiller, Angew. Chem., Int. Ed., 2006, 45, 6104–6117 CrossRef CAS PubMed.
- H. Fenniri, L. Ding, A. E. Ribbe and Y. Zyrianov, J. Am. Chem. Soc., 2001, 123, 8151–8152 CrossRef CAS PubMed.
- H. Fenniri, S. Chun, L. Ding, Y. Zyrianov and K. Hallenga, J. Am. Chem. Soc., 2003, 125, 10546–10560 CrossRef CAS PubMed.
- M. Trau and B. J. Battersby, Adv. Mater., 2001, 13, 975–979 CrossRef CAS.
- B. J. Battersby, G. A. Lawrie, A. P. Johnston and M. Trau, Chem. Commun., 2002, 1435–1441 RSC.
- L. Marcon, B. J. Battersby, A. Ruhmann, K. Ford, M. Daley, G. A. Lawrie and M. Trau, Mol. BioSyst., 2010, 6, 225–233 RSC.
- M. Han, X. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631 CrossRef CAS PubMed.
- P. S. Eastman, W. Ruan, M. Doctolero, R. Nuttall, G. de Feo, J. S. Park, J. S. F. Chu, P. Cooke, J. W. Gray, S. Li and F. F. Chen, Nano Lett., 2006, 6, 1059–1064 CrossRef CAS PubMed.
- J. H. Kim, H. Kang, S. Kim, B. H. Jun, T. Kang, J. Chae, S. Jeong, J. Kim, D. H. Jeong and Y. S. Lee, Chem. Commun., 2011, 47, 2306–2308 RSC.
- Y. Lai, S. Sun, T. He, S. Schlücker and Y. Wang, RSC Adv., 2015, 5, 13762–13767 RSC.
- D. Zhu, Z. Wang, S. Zong, H. Chen, P. Chen and Y. Cui, RSC Adv., 2014, 4, 60936–60942 RSC.
- C. Schmuck, P. Wich, B. Kustner, W. Kiefer and S. Schlucker, Angew. Chem., Int. Ed., 2007, 46, 4786–4789 CrossRef CAS PubMed.
- R. Perez-Pineiro, M. A. Correa-Duarte, V. Salgueirino and R. A. Alvarez-Puebla, Nanoscale, 2012, 4, 113–116 RSC.
- C. E. Talley, J. B. Jackson, C. Oubre, N. K. Grady, C. W. Hollars, S. M. Lane, T. R. Huser, P. Nordlander and N. J. Halas, Nano Lett., 2005, 5, 1569–1574 CrossRef CAS PubMed.
- H. Kang, S. Jeong, Y. Koh, M. Geun Cha, J.-K. Yang, S. Kyeong, J. Kim, S.-Y. Kwak, H.-J. Chang, H. Lee, C. Jeong, J.-H. Kim, B.-H. Jun, Y.-K. Kim, D. Hong Jeong and Y.-S. Lee, Sci. Rep., 2015, 5, 10144 CrossRef PubMed.
- B. Mir-Simon, I. Reche-Perez, L. Guerrini, N. Pazos-Perez and R. A. Alvarez-Puebla, Chem. Mater., 2015, 27, 950–958 CrossRef CAS.
- K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82–84 CrossRef CAS PubMed.
- D.-S. Shin, K.-N. Lee, B.-W. Yoo, J. Kim, M. Kim, Y.-K. Kim and Y.-S. Lee, J. Comb. Chem., 2010, 12, 463–471 CrossRef CAS PubMed.
- D.-H. Kim, D.-S. Shin and Y.-S. Lee, J. Pept. Sci., 2007, 13, 625–633 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cc10224d |
| ‡ Current address: Wellman Center for Photomedicine, Department of Dermatology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA. |
| § Current address: Gordon Center for Medical Imaging, Department of Radiology, Boston, MA 02114, USA. |
| This journal is © The Royal Society of Chemistry 2019 |