Synthesis and characterization of a 3-hydroxybutyrate and 3-hydroxy-9-octadecenoate copolymer from engineered Halomonas†
Xinyu
Chen
ab,
Jiangnan
Chen
ab,
Xu
Yan
a,
Qiong
Wu
a,
Fuqing
Wu
ac and
Guo-Qiang
Chen
 *abcde
*abcde
aSchool of Life Sciences, Tsinghua University, Beijing 100084, China
bTsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing 100084, China
cCenter for Synthetic and Systems Biology, Tsinghua University, Beijing 100084, China
dMOE Key Lab of Industrial Biocatalysis, Department Chemical Engineering, Tsinghua University, Beijing 100084, China
eState Key Laboratory of Biomanufactoring, Tsinghua University, Beijing 100084, China
First published on 25th July 2025
Abstract
Polyhydroxyalkanoates (PHAs) are a family of biodegradable polyesters accumulated by various microorganisms. The halophilic microorganism Halomonas bluephagenesis has been developed as a production platform strain for various PHAs, chemicals and proteins under open nonsterile and continuous conditions. For the first time, H. bluephagenesis has been engineered to synthesize a copolymer of a short chain length PHA (SCL PHA) and a long chain length PHA (LCL PHA) called, SCL-co-LCL PHA. This study engineered H. bluephagenesis for synthesizing SCL-co-LCL PHA via β-oxidation inhibition and expression of heterologous PHA synthase (PhaC). Sodium oleate was utilized as a structurally related carbon source due to its low cost, long carbon chain length, unsaturated bonds and sustainability. The engineered H. bluephagenesis successfully synthesized copolymers of 3-hydroxybutanoic acid (3HB) and 3-hydroxy-9-octadecenoic acid (3H9Od) on sodium oleate. It could be grown to 8 g L−1 cell dry weight (CDW) containing 51 wt% P(3HB-co-12 mol% 3H9Od) in shake flasks, and 28 g L−1 cell dry weight (CDW) containing 53 wt% P(3HB-co-6 mol% 3H9Od) in 7 L fermenters. Subsequently, the thermal properties of this new PHA were characterized. Finally, chemical click reactions were applied to modify P3HB3H9Od into an organogel, and rare-earth modified fluorescent PHA was successfully prepared.
Green foundation1. Our work advances green chemistry by engineering Halomonas to synthesize PHA from sustainable resources, namely glucose and oleate, offering a biodegradable alternative to traditional plastics and reducing reliance on fossil fuels.2. We achieved green chemistry objectives by engineering Halomonas to produce P3HB3H9Od. We enabled the strain to utilize sustainable carbon sources and synthesize PHA with novel monomers. In shake flask studies, the engineered strain reached 8.1 g L−1 CDW with 51.0 wt% P(3HB-co-11.6 mol% 3H9Od). In batch-fed study, the fermentation can be carried out under non sterile open conditions, thus reducing energy consumption. 3. To make our work greener, we could further optimize the fermentation process to improve yields and reduce energy consumption. Additionally, exploring more strategies could enhance the synthesis of P3HB3H9Od and expand its applications in various fields, thereby amplifying its positive impact on green chemistry. |
1. Introduction
Current environmental challenges include industrial pollution, resource scarcity and climate change. Synthetic biology offers innovative solutions for sustainable industrial development, as it utilizes the design and optimization of metabolic pathways for microbial production. This reduces fossil fuel dependence and operates under mild conditions.1–4 Engineered microorganisms have been developed to produce polyhydroxyalkanoate (PHA), a biodegradable polyester material from renewable resources, thereby minimizing environmental impact.5–9PHAs, a family of natural polyesters synthesized by microorganisms for carbon and energy storage as well as microbial stress resistance,10–14 possess biodegradability, biocompatibility, optical activity and piezoelectricity, with potential applications in packaging, biomedicine, agriculture, and biofuels.8,15–19 Classified by monomer length, PHAs with monomers containing over 14 carbon atoms are termed long chain length PHAs (LCL PHAs) and are expected to be more flexible than short chain length PHAs (SCL PHAs) composed of C2–C5 monomers. Copolymers of SCL and LCL monomers may surpass homopolymers with better properties. Furthermore, PHAs with functional groups in monomers can be chemically modified for additional properties.20–24
PHA synthase (PhaC) is responsible for the polymerization of the monomers. It exhibits diverse substrate specificities across species and hosts.1,7,25 Some broad-specificity PhaCs, like PhaC1437 and PhaCPs61–3, can synthesize various PHA chain-lengths.7,26,27 PhaC polymerization ability can also be altered by specific mutations.
Microbial metabolism impacts monomer supply and thus PHA types. Metabolic engineering has enabled PHA production in organisms like Escherichia coli and yeast.28–32Cupriavidus necator, a model PHA producer, has long been engineered to synthesize diverse PHAs with or without additional monomer precursors.18,33–37 Notably, fatty acids can be easily converted into structurally related LCL PHA precursors.38–40
Extremophiles hold great promise for industrial production due to their ability to thrive under harsh conditions, reducing contamination risks and energy consumption. Halomonas bluephagenesis TD01, a moderately halophilic bacterium isolated from the Aydingkol Lake in Xinjiang, China, grows optimally in 60 g L−1 NaCl at pH 8.5.41,42 These conditions effectively suppress growth of other environmental bacteria, reducing production costs and energy consumption.41,43 Additionally, H. bluephagenesis can be grown to high cell dry weight (CDW), and it has been engineered for enhanced bioproduction of various chemicals.43–46
This study aimed to engineer H. bluephagenesis to produce SCL-co-LCL PHA containing unsaturated bonds from sodium oleate and glucose via inhibiting β-oxidation and expressing heterologous PhaCPs-STQK replacing native PhaCTD. The thermal properties and molecular weight of the PHA material were characterized. Finally, it was chemically modified into an organogel and rare-earth modified fluorescent PHA.
2. Materials and methods
2.1 Bacterial strains and cultivation conditions
All bacterial strains and plasmids used in this study are listed in Table S1.† Wild type H. bluephagenesis TD01 was isolated from the Aydingkol Lake, Xinjiang, China and stored at the China General Microbiological Culture Collection Center under collection number 4353. Escherichia coli S17-1 was employed for plasmid construction and vector donation during conjugation.For cell growth, E. coli was cultivated in 10 LB medium consisting of 5 g L−1 yeast extract, 10 g L−1 tryptone and 10 g L−1 NaCl. H. bluephagenesis TD01 and its derivatives were cultured in 60 LB medium consisting of 5 g L−1 yeast extract, 10 g L−1 tryptone and 60 g L−1 NaCl. Both E. coli and H. bluephagenesis were cultivated at 37 °C and 200 rpm. For solid medium, 15 g L−1 agar was added on the basis of the liquid medium.
For PHA accumulation studies, H. bluephagenesis was cultivated in minimal medium (MM) consisting of 1 g L−1 yeast extract, 50 g L−1 NaCl, 0.5 g L−1 CO(NH2)2, 0.2 g L−1 MgSO4, 3.83 g L−1 Na2HPO4, 1.5 g L−1 KH2PO4, 10 mL L−1 trace element solution III and 1 mL L−1 trace element solution IV. CO(NH2)2 and MgSO4 were mixed to prepare solution I (50×), while Na2HPO4 and KH2PO4 were mixed to prepare solution II (50×). The composition of trace element solution III was: 5 g L−1 Fe(III)-NH4-citrate, 2 g L−1 CaCl2 and 1 M HCl. The composition of trace element solution IV was: 100 mg L−1 ZnSO4·7H2O, 30 mg L−1 MnCl2·4H2O, 300 mg L−1 H3BO3, 200 mg L−1 CoCl2·6H2O, 10 mg L−1 CuSO4·5H2O, 20 mg L−1 NiCl2·6H2O and 30 mg L−1 NaMoO4·2H2O. Trace element solution III and trace element solution IV were mixed with water in a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, respectively, to become mixture solutions III and IV. All solutions were sterilized and cooled to room temperature before inoculation. A corresponding amount of glucose was added, and the pH of the medium was adjusted by 5 M NaOH according to the experimental design. Before the NaCl concentration optimization, the NaCl concentration was 50 g L−1 (50 MM), and it was optimized to 35 g L−1 (35 MM). The cultivation temperature was set to 37 °C and changed to 34 °C after optimization.
9, respectively, to become mixture solutions III and IV. All solutions were sterilized and cooled to room temperature before inoculation. A corresponding amount of glucose was added, and the pH of the medium was adjusted by 5 M NaOH according to the experimental design. Before the NaCl concentration optimization, the NaCl concentration was 50 g L−1 (50 MM), and it was optimized to 35 g L−1 (35 MM). The cultivation temperature was set to 37 °C and changed to 34 °C after optimization.
Specifically, 25 mg L−1 chloramphenicol, 50 mg L−1 kanamycin and/or 100 mg L−1 spectinomycin were added to maintain the stability of intracellular plasmids.
2.2 Plasmid construction and conjugation
Q5 high-fidelity DNA polymerase (New England Biolabs, Inc., USA) was employed for PCR amplification of DNA fragments. A gel purification kit (Omega, USA) was used to purify DNA fragments. Subsequently, fragments were cyclized using a Gibson Assembly (New England Biolabs, Inc., USA). Primers were synthesized by Beijing Xianghong Biotechnology Co., Ltd (China) and Beijing Tsingke Biotech Co., Ltd (China). All DNA was sequenced by Beijing Tsingke Biotech Co., Ltd (China). All gene sequences mentioned in this study are listed in Table S2.† All the plasmids were derived from a pSEVA341 backbone,47 and the genes were controlled by the phaC promoter from Ralstonia eutropha (or Cupriavidus necator).Plasmids were firstly introduced into E. coli S17-1 via chemical transformation, followed by conjugation to H. bluephagenesis for plasmid transfers. Firstly, donor E. coli and recipient H. bluephagenesis were cultured in 10 LB and 60 LB media, respectively, to the logarithmic phase. Cells were collected via centrifugation at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400g for 3 min, then resuspended and mixed in 35 μL 10 LB. Subsequently, the mixture was placed on a 20 LB plate (containing 20 g L−1 NaCl). Finally, after 6 h of growth, the cells were spread on a 60 LB plate containing chloramphenicol and/or spectinomycin depending on the resistance gene present on the plasmid.
400g for 3 min, then resuspended and mixed in 35 μL 10 LB. Subsequently, the mixture was placed on a 20 LB plate (containing 20 g L−1 NaCl). Finally, after 6 h of growth, the cells were spread on a 60 LB plate containing chloramphenicol and/or spectinomycin depending on the resistance gene present on the plasmid.
2.3 Gene editing
Gene editing of H. bluephagenesis was achieved via a CRISPR/Cas9 system.48 Plasmids harboring the Cas9 coding sequence and homologous donor sequence were introduced into H. bluephagenesis via conjugation, respectively. The formed colonies were verified by PCR and sequencing. Finally, plasmids were removed by repeatedly subculturing in antibiotic-free 60LB medium. All primers were synthesized by Xianghong Biotech (China) or Tsingke Biotech (China). All DNA sequences were determined by Tsingke Biotech (China).2.4 Shake flask and fed-batch studies
Cells were firstly activated on a 60 LB plate, followed by two rounds of cultivation in 60 LB at 37 °C to the logarithmic phase as seed culture. Shake flask studies were conducted in Mineral Medium (MM). Bacteria were cultured in 500 mL shake flasks containing 50 mL of mixed culture medium. Solution I, solution II, a mixture of solutions III and IV and sodium oleate were added in the basic medium consisting of NaCl and yeast extract, followed by adjusting the pH to 8.5. Then, 5% of the seed culture was inoculated into the medium. Finally, the bacteria were cultured for 48 h at a rotation rate of 200 rpm and the corresponding temperature according to the experimental design.Cells were grown overnight in 60 LB medium in 500 mL shake flasks to obtain 300 mL seed cultures (OD600 at 3–5) as inocula in a fed-batch study conducted in a 7 L bioreactor (NBS Bioflo3000, New Brunswick, USA). 3 L of batch medium contained 35 g L−1 NaCl, 20 g L−1 glucose, 0.67 g L−1 7H2O·MgSO4, 4.13 g L−1 urea, 3.3 g L−1 K2HPO4, 6.8 g L−1 12H2O·Na2HPO4, 10 mL L−1 trace element solution III and 1 mL L−1 trace element solution IV. Then, 30 g L−1 sodium oleate was added to the medium and thoroughly stirred. A two-stage and intermittent feeding strategy was adopted using different feeding solutions. 250 mL of Fed Solution I containing 560 g L−1 glucose, 30.4 g L−1 urea, 3.16 g L−1 K2HPO4, 8.48 g L−1 12H2O·Na2HPO4, 80 mL L−1 trace element solution III and 12 mL L−1 trace element solution IV was used to increase cell mass accumulation and PHA formation during the first 16 h. Fed Solution II containing 560 g L−1 glucose, 30.4 g L−1 urea, 40 mL L−1 trace element solution III and 12 mL L−1 trace element solution IV was added after exhaustion of Fed Solution I. The flow rates of the fed solutions depended on the residual glucose concentrations that should be controlled between 8 and 12 g L−1 measured using the SANNUO medical glucometer (China). All fermentation media were prepared without sterilization, and all batches were also operated under unsterilized open conditions. The OD600 of the cultured bacteria was measured using a V-5600 visible light spectrophotometer (Shanghai Yuanxi Instrument, China).
2.5 Study on cell dry weight (CDW)
After the fermentation, cells were harvested by centrifugation at 9600g (CR 21GIII, HITACHI, Japan) for 15 min. For each sample, 30 mL of the fermentation broth was centrifuged in a 50 mL volume centrifuge tube that had been weighed in advance. Then the harvested cells were suspended and washed with 75% ethanol and distilled water, respectively. After 12 h of lyophilization, the weight of the cells and the centrifuge tubes was measured. The CDW was calculated from the difference in weights.2.6 PHA analysis by gas chromatography and mass spectrometry (GC-MS)
To assay the PHA, 35–45 mg of lyophilized cells or 15–30 mg of extracted PHA were esterified at 100 °C for 4 h together with 2 mL of esterification mixture (3 vol% sulfuric acid and 2 g L−1 benzoic acid in methanol) and 2 mL of chloroform.49 In this process, the PHA polymer was hydrolyzed, and the monomers were converted into methyl-hydroxyalkanoates for GC analysis. P3HB powder (Sigma-Aldrich, St Louis, MO, USA) and extracted P3HB3H9Od were used as standard samples for composition analysis. After aqueous extraction, the organic phase was analyzed using an Agilent 5977B GC/MSD instrument (Agilent Technologies, Santa Clara, CA, USA) equipped with a DB-5 column. The optimized GC detection procedure designed for 3HB and 3H9Od was as follows: the temperature program began at 80 °C with a hold time of 2.5 min, then increased at a rate of 35 °C min−1 to 140 °C, and finally increased at a rate of 30 °C min−1 to 300 °C with a hold time of 3 min at 300 °C. High-purity helium was used as the carrier gas. The m/z values of the characteristic ions of methylated 3H9Od are 103 and 312.2.7 PHA extraction and purification
Intracellular PHA was extracted using a Soxhlet extractor (Soxtec™ 8000, FOSS, Hilleroed, Denmark) using chloroform as the solvent. The process was conducted at 110 °C for 3 h, followed by elution for 2 h and solvent recovery for 30 min. Subsequently, the extracted PHA was dissolved in chloroform and then precipitated with a six-fold volume of ethanol. After centrifugation at 9600g for 15 min, the obtained PHA was dried and then stored under anaerobic conditions.2.8 NMR assay
Approximately 20–30 mg PHA samples were dissolved in 0.5 mL of deuterated chloroform for 1H NMR and 13C NMR studies in a JNM-ECA600 nuclear magnetic resonance instrument (JEOL, Japan). Chemical shifts were recorded in ppm, calibrated to the signal of 0.03% v/v tetramethylsilane (TMS) in the solvent to serve as an internal standard.2.9 Gel permeation chromatography (GPC)
Sample molar masses and polydispersities were determined by GPC using an LC-20AD instrument equipped with a GPC-804C column and a refractive index detector (SHIMADZU, Japan). PHA samples were dissolved in chromatography-grade chloroform to a final concentration of 2 mg mL−1, followed by filtration through a 0.22 μm nylon membrane filter (Jinglong, China) to eliminate any particulate substance. The mobile phase for the chromatography was HPLC-grade chloroform delivered at a flow rate of 1 mL min−1 and maintained at 40 °C. A 40 μL aliquot of each sample was injected into the column. Calibration of the GPC system was conducted using a series of polystyrene standards with varying number-average molar masses: 1 × 104, 2 × 104, 3 × 104, 7 × 104, 1.5 × 105, 3 × 105, 7 × 105 and 1 × 106, respectively (Sigma-Aldrich, USA).2.10 Differential scanning calorimetry (DSC)
The DSC data of the polymers were acquired utilizing a DSC-Q20 (TA instrument, USA). The experimental temperature profile spanned from −80 °C to 200 °C, with a nitrogen purge rate of 50 mL min−1. The processes were initially cooled to −80 °C and subsequently heated to 180 °C at a heating rate of 10 °C min−1. Subsequently, the samples were quenched and were then reheated within the identical temperature range to complete the second thermal cycle.2.11 Thermal gravimetry analysis (TGA)
A TGA-50 instrument (SHIMADZU, Japan) was utilized to determine the thermal stability of PHA. Approximately 5–10 mg samples were purged with nitrogen and heated at a rate of 10 °C min−1 from ambient temperature to 600 °C.2.12 Material chemical modification
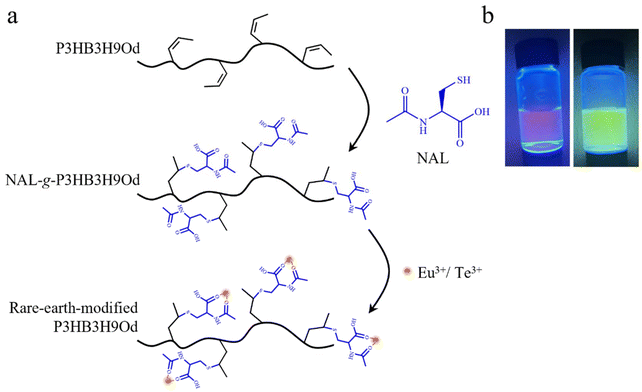
For the crosslinking reaction, 1 g of P3HB3H9Od, 36 μL of 1,2-ethanedithiol and 0.01 g of Irgacure 2959 were dissolved in 10 mL of chloroform. The PHA solution was exposed to UV for 1 h.For the rare-earth modified PHA, 1 g of P3HB3H9Od, 0.17 g of N-acetyl-L-cysteine (NAL) and 0.005 g 2,2-bis(hydroxymethyl)propionic acid (DMPA) were dissolved in a mixed solvent of tetrahydrofuran (THF) and methanol with a volume ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The solution was then exposed to UV for 30 min. After the reaction, the precipitate was washed with methanol three times to obtain purified NAL-grafted PHA. Subsequently, 0.6 g of NAL-grafted PHA and 0.2 g of rare-earth salt [Eu(NO3)3 or Tb(NO3)3] were dissolved in 5 mL of THF and stirred under heating at 70 °C for 4 h. After the reaction, the precipitate was washed using deionized water three times to obtain the rare-earth-modified PHA.
1. The solution was then exposed to UV for 30 min. After the reaction, the precipitate was washed with methanol three times to obtain purified NAL-grafted PHA. Subsequently, 0.6 g of NAL-grafted PHA and 0.2 g of rare-earth salt [Eu(NO3)3 or Tb(NO3)3] were dissolved in 5 mL of THF and stirred under heating at 70 °C for 4 h. After the reaction, the precipitate was washed using deionized water three times to obtain the rare-earth-modified PHA.
3. Results
3.1 Design and construction of the LCL PHA synthetic pathway in H. bluephagenesis
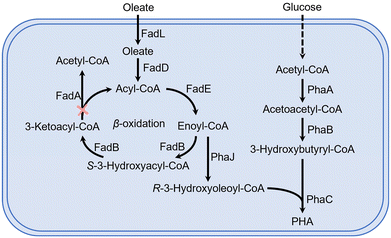
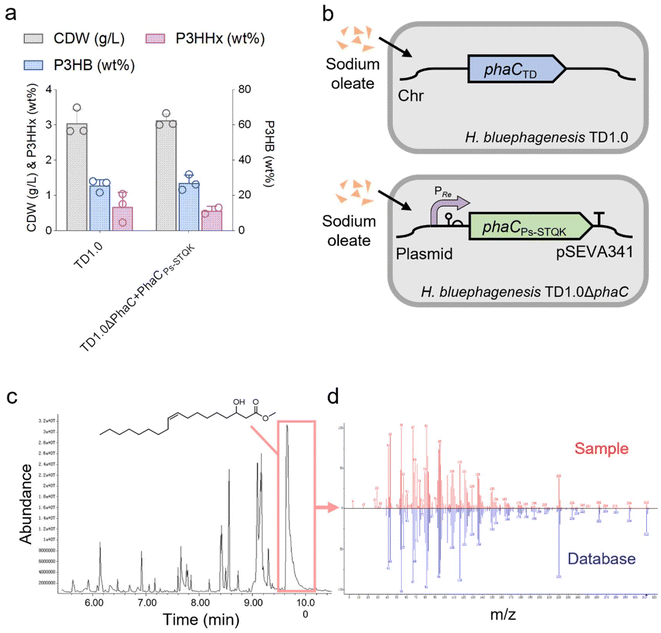
In this study, sodium oleate was used as the structurally related carbon source for the production of unsaturated LCL PHA (Fig. 1). Low-cost sodium oleate was used due to its long carbon chain length and the presence of a double bond in its side chain. The native PhaC of H. bluephagenesis can only polymerize SCL monomers,41,50 thus a new PhaC was required to accomplish the polymerization of LCL PHA in H. bluephagenesis. Moreover, H. bluephagenesis's ability to utilize sodium oleate needs to be confirmed.H. bluephagenesis was found to be able to utilize sodium oleate, and it accumulated 25.6 wt% P3HB and 0.7 wt% P3HHx in 3.05 g L−1 cell dry weight (CDW) from 5 g L−1 sodium oleate when grown in shake flasks (Fig. 2a). Subsequently, native PhaCTD in H. bluephagenesis was deleted, and a plasmid harboring PhaCPs-STQK, a PHA synthase with broad substrate specificity mutated (S325T, Q481K) from PhaCPs of Pseudomonas sp. 61-3, was introduced into H. bluephagenesis (Fig. 2b). PhaCPs-STQK has been previously used to produce LCL PHA in P. entomophila.26,27 The recombinant H. bluephagenesis was grown to 3.12 g L−1 CDW containing 27.1 wt% P3HB and 0.6 wt% P3HHx from 5 g L−1 sodium oleate (Fig. 2a), and no PHA monomer with a longer carbon chain length was detected. The results implied that H. bluephagenesis possesses very strong β-oxidation that can convert most of the sodium oleate to acetyl-CoA, butyryl-CoA and hexanoyl-CoA. Thus, FadA in H. bluephagenesis was deleted to inhibit the β-oxidation. The PhaCTD and FadA deficient strain was named H. bluephagenesis TXY21, and it was unable to grow normally on sodium oleate as the sole carbon source. After the shake flask study, a trace amount of methyl 3-hydroxy-9-octadecenoate was detected in this FadA deficient strain via gas chromatography–mass spectrometry (GC-MS) (Fig. 2c and d). To identify the source of the signal detected by GC-MS (Fig. S1†), intracellular PHA was extracted to confirm the presence of P3HB3H9Od (Fig. S2a and S2b†). The result confirmed that P3HB3H9Od was synthesized by recombinant H. bluephagenesis.
3.2 Engineering H. bluephagenesis for enhanced P3HB3H9Od synthesis
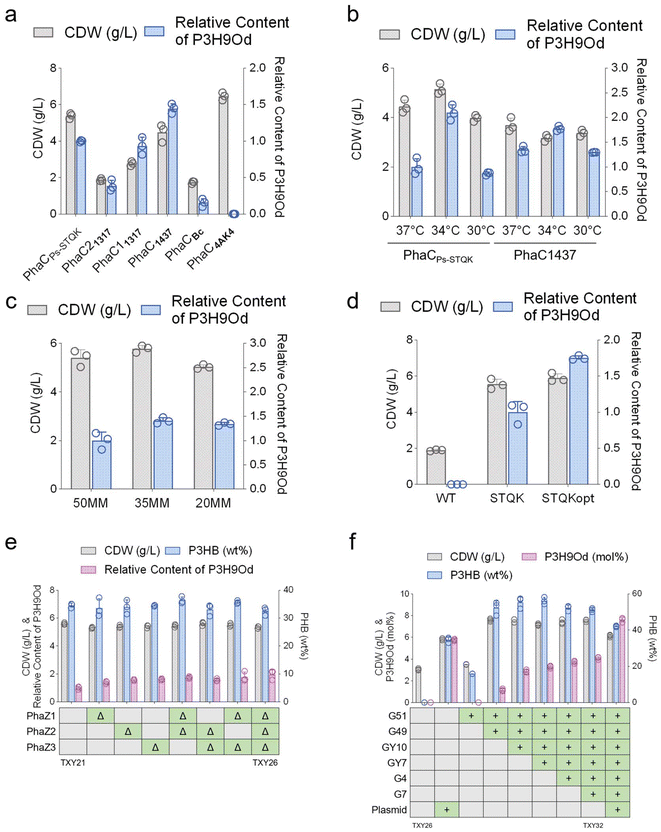
To achieve a higher proportion of 3H9Od in PHA, a broader range of PhaC variants with higher condensing ability should be screened. Thus, six PhaCs reported to exhibit polymerization of MCL and/or LCL PHA were expressed in H. bluephagenesis for P3HB3H9Od synthesis, respectively. Their protein sequences and homology are shown (Fig. S3†). Five of them were found to produce P3HB3H9Od in H. bluephagenesis (Fig. 3a). H. bluephagenesis expressing PhaCPs-STQK or PhaC1437 were grown to a higher CDW and accumulated with a higher proportion of 3H9Od in the PHA. Both PhaCPs-STQK and PhaC1437 originated from Pseudomonas spp. grown at an optimal 30 °C, compared to H. bluephagenesis, that grows best at 37 °C. This temperature difference may have a significant negative impact on the PhaC activity. It was found that PhaCPs-STQK demonstrated superior performance compared to PhaC1437 at 34 °C (Fig. 3b), and the performances of both PhaC variants were enhanced at this temperature. Similarly, Pseudomonas spp. grows at around 10 g L−1 NaCl, compared to H. bluephagenesis, that grows best at 50–60 g L−1 NaCl. The difference in growth under various NaCl concentrations may affect the PhaC activity. In 35 g L−1 NaCl-containing mineral medium (35 MM), the proportion of 3H9Od in PHA was approximately 1.4 times higher than that in 50 MM, and the CDW was also higher (Fig. 3c), suggesting that conditions between the optimal growth environments of Halomonas and Pseudomonas were more conducive to P3H9Od accumulation. Additionally, the codon-optimized PhaCPs-STQKopt exhibited a 1.7-fold increase in 3H9Od content in PHA compared to PhaCPs-STQK, and the wild type PhaCPs lacked the ability to condense 3H9Od in H. bluephagenesis (Fig. 3d).In addition to deleting FadA to block the β-oxidation of fatty acid salts, several other β-oxidation enzymes in H. bluephagenesis were also deleted (Table S3†). However, none of these additional deletions resulted in a statistically significant increase in the proportion of 3H9Od (Fig. S4a and S4b†). Therefore, it was considered that FadA deletion was sufficient for P3HB3H9Od production in H. bluephagenesis.
The accumulation of PHA is a dynamic process involving its synthesis and degradation. Enhanced PHA synthesis can also be achieved by reducing its degradation. There are three PHA depolymerases in H. bluephagenesis, namely, PhaZ1424, PhaZ2133, and PhaZ3536, abbreviated as PhaZ1, PhaZ2 and PhaZ3. The deletion of these depolymerases had no significant impact on P3HB accumulation.51 This may be due to the rate of P3HB synthesis being much faster than its degradation rate, or due to PhaZs possibly having specificity for other PHAs with different chain lengths. The synthesis of P3H9Od was inefficient, and the depolymerization of this new PHA has not been studied. Consequently, these three PhaZs were deleted individually or in combination. The content of P3HB did not show significant changes, while the content of P3H9Od gradually increased (Fig. 3e), demonstrating that the three PHA depolymerases were indeed able to hydrolyze P3H9Od. The strain with all three PhaZs deleted was named H. bluephagenesis TXY26.
PhaC is the most critical enzyme in PHA synthesis, and both its quality and quantity have a significant impact on the synthesized PHA. In the study of the key enzymes involved in the P3HB3H9Od synthesis pathway, overexpressing PhaC was found to exhibit the highest enhancement of the proportion of 3H9Od (Fig. S5†). To enhance its expression beyond the levels achievable with high-copy plasmids and strong promoters, and to realize plasmid-free fermentation, we integrated the phaCPs-STQKopt gene into multiple chromosomal loci. After inserting one copy of the gene at each selected locus, the highest-performing strain was selected and used as the basis for subsequent insertions at other loci.
Insertions of phaC were found to increase the proportion of 3H9Od in the cells gradually (Fig. 3f). After inserting six copies of phaCPs-STQKopt into the chromosome, H. bluephagenesis TXY32 harboring a plasmid expressing phaCPs-STQKopt was grown to 6.1 g L−1 CDW containing 42.1 wt% P(3HB-co-7.6 mol% 3H9Od) (Fig. 3f). More insertions of phaCs may further lead to a higher proportion of 3H9Od in the PHA.
3.3 Medium optimization for enhanced P3HB3H9Od synthesis
Concentrations of glucose, urea and sodium oleate were optimized for improved P3HB3H9Od production. Single-factor gradient experiments were conducted to assess optimized concentrations of urea (beyond what was included in solution I) and sodium oleate, aiming to establish a preliminary range for their optimal levels (Fig. 4a and b). The optimal dosage ranges for additional urea and sodium oleate were found to be 0.5 g L−1 and 8 g L−1, respectively. Based on these results, three concentration gradients were selected for glucose, urea, and sodium oleate to conduct orthogonal experiments. Suitable compositions in the shake flask medium included 30 g L−1 glucose, 10 g L−1 sodium oleate and 0.5 g L−1 additional urea (Fig. 4c), which allowed growth to 6.7 g L−1 CDW containing 50.4 wt% P(3HB-co-10.8 mol% 3H9Od).3.4 P3HB3H9Od production by H. bluephagenesis in a 7-liter bioreactor
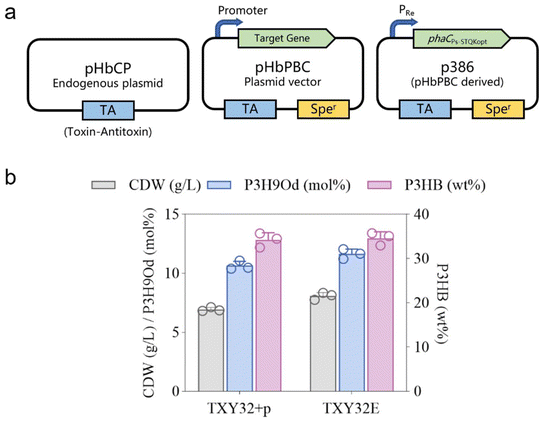
To combine plasmid stability, high gene expression level, and antibiotic-free conditions in fed-batch studies, a plasmid backbone named pHbPBC was utilized to express phaC gene.52 This plasmid backbone was constructed based on an endogenous toxin–antitoxin system (Fig. 5a), enabling stable maintenance without antibiotic selection and allowing high-level gene expression.52 The plasmid was introduced into the H. bluephagenesis TXY32 with the endogenous toxin–antitoxin system knocked out, and the recombinant strain was named H. bluephagenesis TXY32E. The best performance of this strain in the shake flask reached 8.1 g L−1 CDW containing 51.0 wt% P(3HB-co-11.6 mol% 3H9Od) (Fig. 5b).P3HB3H9Od was produced in a 7-liter fermenter incubated with H. bluephagenesis TXY32E under open unsterile conditions using sodium oleate and glucose. H. bluephagenesis TXY32E produced 28.1 g L−1 CDW containing 52.7 wt% P(3HB-co-6.03 mol% 3H9Od) (Fig. 6), with an overall PHA productivity of 0.31 g L−1 h−1. This result suggested the feasibility of industrial production of P3HB3H9Od using H. bluephagenesis.
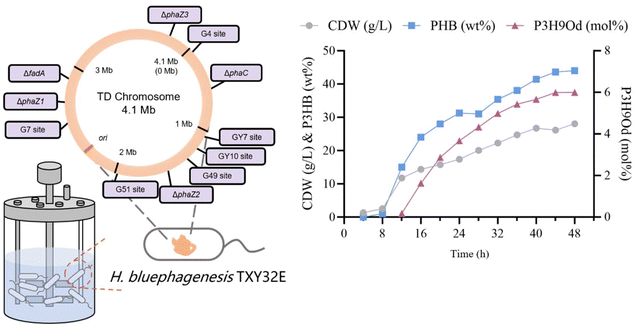 | ||
| Fig. 6 Growth and PHA production using H. bluephagenesis TXY32E cultured in a 7-liter bioreactor. Culture condition: the culture medium is described in section 2.4, pH = 8.5, 34 °C. | ||
3.5 Characterization of P3HB3H9Od produced by H. bluephagenesis
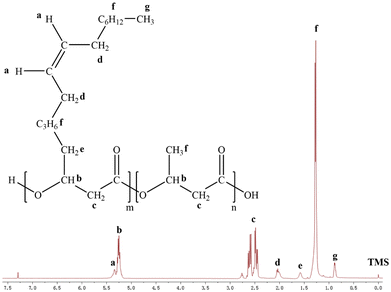
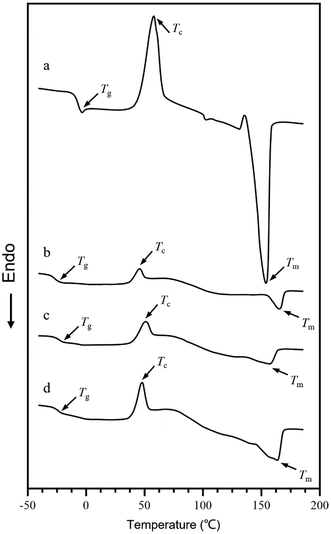
The 1H nuclear magnetic resonance result of the high P3H9Od content containing PHA was studied (Fig. 7). The average molecular weight of P3HB3H9Od studied via gel permeation chromatography (GPC) was found to be between 6 × 104 and 8 × 104 Da with a polydispersity index (PDI) of around 1.8 (Table 1). Since 3H9Od is not the common substrate of PHA polymerase, with the increasing proportion of 3H9Od components in the P3HB3H9Od, the molecular weight of the polymer decreased gradually. This may be due to the difference in the catalytic activity of PhaC toward different substrates. That is, the addition of the 3H9Od monomer affected the PHA polymerization process, thereby hindering the elongation of the polymer chain. Additionally, the molecular weight of PHA synthesized by type II PhaC was lower than that of other PhaC types,53,54 and the molecular weight of P3HB3H9Od was observed to be relatively low. DSC and TGA showed that the glass transition temperature (Tg), crystallization temperature (Tc), and melting temperature (Tm) of P(3HB-co-10 mol% 3H9Od) were −18.7 °C, 49.6 °C, and 166.0 °C, respectively (Fig. 8 and Table 1). TGA showed that the decomposition temperature (Td) of P(3HB-co-10 mol% 3H9Od) was 283.5 °C (Table 1).| Samples | Molecular weight | Thermal properties | Ref./sources | ||||
|---|---|---|---|---|---|---|---|
| M w (×104 Da) | PDI | T g (°C) | T c (°C) | T m (°C) | T d (°C) | ||
| PHB | 7.91 | 1.58 | −1.52 | 52.43 | 154.8 | 180 | This study |
| P(3HB-co-5 mol% 3H9Od) | 7.84 | 1.76 | −20.52 | 47.46 | 168.0 | 282.9 | |
| P(3HB-co-7.5 mol% 3H9Od) | 6.99 | 1.79 | −18.58 | 52.76 | 159.7 | 281.9 | |
| P(3HB-co-10 mol% 3H9Od) | 5.94 | 1.88 | −18.66 | 49.63 | 166.0 | 283.5 | |
| P(3HB-co-11.17 mol% 3HD) | 7.05 | 1.66 | −2.07 | 154.71 | 27 | ||
| P(3HB-co-15.96 mol% 3HDD) | 6.61 | 1.76 | −1.47 | 155.84 | |||
| P(3HB-co-13.74 mol% 3HTD) | 7.16 | 1.64 | −3.49 | 149.56 | |||
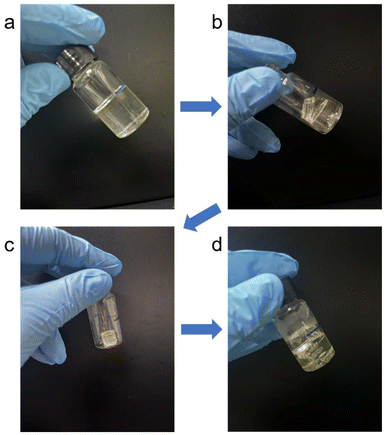
3.6 Chemical modification of P3HB3H9Od
4. Conclusions and discussion
Recombinant H. bluephagenesis with deletions of native PhaCTD, FadA and PhaZ123 combined with the expression of heterogenous PhaCPs-STQKopt was successfully constructed for the synthesis of P3HB3H9Od from glucose and sodium oleate. The highest 3H9Od proportion in P3HB3H9Od reached was 11.6 mol%.This study expanded PHA diversity and laid the foundation for MCL/LCL PHA synthesis by H. bluephagenesis. Future studies should focus on optimizing the production process to improve the P3HB3H9Od yield, exploring more double bond modifications to tailor its properties for specific applications, and conducting in-depth studies on its biodegradability and environmental impact to promote its commercialization and sustainable development.
This study proposed four green advances that collectively established a greener paradigm for unconventional PHA biosynthesis compared with conventional ones. First, sodium oleate was employed, as it is a low-cost, bio-derived feedstock with long aliphatic chains and inherent unsaturation, eliminating the need for chemical pre-modification. This is an improvement over traditional processes that rely on expensive purified substrates or non-renewable petrochemical derivatives. Second, this study introduced Halomonas bluephagenesis as a microbial chassis for the biosynthesis of LCL PHA. Notably, this platform operates under harsh conditions and can utilize seawater for fermentation, reducing freshwater consumption, energy requirements and the environmental footprint compared to conventional systems.42 In previous studies, Pseudomonas entomophila was utilized to produce LCL PHA,26,27 which typically required strictly controlled sterile conditions, leading to higher freshwater and energy demands. Additionally, the Halomonas strain achieved higher cell density and better PHA yield, showing superior incubation efficiency. Third, the chromosomal integration of phaC coupled with toxin–antitoxin stabilized plasmids achieved high-level gene expression without the use of antibiotics. This eliminated the risk of antibiotic release into the environment, reducing potential ecological harm. Finally, click chemistry was used for PHA modification. The thiol–ene click reaction-based modification of PHA contributed to green chemistry by enabling efficient functionalization under mild conditions with high selectivity and atom economy, minimizing energy consumption and waste generation.58 However, the use of an organic solvent remained a drawback in terms of green chemistry. These integrated innovations collectively created an eco-efficient production system for unconventional PHA and new applications.
Author contributions
Xinyu Chen: data analysis, validation, investigation, visualization, methodology, software, data curation, and writing original draft. Jiangnan Chen: visualization, software, and writing original draft. Xu Yan: software, data analysis, and methodology. Qiong Wu: resources and supervision. Fuqing Wu: resources and supervision. Guo-Qiang Chen: conceptualization, resources, supervision, funding acquisition, writing original draft, revision, data analysis, and validation.Conflicts of interest
All of the authors in this study declared no conflicts of interest.Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.Acknowledgements
This work was supported by the Ministry of Science and Technology of the People's Republic of China (Grant No. 2025YFA0921100) and the National Natural Science Foundation of China (Grant No. 32130001). This work was also supported by the grants from PhaBuilder and the Tsinghua-Peking Center for Life Sciences.References
- S. Y. Choi, M. N. Rhie, H. T. Kim, J. C. Joo, I. J. Cho, J. Son, S. Y. Jo, Y. J. Sohn, K.-A. Baritugo, J. Pyo, Y. Lee, S. Y. Lee and S. J. Park, Metab. Eng., 2020, 58, 47–81 CrossRef CAS PubMed.
- C. DeLisi, A. Patrinos, M. MacCracken, D. Drell, G. Annas, A. Arkin, G. Church, R. Cook-Deegan, H. Jacoby, M. Lidstrom, J. Melillo, R. Milo, K. Paustian, J. Reilly, R. J. Roberts, D. Segrè, S. Solomon, D. Woolf, S. D. Wullschleger and X. Yang, Biodes. Res., 2020, 2020, 1016207 CrossRef PubMed.
- F. Hassard, T. P. Curtis, G. C. Dotro, P. Golyshin, T. Gutierrez, S. Heaven, L. Horsfall, B. Jefferson, D. L. Jones, N. Krasnogor, V. Kumar, D. J. Lea-Smith, K. Le Corre Pidou, Y. Liu, T. Lyu, R. R. McCarthy, B. McKew, C. Smith, A. Yakunin, Z. Yang, Y. Zhang and F. Coulon, ACS Synth. Biol., 2024, 13, 1586–1588 CrossRef CAS PubMed.
- J. Symons, T. A. Dixon, J. Dalziell, N. Curach, I. T. Paulsen, A. Wiskich and I. S. Pretorius, Nat. Commun., 2024, 15, 2669 CrossRef CAS PubMed.
- S. Kind, S. Neubauer, J. Becker, M. Yamamoto, M. Völkert, G. v. Abendroth, O. Zelder and C. Wittmann, Metab. Eng., 2014, 25, 113–123 CrossRef CAS PubMed.
- C. G. Chae, Y. J. Kim, S. J. Lee, Y. H. Oh, J. E. Yang, J. C. Joo, K. H. Kang, Y.-A. Jang, H. Lee, A. R. Park, B. K. Song, S. Y. Lee and S. J. Park, Biotechnol. Bioprocess Eng., 2016, 21, 169–174 CrossRef CAS.
- S. Y. Choi, S. J. Park, W. J. Kim, J. E. Yang, H. Lee, J. Shin and S. Y. Lee, Nat. Biotechnol., 2016, 34, 435–440 CrossRef CAS PubMed.
- H. Park, H. T. He, X. Yan, X. Liu, N. S. Scrutton and G. Q. Chen, Biotechnol. Adv., 2024, 71, 108320 CrossRef CAS PubMed.
- D. Pal, R. Prabhakar, V. B. Barua, I. Zekker, J. Burlakovs, A. Krauklis, W. Hogland and Z. Vincevica-Gaile, Environ. Sci. Pollut. Res., 2025, 32, 56–88 CrossRef PubMed.
- M. Koller, L. Maršálek, M. M. de Sousa Dias and G. Braunegg, New Biotechnol., 2017, 37, 24–38 CrossRef CAS PubMed.
- K. Sudesh, H. Abe and Y. Doi, Prog. Polym. Sci., 2000, 25, 1503–1555 CrossRef CAS.
- M. Lakshmanan, C. P. Foong, H. Abe and K. Sudesh, Polym. Degrad. Stab., 2019, 163, 122–135 CrossRef CAS.
- S. Y. Lee, Biotechnol. Bioeng., 1996, 49, 1–14 CrossRef CAS PubMed.
- S. Obruca, P. Sedlacek, E. Slaninova, I. Fritz, C. Daffert, K. Meixner, Z. Sedrlova and M. Koller, Appl. Microbiol. Biotechnol., 2020, 104, 4795–4810 CrossRef CAS PubMed.
- Z. B. Li, J. Yang and X. J. Loh, NPG Asia Mater., 2016, 8, e265 CrossRef CAS.
- S. Behera, M. Priyadarshanee, Vandana and S. Das, Chemosphere, 2022, 294, 133723 CrossRef CAS PubMed.
- D. M. Miu, M. C. Eremia and M. Moscovici, Materials, 2022, 15, 1410 CrossRef CAS PubMed.
- L. Getino, J. L. Martín and A. Chamizo-Ampudia, Microorganisms, 2024, 12, 2028 CrossRef CAS PubMed.
- A. M. Yousefi and G. E. Wnek, Biomed. Mater. Devices, 2025, 3, 19–44 CrossRef.
- L. P. Yu, X. Zhang, D. X. Wei, Q. Wu, X. R. Jiang and G. Q. Chen, Biomacromolecules, 2019, 20, 3233–3241 CrossRef CAS PubMed.
- X. M. Liang, D. K. Cha and Q. Q. Xie, Resour. Conserv. Recycl. Adv., 2024, 21, 200206 Search PubMed.
- K. Prajapati, R. Nayak, A. Shukla, P. Parmar, D. Goswami and M. Saraf, J. Polym. Environ., 2021, 29, 2013–2032 CrossRef CAS.
- Z. A. Raza, S. Riaz and I. M. Banat, Biotechnol. Prog., 2018, 34, 29–41 CrossRef CAS PubMed.
- D. Y. Kim, H. W. Kim, M. G. Chung and Y. H. Rhee, J. Microbiol., 2007, 45, 87–97 CAS.
- S. Z. Neoh, M. F. Chek, H. T. Tan, J. A. Linares-Pastén, A. Nandakumar, T. Hakoshima and K. Sudesh, Curr. Res. Biotechnol., 2022, 4, 87–101 CrossRef.
- M. Y. Li, X. B. Chen, X. M. Che, H. Q. Zhang, L. P. Wu, H. T. Du and G. Q. Chen, Metab. Eng., 2019, 52, 253–262 CrossRef CAS PubMed.
- M. Y. Li, Y. Y. Ma, X. Zhang, L. Z. Zhang, X. Y. Chen, J. W. Ye and G. Q. Chen, Adv. Mater., 2021, 33, 2102766 CrossRef CAS PubMed.
- C. J. Gao, Q. S. Qi, C. Madzak and C. S. K. Lin, J. Ind. Microbiol. Biotechnol., 2015, 42, 1255–1262 CrossRef CAS PubMed.
- A. G. Sandström, A. Muñoz de las Heras, D. Portugal-Nunes and M. F. Gorwa-Grauslund, AMB Express, 2015, 5, 14 CrossRef PubMed.
- Q. Q. Zhuang and Q. S. Qi, Microb. Cell Fact., 2019, 18, 135 CrossRef PubMed.
- Y. J. Sohn, H. T. Kim, K. A. Baritugo, H. M. Song, M. H. Ryu, K. H. Kang, S. Y. Jo, H. Kim, Y. J. Kim, J. I. Choi, S. K. Park, J. C. Joo and S. J. Park, Int. J. Biol. Macromol., 2020, 149, 593–599 CrossRef CAS PubMed.
- A. Ylinen, H. Maaheimo, A. Anghelescu-Hakala, M. Penttilä, L. Salusjärvi and M. Toivari, J. Ind. Microbiol. Biotechnol., 2021, 48, kuab028 CrossRef CAS PubMed.
- H. E. Valentin, D. L. Broyles, L. A. Casagrande, S. M. Colburn, W. L. Creely, P. A. DeLaquil, H. M. Felton, K. A. Gonzalez, K. L. Houmiel, K. Lutke, D. A. Mahadeo, T. A. Mitsky, S. R. Padgette, S. E. Reiser, S. Slater, D. M. Stark, R. T. Stock, D. A. Stone, N. B. Taylor, G. M. Thorne, M. Tran and K. J. Gruys, Int. J. Biol. Macromol., 1999, 25, 303–306 CrossRef CAS PubMed.
- S. J. Park, Y.-A. Jang, H. Lee, A. R. Park, J. E. Yang, J. Shin, Y. H. Oh, B. K. Song, J. Jegal, S. H. Lee and S. Y. Lee, Metab. Eng., 2013, 20, 20–28 CrossRef CAS PubMed.
- S. J. Park, K.-H. Kang, H. Lee, A. R. Park, J. E. Yang, Y. H. Oh, B. K. Song, J. Jegal, S. H. Lee and S. Y. Lee, J. Biotechnol., 2013, 165, 93–98 CrossRef CAS PubMed.
- P. Thamarai, A. S. Vickram, A. Saravanan, V. C. Deivayanai and S. Evangeline, Bioresour. Technol. Rep., 2024, 27, 101957 CrossRef CAS.
- S. González-Rojo, A. I. Paniagua-García and R. Díez-Antolínez, Microorganisms, 2024, 12, 1668 CrossRef PubMed.
- J. A. Pérez Aguilar, J. M. Franco, I. D. Otero and R. Benítez Benítez, Waste Biomass Valorization, 2024, 15, 4221–4233 CrossRef.
- J. B. Silva, J. R. Pereira, B. C. Marreiros, M. A. M. Reis and F. Freitas, Proc. Biochem., 2021, 102, 393–407 CrossRef CAS.
- X. J. Jiang, Z. Sun, J. A. Ramsay and B. A. Ramsay, AMB Express, 2013, 3, 50 CrossRef PubMed.
- D. Tan, Y. S. Xue, G. Aibaidula and G. Q. Chen, Bioresour. Technol., 2011, 102, 8130–8136 CrossRef CAS PubMed.
- D. Tan, Q. Wu, J. C. Chen and G. Q. Chen, Metab. Eng., 2014, 26, 34–47 CrossRef CAS PubMed.
- X. Z. Fu, D. Tan, G. Aibaidula, Q. Wu, J. C. Chen and G. Q. Chen, Metab. Eng., 2014, 23, 78–91 CrossRef CAS PubMed.
- X. B. Chen, J. Yin, J. W. Ye, H. Q. Zhang, X. M. Che, Y. M. Ma, M. Y. Li, L. P. Wu and G. Q. Chen, Bioresour. Technol., 2017, 244, 534–541 CrossRef CAS PubMed.
- H. Ma, Y. Q. Zhao, W. Z. Huang, L. Z. Zhang, F. Q. Wu, J. W. Ye and G. Q. Chen, Nat. Commun., 2020, 11, 3313 CrossRef CAS PubMed.
- Y. N. Lin, Y. Y. Guan, X. Dong, Y. Y. Ma, X. Wang, Y. C. Leng, F. Q. Wu, J. W. Ye and G. Q. Chen, Metab. Eng., 2021, 64, 134–145 CrossRef CAS PubMed.
- R. Silva-Rocha, E. Martínez-García, B. Calles, M. Chavarría, A. Arce-Rodríguez, A. de las Heras, A. D. Páez-Espino, G. Durante-Rodríguez, J. Kim, P. I. Nikel, R. A. Platero and V. c. de Lorenzo, Nucleic Acids Res., 2012, 41, 666–675 CrossRef PubMed.
- Q. Qin, C. Ling, Y. Q. Zhao, T. Yang, J. Yin, Y. Y. Guo and G. Q. Chen, Metab. Eng., 2018, 47, 219–229 CrossRef CAS PubMed.
- G. Braunegg, B. Sonnleitner and R. M. Lafferty, Eur. J. Appl. Microbiol. Biotechnol., 1978, 6, 29–37 CrossRef CAS.
- L. Cai, D. Tan, G. Aibaidula, X. R. Dong, J. C. Chen, W. D. Tian and G. Q. Chen, Microb. Cell Fact., 2011, 10, 88 CrossRef CAS PubMed.
- D. Tan, Doctoral dissertation, Tsinghua University, 2014.
- K. Ren, Y. Q. Zhao, G. Q. Chen, X. Ao and Q. Wu, ACS Synth. Biol., 2024, 13, 61–67 CrossRef CAS PubMed.
- B. H. A. Rehm, Biochem. J., 2003, 376, 15–33 CrossRef CAS PubMed.
- B. H. A. Rehm and A. Steinbüchel, Int. J. Biol. Macromol., 1999, 25, 3–19 CrossRef CAS PubMed.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11–18 CrossRef CAS PubMed.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Nat. Methods, 2008, 5, 763–775 CrossRef CAS PubMed.
- S. S. Zhou, X. Xue, J. F. Wang, Y. Dong, B. Jiang, D. Wei, M. L. Wan and Y. Jia, J. Mater. Chem., 2012, 22, 22774–22780 RSC.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5gc02422f |
| This journal is © The Royal Society of Chemistry 2025 |