Coal waste-derived synthesis of yellow oxidized graphene quantum dots with highly specific superoxide dismutase activity: characterization, kinetics, and biological studies†
Tushar
Das
 a,
Subrata
Das
a,
Subrata
Das
 *a,
Prakash
Kumar
b,
Betty
C.A.
cd and
Debabrata
Mandal
*a,
Prakash
Kumar
b,
Betty
C.A.
cd and
Debabrata
Mandal
 *b
*b
aDepartment of Chemistry, National Institute of Technology Patna, Bihar 800005, India. E-mail: subrataorgchem@gmail.com; subratad@nitp.ac.in
bDepartment of Biotechnology, National Institute of Pharmaceutical Education and Research Hajipur, Vaishali 844102, India. E-mail: debabrataman@gmail.com
cChemistry Division, Bhabha Atomic Research Centre, Mumbai 400085, India
dHomi Bhabha National Institute, Mumbai 400085, India
First published on 10th October 2023
Abstract
The disintegration of coal-based precursors for the scalable production of nanozymes relies on the fate of solvothermal pyrolysis. Herein, we report a novel economic and scalable strategy to fabricate yellow luminescent graphene quantum dots (YGQDs) by remediating unburnt coal waste (CW). The YGQDs (size: 7–8 nm; M.W: 3157.9 Da) were produced using in situ “anion-radical” assisted bond cleavage in water (within 8 h; at 121 °C) with yields of ∼87%. The presence of exposed surface and edge groups, such as COOH, C–O–C, and O–H, as structural defects accounted for its high fluorescence with εmax ∼530 nm at pH 7. Besides, these defects also acted as radical stabilizers, demonstrating prominent anti-oxidative activity of ∼4.5-fold higher than standard ascorbic acid (AA). In addition, the YGQDs showed high biocompatibility towards mammalian cells, with 500 μM of treatment dose showing <15% cell death. The YGQDs demonstrated specific superoxide dismutase (SOD) activity wherein 15 μM YGQDs equalled the activity of 1-unit biological SOD (bSOD), measured using the pyrogallol assay. The Km for YGQDs was ∼10-fold higher than that for bSOD. However, the YGQDs retained their SOD activity in harsh conditions like high temperatures or denaturing reactions, where the activity of bSOD is completely lost. The binding affinity of YGQDs for superoxide ions, measured from isothermal calorimetry (ITC) studies, was only 10-fold lower than that of bSOD (Kd of 586 nM vs. 57.3 nM). Further, the pre-treatment of YGQDs (∼10–25 μM) increased the cell survivability to >75–90% in three cell lines during ROS-mediated cell death, with the highest survivability being shown for C6-cells. Next, the ROS-induced apoptosis in C6-cells (model for neurodegenerative diseases study), wherein YGQDs uptake was confirmed by confocal microscopy, showed ∼5-fold apoptosis alleviation with only 5 μM pretreatment. The YGQDs also restored the expression of pro-inflammatory Th1 cytokines (TNF-α, IFN-γ, IL-6) and anti-inflammatory Th2 cytokines (IL-10) to their basal levels, with a net >3-fold change observed. This further explains the molecular mechanism for the antioxidant property of YGQDs. The high specific SOD activity associated with YGQDs may provide the cheapest alternative source for producing large-scale SOD-based nanozymes that can treat various oxidative stress-linked disorders/diseases.
Introduction
Graphene quantum dots (GQDs) represent an arsenal class of fragmented zero-band gap material with a size range usually below ∼15 nm. The diverse physio-chemical properties from its quantum Hall and confinement effects prospect its applications in nano-biotechnology-driven research.1 Chemically, GQDs can be produced from a variety of biogenic precursors like glucose, graphitic derivatives, carbon nanotubes (CNT), carbon nanofibers (CNF), and other carbon-rich sources either through “top–down” or “bottom–up” routes.2–9 However, using such manifested precursors is often bounded by their expensiveness and operational instabilities, resulting in high manufacturing costs and lower practical yields. Moreover, precision techniques, such as critical water treatment,10 pulse laser ablation in liquid,11 and material deflagration methods,12 require complicated inert assemblies for production. A considerable switch to sustainable precursors with facile methodologies might provide a profitable and scalable alternative to solve such limitations.13 In this regard, stable GQDs were produced from trifling varieties of coal through simplistic methods, offering good aqueous dispersibility and providing newer developments in coal-based research.14–17 The natural process of thermal cracking that entirely relies on carbonization phenomena arising from the differences in the atomic composition of materials can be a promising approach to produce GQDs.18 Besides, the abundance of stacked aliphatic domains and different oxygenated compositions of coal allow additional sites for oxidative cleavage.19 In addition, the logarithmic increase in the rapid utilization of coal-based energy has led to an alarming rise in different environmental pollutions.20–22 Hence, we envisioned our strategy of remediating harmful coal remains from nature by converting them into less toxic GQDs with a nano-enzymatic activity that can be used for alleviating free radical-induced oxidative disease.Biologically, the inherent properties of GQDs, like controllable tunability in photoluminescence, high loading capacity, and radiation capture ability, make them suitable for photodynamic therapies, biosensors, bio-imaging, and drug carriers.23 Moreover, the abundance of surface functional groups like COOH, OH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and C–O–C serve as catalytic sites, similar to that in enzymes, to carry out various bio-transformation reactions.24 Unlike other inorganic nanomaterials, GQDs offer much higher cellular biocompatibility, for which they are used as a therapy replacement for natural enzymes.25,26 In addition, GQDs provide a much cheaper production cost, better shelf-life, and reusability than natural enzymatic supplements.27 In this regard, Sun et al. used SOD/CAT-mediated anti-oxidative and protective oxidized GQDs against alcohol-intoxicated cell lines.28 Kim et al. showed the blood–brain barrier permeability of GQDs, and its specific interactions with α-synuclein can prevent its accumulation and aggregation during Parkinson's disease progression.29 Hu et al. used GQDs/semiconducting polymer nanocomposites as a synergistic photothermal and nano-enzymatic agent for better tumor therapy by combining the intrinsic peroxidase activity of GQDs and temperature-sensitive production of singlet oxygen (1O2).30 So far, most of the nanoenzymatic roles of GQDs reported are linked to its natural peroxidase-like behavior. For example Xin et al. synthesized histidine-functionalized GQDs/hemin complex with high peroxidase turnover, with the hemin group acting as natural peroxidase mimics.31 Sun et al. illustrated the catalytic mechanism of peroxidase-like activity in GQDs by the selective activation and deactivation of different functional groups, wherein the surface-exposed C
C, and C–O–C serve as catalytic sites, similar to that in enzymes, to carry out various bio-transformation reactions.24 Unlike other inorganic nanomaterials, GQDs offer much higher cellular biocompatibility, for which they are used as a therapy replacement for natural enzymes.25,26 In addition, GQDs provide a much cheaper production cost, better shelf-life, and reusability than natural enzymatic supplements.27 In this regard, Sun et al. used SOD/CAT-mediated anti-oxidative and protective oxidized GQDs against alcohol-intoxicated cell lines.28 Kim et al. showed the blood–brain barrier permeability of GQDs, and its specific interactions with α-synuclein can prevent its accumulation and aggregation during Parkinson's disease progression.29 Hu et al. used GQDs/semiconducting polymer nanocomposites as a synergistic photothermal and nano-enzymatic agent for better tumor therapy by combining the intrinsic peroxidase activity of GQDs and temperature-sensitive production of singlet oxygen (1O2).30 So far, most of the nanoenzymatic roles of GQDs reported are linked to its natural peroxidase-like behavior. For example Xin et al. synthesized histidine-functionalized GQDs/hemin complex with high peroxidase turnover, with the hemin group acting as natural peroxidase mimics.31 Sun et al. illustrated the catalytic mechanism of peroxidase-like activity in GQDs by the selective activation and deactivation of different functional groups, wherein the surface-exposed C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups served as active sites.32 Nirala et al. reported an electrochemical preparation of GQDs having natural peroxidase activity that were further used as natural colorimetric probes to detect trace levels of glucose and H2O2.33 Amongst the different bio-properties, GQDs with inherent radical scavenging activity (RSA) are most widely studied due to its association with different therapies to counter elevated levels of oxidative stress (OS) under a diseased state. In this regard, the explicit report of Wang et al. showed that hydrogen bond donation and radical adduct formation ability of GQDs initiated by different surface speciation of oxygenated functional groups are the main reason for the quenching of free radicals.34 Nilewski et al. showed that polyethylene glycol-functionalized GQDs can rescue murine endothelioma cells from severe H2O2 stress through SOD-linked activity.35 Krunić et al. explored the anti-oxidative property of GQDs against oxidative and nitrative damages in SH-SY5Y neuroblastoma cells generated through sodium nitroprusside, with GQDs acting as non-selective antioxidants and iron chelators, with no enzymatic activity.36 More interestingly, Wang et al. showed that upon halogen doping, GQDs shows a switch between its anti- and pro-oxidant behavior with good antibacterial effect mediated through generation (1O2).37 Although multiple attempts are made to use GQDs-like enzyme mimics to treat various diseases/disorders associated with stress, its exact treatment mechanism and administrative dose for human applications is yet to be resolved. We strongly believe that even before switching to in vivo models, determining the dose-dependent RSA roles of such nanozymes through ex vivo studies will allow for fabricating much more cost-effective and precision-based formulations with lower cellular accumulation toxicities.
O groups served as active sites.32 Nirala et al. reported an electrochemical preparation of GQDs having natural peroxidase activity that were further used as natural colorimetric probes to detect trace levels of glucose and H2O2.33 Amongst the different bio-properties, GQDs with inherent radical scavenging activity (RSA) are most widely studied due to its association with different therapies to counter elevated levels of oxidative stress (OS) under a diseased state. In this regard, the explicit report of Wang et al. showed that hydrogen bond donation and radical adduct formation ability of GQDs initiated by different surface speciation of oxygenated functional groups are the main reason for the quenching of free radicals.34 Nilewski et al. showed that polyethylene glycol-functionalized GQDs can rescue murine endothelioma cells from severe H2O2 stress through SOD-linked activity.35 Krunić et al. explored the anti-oxidative property of GQDs against oxidative and nitrative damages in SH-SY5Y neuroblastoma cells generated through sodium nitroprusside, with GQDs acting as non-selective antioxidants and iron chelators, with no enzymatic activity.36 More interestingly, Wang et al. showed that upon halogen doping, GQDs shows a switch between its anti- and pro-oxidant behavior with good antibacterial effect mediated through generation (1O2).37 Although multiple attempts are made to use GQDs-like enzyme mimics to treat various diseases/disorders associated with stress, its exact treatment mechanism and administrative dose for human applications is yet to be resolved. We strongly believe that even before switching to in vivo models, determining the dose-dependent RSA roles of such nanozymes through ex vivo studies will allow for fabricating much more cost-effective and precision-based formulations with lower cellular accumulation toxicities.
Current neuro-epidemiology suggests that 61% of the global mortality rate increase is due to different neurological disorders originating from the imbalance of molecular oxygen.38,39 Perhaps almost all neurological disorders are activated by in situ elevated levels of reactive radical species that cause oxidative harm to both growing and mature neural cells, leading to their progression.40,41 Ascertained by its harmful health effects, notable interest in nano-based antioxidants has shown prolific results in down-regulating the spread of tissue, protein, and DNA damages when supplemented.42,43 Generally, inherent detoxification enzymes like superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPx) act in tandem to counter these superoxide radical anions and hydrogen peroxides (O2˙− and H2O2) to protect O2 and H2O in biological systems.44 However, irreversible enzymatic damage resulting from a prolonged disorder/disease state requires enzymatic supplements for compensating such a loss. Till now, therapeutic replacements for natural enzymes have been quite challenging due to their high instability, low immunogenicity, and low cellular diffusibility, alongwith very low circulation half-lives. Therefore, nanozymes from non-biological sources like QDs, GQDs, and inorganic materials are considered an alternative due to their lower production cost and high scalability, besides being highly active and stable in harsh environments wherein biological enzymes become entirely inactive.
This is the first report wherein a simple fabrication strategy is reported for the large-scale production of YGQDs by CW remediation interceded through the generation of in situ anion radicals directly in water. The presence of different oxygenated functional groups (like COOH, OH, and C–O–C) as surface and edge defects in YGQDs showed the stabilization of free radicals, both in ex vivo and in vitro, in a dose-dependent order. More importantly, the RSA in YGQDs was associated with its high specific SOD activity. Having demonstrated its excellent stability, the binding affinity towards O2˙− (Kd ∼586 nM) and high biocompatibility, the YGQDs were envisioned for mitigating ROS under ex vivo conditions. Optimal results showed amongst the different cell lines studied; YGQDs showed the highest survivability/reversal of C6-cells (∼90%) from severe ROS. Furthermore, the complete normalization of different pro- and anti-inflammatory cytokines levels explains the plausible anti-apoptotic and cell survivability mechanism of YGQDs in C6-cells alongwith its time-dependent cytosolic and nuclear localization. A brief overview of the entire work is illustrated in Scheme 1. The present results emphasize an easy and cost-effective fabrication of YGQDs by remediating CW intended to countering O2˙−-driven neuronal stress.
Results
Preparation and characterization of GO and YGQDs
We first began with the synthesis of GO from a large molecular fragment of CW (isolation process mentioned in Fig. S1, ESI†), based on a safer methodology of improved Hummers method,45 using a mixture of concentrated orthophosphoric acid and sulfuric acid (VH3PO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) VH2SO4 = 4
VH2SO4 = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 v/v) and potassium permanganate (KMnO4 ∼6 eq.) as the oxidant. The presence of disorders and small crystallizable domains in CW allowed for faster exfoliation and the formation of surface and edge defects. The fabrication of YGQDs was done using a simplified method of in situ generation of anion radical (NaO2˙−)-assisted bond cleavage in water.46 The present method exempts the use of commercial radicals that suffer instability and requires special handling; hence, the in situ generation of radicals was preferred for synthesis.47 The effective concentration of H2O2 and NaOH was determined to be 3
6 v/v) and potassium permanganate (KMnO4 ∼6 eq.) as the oxidant. The presence of disorders and small crystallizable domains in CW allowed for faster exfoliation and the formation of surface and edge defects. The fabrication of YGQDs was done using a simplified method of in situ generation of anion radical (NaO2˙−)-assisted bond cleavage in water.46 The present method exempts the use of commercial radicals that suffer instability and requires special handling; hence, the in situ generation of radicals was preferred for synthesis.47 The effective concentration of H2O2 and NaOH was determined to be 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (mmol) for converting 100 mg GO efficiently to YGQDs, with practical yields reaching ∼87.4%. Further, the purification of the formed YGQDs was done using a dialysis bag of pore size 3000 Da for 12 h, wherein we noticed that all the YGQDs remained inside the dialysis bag, while the same upon dialysis through higher pore size bags ∼5000 Da bag were completely lost. This indicates the possibility of YGQDs being in the size range of 3000–5000 Da. Furthermore, CHO elemental analysis supported with TOF-mass analysis was done to estimate the M.W. of YGQDs accurately. The details of the results of TOF-mass are shown in Fig. S2.† The elemental percentage of purified YGQDs shows 68.89% C, 4.82% H, and 24.87% O. The positive mode TOF-mass taken in the range 3–5 kDa shows molecular components at m/z = 2999.858, 3157.9063, 3576.7517, and 3708.513. Among the four, the highest molecular component is 3157.9063 [M + Na]+ with calc. mass m/z = 3157.9177 and an empirical formula of C184H157O48Na+. Based on the findings, we speculate the appropriate mass of YGQDs to be 3157.94 Da. Besides, the determined structure of YGQDs also shows higher surface oxygenated surface groups and lower sp2 centers. Next, we optimized the reaction condition by increasing the radical concentration, where we noticed that beyond the optimum range, trace levels of black precipitate were observed. The temperature optimization suggests that YGQDs formation occurs more efficiently at 121 °C, beyond which the formation of a black precipitate with diminished yellow and green fluorescence with practical yields > 50–60% are seen. This indicates that the degradation of the GO matrix occurs at high temperatures and due to unreacted radical induction. The results allowed us to select YGQDs for further study. The optimized conditions are summarized in Table S1.†
2 (mmol) for converting 100 mg GO efficiently to YGQDs, with practical yields reaching ∼87.4%. Further, the purification of the formed YGQDs was done using a dialysis bag of pore size 3000 Da for 12 h, wherein we noticed that all the YGQDs remained inside the dialysis bag, while the same upon dialysis through higher pore size bags ∼5000 Da bag were completely lost. This indicates the possibility of YGQDs being in the size range of 3000–5000 Da. Furthermore, CHO elemental analysis supported with TOF-mass analysis was done to estimate the M.W. of YGQDs accurately. The details of the results of TOF-mass are shown in Fig. S2.† The elemental percentage of purified YGQDs shows 68.89% C, 4.82% H, and 24.87% O. The positive mode TOF-mass taken in the range 3–5 kDa shows molecular components at m/z = 2999.858, 3157.9063, 3576.7517, and 3708.513. Among the four, the highest molecular component is 3157.9063 [M + Na]+ with calc. mass m/z = 3157.9177 and an empirical formula of C184H157O48Na+. Based on the findings, we speculate the appropriate mass of YGQDs to be 3157.94 Da. Besides, the determined structure of YGQDs also shows higher surface oxygenated surface groups and lower sp2 centers. Next, we optimized the reaction condition by increasing the radical concentration, where we noticed that beyond the optimum range, trace levels of black precipitate were observed. The temperature optimization suggests that YGQDs formation occurs more efficiently at 121 °C, beyond which the formation of a black precipitate with diminished yellow and green fluorescence with practical yields > 50–60% are seen. This indicates that the degradation of the GO matrix occurs at high temperatures and due to unreacted radical induction. The results allowed us to select YGQDs for further study. The optimized conditions are summarized in Table S1.†
The FTIR spectrum, Fig. 1A, of GO shows absorption bands at 3335 cm−1 for stretching vibrations of O–H groups; the prominent bands appearing at ∼1740 cm−1 and ∼1628 cm−1 are ascribed to the stretching of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups, respectively. The peaks at ∼1223 cm−1 and ∼1034 cm−1 are due to –C–OH and –C–O–C stretching and bending vibrations, respectively.45 For YGQDs, the peaks at ∼3335 cm−1 and ∼1628 cm−1 represent the stretching vibrational patterns of OH and C
C groups, respectively. The peaks at ∼1223 cm−1 and ∼1034 cm−1 are due to –C–OH and –C–O–C stretching and bending vibrations, respectively.45 For YGQDs, the peaks at ∼3335 cm−1 and ∼1628 cm−1 represent the stretching vibrational patterns of OH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, respectively. The sharp peaks at 1416 cm−1 indicate the in-plane vibrations of the carbon skeleton, which is common to graphitic materials. Finally, the low intense peaks at 1034 cm−1 are accounted for –C–OH stretching vibrations. The comparative spectral changes for both GO and YGQDs are shown in Fig. 1A. The X-ray photoelectron spectroscopy (XPS) patterns show the distribution of surface components in YGQDs with two signatures regions, C 1s and O 1s appearing at ∼282 eV and ∼584 eV, respectively (Fig. 1B). The deconvolution of O 1s spectrum shows three prominent peaks at 530.9 eV and 532 eV, representing the existence of C
O groups, respectively. The sharp peaks at 1416 cm−1 indicate the in-plane vibrations of the carbon skeleton, which is common to graphitic materials. Finally, the low intense peaks at 1034 cm−1 are accounted for –C–OH stretching vibrations. The comparative spectral changes for both GO and YGQDs are shown in Fig. 1A. The X-ray photoelectron spectroscopy (XPS) patterns show the distribution of surface components in YGQDs with two signatures regions, C 1s and O 1s appearing at ∼282 eV and ∼584 eV, respectively (Fig. 1B). The deconvolution of O 1s spectrum shows three prominent peaks at 530.9 eV and 532 eV, representing the existence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–OH groups, respectively. The low-order peaks at 535.2 eV represent C–O–C groups. The deconstruction of C 1s shows three major areas for C
O and C–OH groups, respectively. The low-order peaks at 535.2 eV represent C–O–C groups. The deconstruction of C 1s shows three major areas for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (sp2), C–O (sp3), and C
C (sp2), C–O (sp3), and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, shown at 284.5 eV, 286.5 eV, and ∼287.7 eV, respectively. The results of the deconvoluted regions of C 1s and O 1s shown in Fig. 1C and D were found to be as per earlier published reports.48,49 The Raman spectra were used to illustrate the nature of defects. In GO, the peaks shown at 1329 cm−1 are assigned to the D bands, while the latter peak at 1585 cm−1 symbolizes G bands, a characteristic of graphene derivatives. Moreover, the minimal rise in the G band intensity compared to the D band represents greater sp2 links in GO; the calculated ID/IG ∼0.85 suggests a larger sp2 domain and the presence of a multilayer.50,51 In contrast, YGQDs showed highly intense D bands and low intense G bands appearing at ∼1340 cm−1 and ∼1552 cm−1 due to the changes in the oxidation levels. Moreover, the calc. ID/IG value of 0.91 suggests the high probability of defects and partial distortions in YGQDs.52 The details of the Raman spectrum of GO and YGQDs are shown in Fig. 1D. Finally, the p-XRD patterns of GO showed peaks centered at (2θ ∼10.18°, d = 0.8 nm), attributed to the (002) facet of GO.53 In contrast, YGQDs peaks appeared at (23.8°, d = 0.316), attributed to the (002) graphitic facets, indicating the restoration of hindered networks (Fig. 1F). We also observed a typical larger d spacing corresponding to the existence of more surface oxygenated groups. Next, using the full width at half maximum (FWHM) of YGQDs diffraction peak and the Debye–Scherrer formula, the average crystalline size of YGQDs was estimated to be ∼10 nm.54,55
O groups, shown at 284.5 eV, 286.5 eV, and ∼287.7 eV, respectively. The results of the deconvoluted regions of C 1s and O 1s shown in Fig. 1C and D were found to be as per earlier published reports.48,49 The Raman spectra were used to illustrate the nature of defects. In GO, the peaks shown at 1329 cm−1 are assigned to the D bands, while the latter peak at 1585 cm−1 symbolizes G bands, a characteristic of graphene derivatives. Moreover, the minimal rise in the G band intensity compared to the D band represents greater sp2 links in GO; the calculated ID/IG ∼0.85 suggests a larger sp2 domain and the presence of a multilayer.50,51 In contrast, YGQDs showed highly intense D bands and low intense G bands appearing at ∼1340 cm−1 and ∼1552 cm−1 due to the changes in the oxidation levels. Moreover, the calc. ID/IG value of 0.91 suggests the high probability of defects and partial distortions in YGQDs.52 The details of the Raman spectrum of GO and YGQDs are shown in Fig. 1D. Finally, the p-XRD patterns of GO showed peaks centered at (2θ ∼10.18°, d = 0.8 nm), attributed to the (002) facet of GO.53 In contrast, YGQDs peaks appeared at (23.8°, d = 0.316), attributed to the (002) graphitic facets, indicating the restoration of hindered networks (Fig. 1F). We also observed a typical larger d spacing corresponding to the existence of more surface oxygenated groups. Next, using the full width at half maximum (FWHM) of YGQDs diffraction peak and the Debye–Scherrer formula, the average crystalline size of YGQDs was estimated to be ∼10 nm.54,55
The crystallinity and surface morphology of YGQDs observed through HR-TEM show the presence of a uniform distribution of circular dots at 100 nm (Fig. 2A), while the larger view of the dots can be seen at 50 nm magnified scale (Fig. 2B). Fig. 2C and D shows the presence of uniform circular shaped dots at 10 nm scale with the average size of YGQDs in the range of 7 ± 1.5 nm. Further, the magnified view of the dots in Fig. 2E shows a prominent lattice fringe with a d value of ∼0.314 nm. The small deviation of the d value ∼0.036 nm from ideal graphite is attributed to the abundance of oxygenated groups. Finally, the SAED taken under perpendicular electron beam projection showed a crystalline pattern in the basal plane of YGQDs (Fig. 2F). The EDS map taken for YGQDs shows a higher oxygen content of 11.52% and a lower carbon content of 88.74% when contrasted with that of GO, as shown in Fig. S3.†
 | ||
| Fig. 2 Surface morphology of YGQDs captured by HR-TEM at (A) 100 nm, (B) 50 nm, (C) 10 nm, (D) 10 nm scale, (E) zoomed YGQDs and (F) SAED patterns of YGQDs. | ||
Photophysical properties of YGQDs
Upon testing for solubility in polar and non-polar solvents, the YGQDs were found to be only soluble in water while they remained insoluble in other solvents (Fig. S4†). Such altered dispersibility in YGQDs arises from its intrinsic confinement effect and edge states that show altered interactions with solvent molecules. Furthermore, the YGQDs demonstrated stability with no aggregation observed after 6 months of long-term storage at room temperature. The UV-vis spectra of YGQDs recorded in Milli-Q water show two distinct absorption bands at ∼235 nm and ∼274 nm, representing the π–π* transition arising from sp2 links and n–π* transitions arising from defects, respectively (Fig. 3A). The UV-vis spectra of synthesized GO from CW and its related explanation are shown in Fig. S5, ESI.† Next, pH-triggered fluorescence studies were performed to study the effect of pH on the photo-controlled stability of YGQDs. The results show shifting in the UV-vis spectra at different pH (Fig. S6A, ESI†). We also noticed significant shifting in the photo-luminescence (PL) spectra upon moving from pH 3 to 10 along with the peak broadening effect beyond pH 8 due to the edge overlap and large domain size in YGQDs. Moreover, the protonation and de-protonation effect of the functional groups at different pH values shows a typical change in the rate of transition and the inherent property of YGQDs that results in the shifting of emission maxima to higher wavelengths, i.e., giving rise to an excitation-independent phenomenon.56 The results of the PL shifts occurring at different pH values are shown in Fig. S6B–F, ESI.† In addition, we noticed an acute emission peak with εmax ∼530 nm at pH 7, which arises due to radiative transitions, while the same under an acidic environment is nearly reduced to half. The formation of new peaks beyond 530 nm indicates plausible non-radiative intra-band shifts in YGQDs.Next, time-resolved fluorescence (TRF) measurements were carried out to quantify the role of carriers in maintaining photo-dynamics. The results are summarized in Table S2.† The tri-exponential curve fitting along with the fluorescence decay shows that relaxation occurs, viz., three different pathways (eqn (i)57), with B1, B2, and B3, indicating the amplitudes of a lifetime. τ1 signifies the intrinsic aromatic sp2 domains, and τ2 and τ3 indicate the lifetime due to the irrelevant surface groups. The higher value for τ2 and B2 indicates emission to be controlled by edge/basal states contributing toward a longer lifetime. The lifetime decay curve of YGQDs is shown in Fig. 3B. The calculated average lifetime for YGQDs was found to be 0.7 ± 0.26 ns and 1.17 ± 0.29 ns for (εmax ∼440 nm and εmax ∼530 nm, respectively) with χ2 deviations of 1.06 and 1.11, respectively. The ns lifetime in YGQDs indicates the singlet nature of emission, with the luminescence dominated by surface defects.
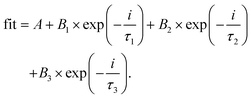 | (i) |
Radical scavenging activity (RSA) and biocompatibility studies of YGQDs
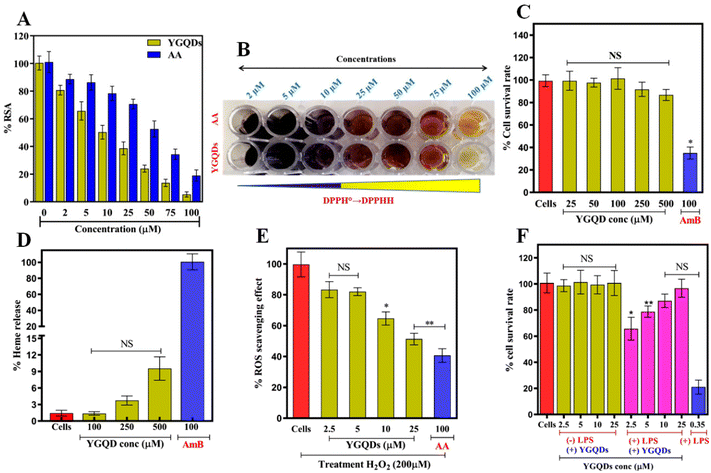
We have seen earlier that nanozymes’ catalytic performance primarily depends on their structural properties like size, morphology, functional groups, and presence of defects.58–60 Unlike other classes of inorganic nanoparticles and carbon quantum dots (CDs), GQDs offer high C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C conjugation and charge transfer mobility with additional surface-oriented structural defects that allow them to act as hydrogen bond donors and radical stabilizers to quench free radicals.61 Hence, to investigate the RSA and its associated enzymatic roles, we first analyzed the antioxidant properties of YGQDs in vitro using the DPPH radical assay. Under optimal conditions, DPPH represents a stable free radical (DPPH°) that shows a characteristic λmax at 517 nm. The decrease in the absorbance of DPPH° due to the formation of stable DPPHH estimates the RSA. The RSA of YGQDs (0–100 μM) was measured and compared to standard AA. We noticed a prominent RSA in YGQDs in a dose-dependent order and with higher efficiency than AA (Fig. 4A). The calculated EC50 was found to be ∼12 μM for YGQDs and ∼55 μM for AA, i.e., a net 4.5-fold better activity for YGQDs. The colorimetric changes for RSA estimation by the DPPH assay are shown in Fig. 4B. The high RSA activity of YGQDs accounted for more hydrogen donor and sp2 center participation toward radial stabilization by forming a stable crystal plane. These preliminary results of the DPPH assay further encouraged us to evaluate the anti-oxidative role of these YGQDs in different cellular models under the influence of OS.
C conjugation and charge transfer mobility with additional surface-oriented structural defects that allow them to act as hydrogen bond donors and radical stabilizers to quench free radicals.61 Hence, to investigate the RSA and its associated enzymatic roles, we first analyzed the antioxidant properties of YGQDs in vitro using the DPPH radical assay. Under optimal conditions, DPPH represents a stable free radical (DPPH°) that shows a characteristic λmax at 517 nm. The decrease in the absorbance of DPPH° due to the formation of stable DPPHH estimates the RSA. The RSA of YGQDs (0–100 μM) was measured and compared to standard AA. We noticed a prominent RSA in YGQDs in a dose-dependent order and with higher efficiency than AA (Fig. 4A). The calculated EC50 was found to be ∼12 μM for YGQDs and ∼55 μM for AA, i.e., a net 4.5-fold better activity for YGQDs. The colorimetric changes for RSA estimation by the DPPH assay are shown in Fig. 4B. The high RSA activity of YGQDs accounted for more hydrogen donor and sp2 center participation toward radial stabilization by forming a stable crystal plane. These preliminary results of the DPPH assay further encouraged us to evaluate the anti-oxidative role of these YGQDs in different cellular models under the influence of OS.
The biocompatibility of YGQDs was next evaluated before their antioxidative examination in ex vivo models to determine its safety. The cytotoxicity measurements of YGQDs were performed using PBMCs. The results of the cytotoxicity studies are shown in Fig. 4C. We noticed almost insignificant toxicity at high treatment doses of YGQDs, i.e., with 500 μM treatment, showing <15% PBMC killing. Contrastingly, AmB taken as a positive drug standard showed ∼65% killing at only 100 μM. In addition, we also noticed very minimal hemolysis in YGQDs, with only 12% hemolysis (P < 0.001) at 500 μM (Fig. 4D). While AmB, which was used as a positive control, showed 100% hemolysis at only 100 μM, indicating high hemolytic activity and toxicity than YGQDs.62 The pictures of the hemolytic effect of YGQDs and AmB at different concentrations with human blood samples are shown in Fig. S7.† The low cytotoxicity and hemolysis effect revealed that the biocompatibility range of YGQDs was very high. Furthermore, considering its safety, the tested doses of YGQDs in ex vivo assays were taken 4-fold less than the determined toxicity doses.
Ex vivo antioxidant role of YGQDs
Encouraged by the excellent biocompatibility of YGQDs, the step-wise assessment of cell survival ability and RSA of YGQDs in the ex vivo setup were studied as per the earlier established procedure.63 We selected three different cell lines based on their susceptibility toward reactive radical species. The related survivability and RSA were measured using the MTT assay. Primarily, all three cell lines, namely, C6-cells (neural cells), HepG2 (human liver cancer cell line), and Caco-2 (human colorectal adenocarcinoma cells), were inducted with 200 μM H2O2 to induce severe ROS. Next, the ability of YGQDs to rescue cells from ROS was monitored in both the presence and absence of YGQDs at different concentrations (2.5–25 μM). Interestingly, we noticed that the cell survival decreased below 10% upon H2O2 induction, whereas the same upon YGQDs treatment showed a systemic increase in survival. The survival rates were estimated as ∼90%, ∼80%, and ∼75% for C6-cells, HepG2 cells, and Caco-2 cells, respectively, at only 25 μM treated dose (Fig. S8A–C†). Interestingly, amongst all the tested cells, we noticed that the ability of YGQDs to alleviate ROS in C6-cells was more prominent than AA with EC50 ∼25 μM YGQDs and beyond 100 μM for AA (Fig. 4E). The results imply that the YGQDs, irrespective of the nature of cells, can be well internalized and can protect the C6-cells majorly from severe OS-mediated cell death. Based on this high RSA response of YGQDs toward C6-cells, next, we used lipopolysaccharide (LPS)-induced neuro-inflammation and ROS against C6-cells as a model system to evaluate the efficacy of YGQDs. We noticed that the YGQDs were able to scavage intracellularly generated ROS in a dose-dependent manner, measured using H2DCFDA-based fluorescence in LPS-treated C6-cells (Fig. 4F). The results further confirmed the extent of cell survival is an intrinsic function of YGQDs. In the presence of 0.35 μM LPS, cell survival decreased to ∼21% from 100%. However, the same with YGQDs pre-treatment showed increased survival of ∼65%, ∼78%, ∼87%, and 96% for concentrations of 2.5, 5, 10, and 25 μM, respectively. The color change in the MTT plates showing C6-cell survival upon the incubation of YGQDs is illustrated in Fig. S9, ESI.†Nanozyme activity in YGQDs
Following in vitro and ex vivo antioxidative property determination in YGQDs, its associated enzymatic activity was investigated for plausible SOD, GPx, and CAT-like activity due to their active role in balancing the cellular integrity by countering free radical-induced damage. Amongst the various peroxidases seen so far, GPx abundance is very high in mammalian systems due to its catalytic preferences toward oxidized lipid peroxides. Yet another class of enzyme in the biological system that is even more important is SOD due to its catalytic role in converting the primary generated O2˙− to less toxic H2O2 within the cellular system. Further, the H2O2 produced in the later steps is degraded to H2O (by GPx) and H2O + O2 (by CAT). So far, much attention is paid to carbon-based nanozymes with either explicit or implicit natural or induced peroxidase and/or CAT-like acitivity.64 However, progress using GQDs for the same purpose is still in its infancy. Although it can be presumed that the carbon quantum dots (CDs) and GQDs share similar surface architecture and functional group composition, the latter can behave more like an enzyme active site. The circular structure observed through HR-TEM shows a dispersed, narrower size distribution of YGQDs with avg. size ∼7.5 nm. The small size of the nanocrystallites increases the specific surface area and the presence of more functional groups on the crystal surface. More oxygenated functional groups in the peripheral location show more trapping of O2˙−.65,66 Furthermore, theoretically predicted models suggest that the presence of functional groups like COOH and OH can act as the substrate-binding sites.25 Using the above interpretation, we next tested the GPx and CAT activities of YGQDs using 3,3′,5,5′-tetramethylbenzidine (TMB) as a chromogenic probe and H2O2 as a radical initiator. The results of the GPx and CAT effect of YGQDs at different concentrations and its comparison to natural Horseradish peroxidases (HRP) and CAT enzymes are shown in Fig. S10A and B, ESI.† The results show very minimal GPx activity when tested at different concentration of YGQDs. However, HRP(1U), taken as a positive control, showed significant changes in the GPx activity. Similarly, the extent of H2O2 degradation, measured by calculating the reduced absorbance at 240 nm, showed trace CAT activity in YGQDs when compared to natural CAT. With no associated GPx or CAT-like activity, the YGQDs were next screened for plausible SOD-like activity. The results of the SOD activity of YGQDs are shown in Fig. 5A. Surprisingly, we found robust SOD-like activity in YGQDs wherein 15 μM YGQDs equaled the activity of 1U bSOD enzyme. Such high specific SOD-like activity can be related to the generation of more sp3 and lesser sp2 regions in the carbon skeleton resulting from the oxidative conversion of GO to YGQDs. Additionally, the increased specific surface area of the circular dots (earlier discussed) due to fragmentation allows them to act as catalytic sites to mediate the enzymatic reaction. These structural defects initiate the formation of hydrogen bonds and static interactions to counter the O2˙− while the lesser sp2 crystal plane operates through П-bonded interactions to stabilize these radicals.The kinetic studies of SOD activity of YGQDs
We performed SOD enzyme assay using pyrogallol as the substrate and determined the kinetic parameters like Vmax and Km through Michalis–Menten (M–M) kinetics. The decreased absorbance of pyrogallol (at 325 nm) due to its auto-oxidation in the presence of the generated O2˙− radical is a well-established model to study the SOD-like activity; hence, it was used. We noticed the SOD activity of YGQDs to be highly selective and the highest determined so far amongst all other graphene-based nanozymes.28 The findings encouraged us further to calculate the kinetic parameters [Michaelis constant (Km) and maximum reaction velocity, (Vmax)] using Michaelis–Menten (M–M) and Lineweaver Burk (LB) equations for both bSOD and YGQDs tested at different concentrations. The results indicate ∼10-fold higher Km but with a 10-fold lower catalytic turnover (Kcat) for YGQDs than bSOD (Fig. 5B, C and Table 1). While having a lower Kcat value for YGQDs than bSOD, almost comparable Vmax can be obtained using a higher amount of YGQDs in the assay. Moreover, upon testing for the enzymatic performance in denaturants [urea (1–7 M)], we noticed the complete retention of SOD activity for YGQDs at urea doses as high as 7 M, while bSOD's activity drastically decreased below 5% under the same tested condition. The details of the effect of denaturants on the dismutase activity of YGQDs are shown in (Fig. 5D). We also studied the effect of temperature on the enzymatic performance of YGQDs. We noticed almost 99.8% and 100% SOD activity for YGQDs and bSOD, respectively, at 24 °C. Interestingly, at higher temperatures (beyond 45 °C and 70 °C), we noticed only 42.8% and 1.5% of SOD activity for bSOD whereas almost∼ 85% and ∼93% of activity retention was shown by YGQDs. This can be reasoned from the high thermal and colloidal stability of YGQDs arising from the inertness of the carbon skeleton. The results indicate that YGQDs as nanozymes are stable and can maintain their activity under harsh conditions. Additionally, the antioxidant and SOD activities associated with it suggest that possibly the highly oxidized groups in YGQDs make carbon domains electro-deficient, allowing them to act as shuttles to counter O2˙− alongwith its circular structure that helps them entrap more radicals.67 Perhaps it is seen that in the presence of denaturing agents and at temperatures beyond 30 °C, an irreversible reduction of the biological activity of enzymes happens due to the loss of its 3D protein folding. However, such limitations do not apply to YGQDs.| Kinetic parameters obtained from M–M kinetics | ||||
|---|---|---|---|---|
| V max (μmol S−1) | K m (μM) | K cat (S−1) | K cat/Km (S−1) | |
| bSOD | 1943 | 40 | 777 | 1.9 × 107 |
| YGQDs | 1358 | 420 | 77.6 | 1.8 × 105 |
Gene expression studies after YGQDs treatment
Following nanozyme activity measurement, SOD, CAT, and GPx gene expression profiles were measured from the isolated RNA and cDNA from bSOD- and YGQDs-treated C6-cells.44 We noticed that the treatment of C6-cells with H2O2 resulted in a reduction of the expression levels of genes for the enzymes SOD, CAT, and GPx. After pre-incubation with YGQDs, we observed a dose-dependent recovery in the expression of these genes. Here, the change in the expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenases (GAPDH), whose expression remains unaltered, is used as a control. Almost ∼3-fold and ∼2.1-fold recovery of the SOD gene expression was obtained after pre-incubation with 5 μM and 2.5 μM YGQDs, respectively (Fig. 5F). The CAT and GPx gene recovery showed a net ∼1.5-fold change with 5 μM YGQD, as shown in Fig. S11A and B.† Therefore, the gene expression studies correlate well with the SOD activity of YGQDs since a more significant change in gene expression is achieved for bSOD, indicating that the target of YGQDs is SOD and not CAT or GPX.Binding affinity of YGQDs and bSOD toward O2˙−
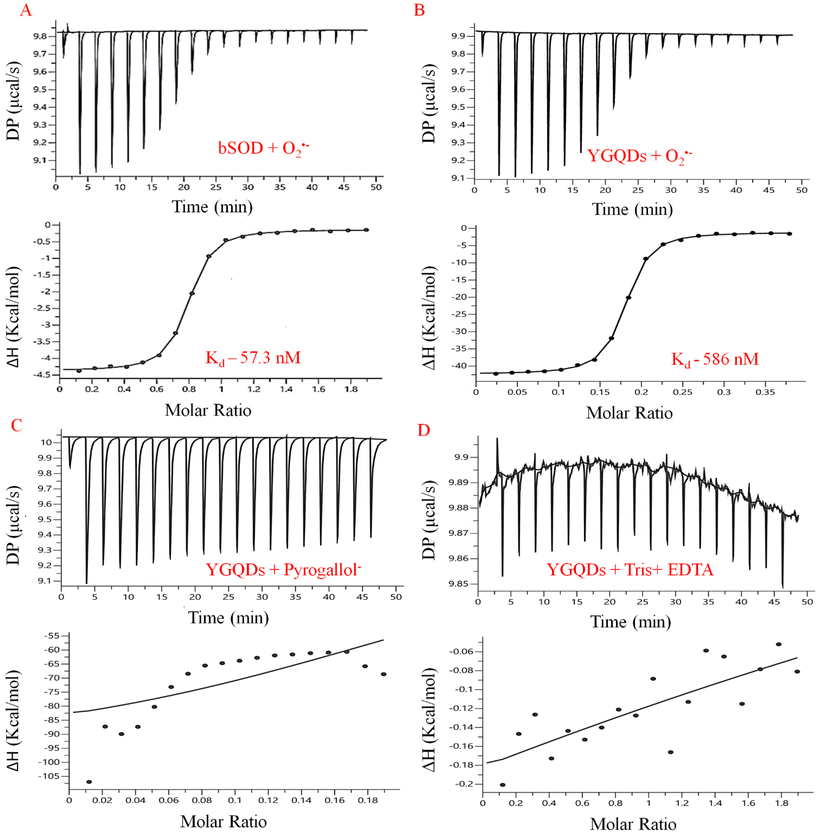
To confirm the high specific binding of O2˙− onto the surface of YGQDs, Isothermal Calorimetry (ITC)-based measurements were done. The thermodynamically-derived parameters are outlined for both YGQDs and bSOD. The details of the ITC-method are illustrated in the ESI.† We took pyrogallol as our model substrate due to its ability to generate O2˙− under milder conditions, high water-solubility, and catalyst-free autoxidation under an alkaline buffer (pH ∼8, Tris-EDTA). The shift of the UV-vis spectrum due to auto-oxidation shows a synchronous increase in the peak at 320 nm, resembling the conversion of pyrogallol to semi-quinone radical and O2˙−. The binding affinity of the formed O2˙− with YGQDs and bSOD were compared. So far, as such, there are no reports wherein direct ITC-based measurements have been employed to estimate the binding kinetics associated with O2˙−quenching using the bSOD enzyme even though its crystal structure has indicated specific interacting residues of bSOD responsible for the binding of O2˙−. The results depict a strong negative enthalpy change (ΔH) determined from the amplitude, indicating an exothermic process. The obtained Kd values from the plot of (ΔH vs. Molar ratio) show 586 nM for YGQDs and 57.3 nM for bSOD (Fig. 6A and B). Even though the YGQDs exhibit slightly lower Kd than bSOD for O2˙−, the binding isotherms follow a similar thermodynamic pattern, representing a highly favorable exothermic reaction. Moreover, having almost similar negative Gibbs free energy (ΔG) for YGQDs and bSOD, it can be considered that the binding of O2˙− to YGQDs or bSOD is spontaneous and endergonic. The determined parameters from the ITC analysis are shown in Table 2. More importantly, we noticed no binding or saturation isotherm when there was no generation of O2˙− (like in alkaline buffer, i.e., Tris + EDTA or in the presence of pyrogallol alone). These results suggest that the binding isotherm is a function of generated O2˙− only (Fig. 6C and D). Besides, the high ΔH and ΔS values calculated for YGQDs indicate a multidomain capture of O2˙− on the YGQDs surface.Anti-apoptotic activity of YGQDs against H2O2-induced C6-cell apoptosis
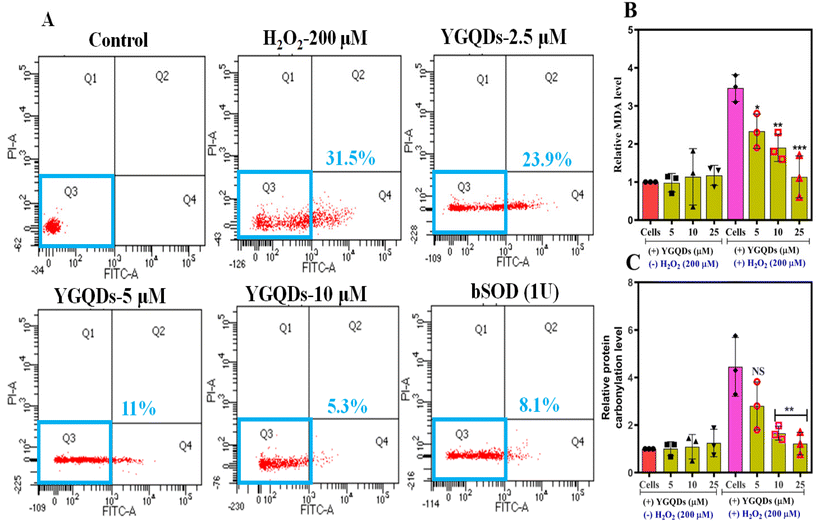
To access the anti-apoptotic roles of YGQDs, H2O2-induced cell apoptosis and death were estimated both in the presence and absence of YGQDs using flow cytometry under ex vivo conditions. The results of the flow cytometry are illustrated in Fig. 7A. As expected, the YGQDs were able to inhibit C6-cell apoptosis in a dose-dependent fashion. Thus, upon treatment with 200 μM H2O2 for 6 h, more than ∼31% cell apoptosis was noticed, determined by the increase in Annexin V-FITC-stained C6-cells. Upon treatment with YGQDs, the same cells showed a gradual decrease in apoptotic cells of ∼23.9%, ∼11%, and 5.3% for the tested concentrations of 2.5, 5, and 10 μM YGQDs, respectively. Furthermore, with bSOD as a positive control, the apoptotic reduction was only ∼8.1%. Moreover, the study did not reveal any C6-cell necrosis with YGQDs treatment, measured from the number of propidium-iodide (PI) stained cells. Together, the results indicate that OS-mediated apoptosis caused by H2O2 can be well suppressed upon treatment with YGQDs and its strong RSA and cytoprotective activity.Next, the lipid peroxidation (LPO) products generated through membrane lipid oxidation and protein carbonylation (PCO) constitute the main markers for measuring OS-mediated cell damage. We presumed that the strong anti-oxidative behavior of YGQDs can also decrease the elevated levels of LPO and PCO under the influence of OS. The assay was conducted considering the conversion of generated LPO to malondialdehyde (MDA), which when reacted with thiobarbituric acid (TBA), produces a colorimetric change with an increased absorbance (λmax) at 532 nm. The MDA and PCO levels estimation findings are shown in Fig. 7B and C. The results show a net ∼3.4-fold increase in the LPO level with 200 μM H2O2 treatment for C6-cells (Fig. 7B). In the presence of 25 μM YGQDs, the LPO levels were reversed to normal. The change in the color intensity during MDA-TBA conversions was studied using ELISA and is shown in Fig. S12, ESI.† In a parallel study, we observed that the PCO was increased to ∼4.3 fold after 200 μM H2O2 treatment but was eventually reversed to normal with only a 25 μM treatment dose of YGQDs. The net reduction in LPO and PCO levels with increasing doses indicates that YGQDs can protect C6-cells from OS injury by acting as a strong radical quencher.
YGQD inhibits the production of pro-inflammatory cytokines
Often, it is seen that prolonged OS in C6-cell and other related cells results in a severe imbalance between released pro-inflammatory and anti-inflammatory cytokines, which causes cellular inflammation, followed by cell/DNA and tissue destruction.68,69 Hence, we explored the possibility of YGQDs controlling the cytokine release pattern under an ex vivo cellular setup. Based on our initial findings, YGQDs are critical in balancing OS and show the highest cell protection toward C6-cells. Next, we used ELISA to measure its controlling effect in maintaining the altered levels of various pro-inflammatory (IFN-γ, TNF-α, IL-6, and IL-1β) and anti-inflammatory (IL-10) cytokines under OS. The result of the multiple cytokines release pattern before and after the induction of YGQDs and bSODs is shown in Fig. 8A–E. We found that the expression of pro-inflammatory cytokines IFN-γ, TNF-α, IL-6, and IL-1β increased to ∼4.6, ∼3.5, ∼3.2, and ∼2.1 fold upon 200 μM treatment of H2O2. These sudden increases in the pro-inflammatory cytokines are physiologically significant determinants of neurodegenerative disease and other associated diseases. Interestingly, upon treatment with C6-cells with only lower concentration of YGQDs (i.e., 2.5 and 5 μM), the expression level of all pro-inflammatory cytokines was significantly reversed to the basal level. The results obtained for YGQDs were comparable to 1 μM bSOD (Fig. 8A–D). Inversely, we displayed the expression of the anti-inflammatory cytokine IL-10, which is highly decreased with H2O2-treatment (by ∼3 fold), were elevated back to its basal level in the presence of YGQDs (Fig. 8E). Considered together, these data of expression levels of different cytokines during cellular OS suggest that the imbalance of pro- and anti-inflammatory cytokines, which are created due to the OS in ex vivo models, are restored to normal with only 5 μM treatment of YGQDs. Interestingly, earlier, during in vitro SOD activity assessment, we noticed that 15 μM treatment dose of YGQDs equaled the activity of 1U bSOD. However, during the ex vivo studies using ELISA, we observed that only 5 μM YGQDs was equivalent to 1 μM bSOD, indicating its better activity upon cellular intake. Earlier, Guo et al. showed a similar study using MoSe4 NPs intended for colitis treatment, but the activity observed was much lower than our YGQDs.70 These findings forecast that YGQD has the potential to be implemented as a therapeutic candidate inside the cell for the treatment of neurological disorders and possibly other diseases that originate due to OS.Cellular localization and cytoprotective activity of YGQDs
The cellular uptake of YGQDs inside C6-cells and its related effect upon internalization were determined using confocal fluorescence microscopy. The results of the time-dependent cellular uptake of YGQDs are shown in Fig. 8F. The YGQDs, due to their intrinsic excellent yellow fluorescence, were used directly without any secondary labels. We noticed that at time t = 0 min, no fluorescence was observed; however, following 1 h and 3 h of treatment with C6-cells, we noticed the complete internalization of YGQDs with almost no change in the fluorescence intensity after 1 h and 3 h (Fig. 8F). The confocal measurements were done within a short exposure time to avoid cellular auto-fluorescence that can interfere with the original fluorescence of YGQDs. In addition, the untreated C6-cells taken as a control showed no fluorescence at any tested time points. This altered sub-cellular distribution of YGQDs can be reasoned from its caveolin-mediated endocytosis of YGQDs. We noticed the bright fluorescence of YGQDs within 1 h of treatment, indicating that the YGQDs were well internalized without damaging the membrane integrity. This can be correlated to its high biocompatibility. Further, the bright fluorescence in the C6-cell interiors suggests its plausible cytosolic localization. The internalization is greatly influenced by the hydrophilic and lipophilic functional groups that allow its permeation. Furthermore, the morphological changes studied using a phase contrast microscope show the resuscitation of ROS-mediated C6-cell damage in the presence of YGQDs (Fig. S13, ESI†). From the internalization studies, we further correlate that the synthesized YGQDs have shown highly biocompatible and increased cell survivability of ROS-inflated C6-cells under ex vivo conditions.Discussion
OS presented by O2˙− directly correlates with the pathophysiology of many diseases, including the progression of neurodegenerative diseases like Alzheimer's disease (AD) that are extensively triggered due to the loss of functions of natural enzymes like SOD, CAT, and GPx.71,72 For example, familial amyotrophic lateral sclerosis (FALS) occurs due to the selective loss of motor neuron activity due to Cu, Zn-SOD gene modification.73 In most cases, it is perceived that the over-expression or supplementation of recombinant SOD (rSOD) levels can prevent such ubiquitous generation of superoxides.74,75 However, the major problem with rSOD is its poor cytoplasmic and nuclear distribution due to its low operational stability, high immunogenicity, low cellular uptake, lesser circulation half-lives, and poor shelf life. So far, only a few SOD-based conjugates have been synthesized that exhibit longer circulation half-lives, high stability, and lesser immunogenicity (polyethylene-liposome conjugated SODs).76 However, using such polymer-liposome-based SOD mimics provides a high production/formulation cost and low clinical success rates. Hence, looking for a synthetic alternative with high stability and increased circulation half-life is still an unfulfilled goal. Carbon-based mimics are mainly used as adsorbents, photo-induced ROS generating agents, and co-delivery vehicles for PDT-based antimicrobial agents apart from some of their multifunctional nano-enzymatic properties, with their enzyme-specific activity still lacking behind.77–79Considering the above challenges, we selected CW as our starting point to subsidize the fabrication cost of synthesizing specific nanozymes. The presence of highly stacked aliphatic domains and different functional groups allows its quicker fragmentation into nanozyme. Moreover, recycling waste materials into meaningful healthcare alternatives is a remarkable commercialization strategy. GQDs are well established for their applications involving biosensors, drug-delivery formulations, bioimaging, etc., due to their excellent structure-related activity and high biocompatibility. However, its use for the treatment of neurodegenerative diseases is very rare.80 Herein, we have shown a simple strategy to synthesize YGQDs from CW. The obtained YGQDs were well characterized and exhibited an avg. size ∼7 nm and M.W. 3197.90 Da. The fabrication mechanism depicted that the in situ formed NaO2˙− acted like “molecular scissors” to cleave across C–O–C linkages of GO to produce circular YGQDs. Further, having shown excellent defect-driven PL, these YGQDs, when tested for in vitro screening anti-oxidative activity, showed prominent RSA that were later analyzed to be linked with high specific SOD activity. Further, upon transcribing to ex vivo conditions, the YGQDs showed high RSA for C6-cells and excellent biocompatibility. These findings allowed us to select C6-cells as our model substrate to explore the further antioxidative-associated role of YGQDs. The stability studies showed YGQDs to be highly stable over a wide range of temperatures (24–70 °C) and denaturants (e.g., 7 M urea), unlike bSOD. Moreover, the strong binding interaction of O2˙− radical for YGQDs estimated for the first time using ITC shows Kd in the nM range. Quite intriguingly, the binding affinity of YGQDs for O2˙− shows a thermodynamically favorable spontaneous process with negative ΔG. With no crystal data available to quantify the binding interaction of CDs, QDs, and GQDs with O2˙−, the only published results from Gao et al. suggest that hydroxyl and carboxyl groups, like the case of our YGQDs, are the main contributors that facilitate O2˙− binding.81 Next, in the ex vivo model, the gene expression studies of YGQDs showed ∼2.1-fold increased SOD expression levels with only 2.5 μM YGQD treatment (i.e., 6-fold lower in vitro dose) alongwith its minimal cytotoxicity toward PBMCs and human cell lines. These preliminary results encouraged us to establish YGQDs as promising candidates for certain new SOD-dependent therapeutics to treat neural diseases.82 In a parallel study, we also found that the progressive markers of neuronal diseases like PCO and LPO levels were normalized with lower treatment doses of YGQDs. Most importantly, the imbalance in the different release patterns of pro-inflammatory and anti-inflammatory cytokines also reverted to its normal levels with YGQDs treatment. Further, using combined confocal and phase contrast microscopy, we show that YGQDs, due to their hydrophobic and hydrophilic groups, were localized in the cytoplasm and nucleus and could protect C6-cells against apoptosis. The increased stability, cellular survival, anti-apoptotic effect, and restored cytokine release under OS estimate YGQDs as an ideal therapeutic nanozyme that can treat various neuronal disorders.
Conclusion
The present work sheds light on the step-wise establishment of biocompatible and SOD-specific YGQDs from their fabrication to neuronal stress suppression in C6-cells. The formed YGQDs were well characterized using different spectroscopic tools (like FTIR, UV, TOF-MS, XPS, Raman, and HR-TEM) and were intended for the suppression of O2˙−. In vitro studies proved YGQDs to be better RSA than AA, showing a dose-dependent radical quenching property. Later, the anti-oxidative role of YGQDs was reasoned for its specific SOD-like activity, determined using the enzyme activity screening. Although the YGQDs demonstrated slightly lesser kinetic control than bSOD, the YGQDs showed excellent stability over a wide range of temperatures and in the presence of strong denaturant concentrations. Furthermore, having demonstrated excellent bio-compatibility, the YGQDs, when transcribed to an ex vivo model, restored damaged the SOD activity at a much lower dose than the in vitro model. Besides its anti-oxidative roles, the YGQDs enhanced cell survivability from H2O2-induced cellular apoptosis. The YGQD's ability to further reverse the elevated PCO and MDA levels and simultaneously normalize the release pattern of various pro- and anti-inflammatory cytokines through cytoplasmic and nuclear internalization indicates its anti-apoptotic roles. Overall, the high antioxidant property and improved cellular revivability from apoptosis using YGQDs may attract future studies for treating neurodegenerative diseases, where the delivery of recombinant SOD is always a challenge. Interestingly, the pro-oxidant effect of QDs/GQDs is available from widespread synthesis. Still, the biologically important antioxidant effect with selective enzyme activity from a natural waste with high productivity is very novel and rare.Materials and methods
General procedure and instrumentation
Sulfuric acid (H2SO4, 98%) (CAS: 7664-93-9), nitric acid (HNO3, 69–72%) (CAS-7697-37-2), hydrochloric acid (HCl, 35–38% v/v) (CAS: 7647-01-0), sodium hydroxide (NaOH, 97%) (CAS: 1310-73-2), and hydrogen peroxide (H2O2, 30% v/v) (CAS: 7722-84-1) were purchased from Fischer scientific, India. Potassium permanganate (KMnO4, extra pure grade) (CAS: 7722-64-7) was purchased from FINAR Chemicals. Double distilled water (DDW) was used for all the experimentation, and Milli-Q grade water was used for spectroscopic and adsorption analysis. Tris-EDTA buffer (SigmaAldrich) GO and YGQDs synthesized were characterized using UV-vis spectroscopy (Shimadzu, UV-1780, 200–500 nm) for determining different transitions. The functional groups, stretching, and bending vibration modes were detected using FTIR spectroscopy in working ATR (Shimadzu, IRAffinity-1S, DLATGS detector, 4000 cm−1 to 500 cm−1). The crystallinity phases and lattice parameters were calculated using powder XRD (Rigaku Smart Lab X-ray diffractometer, 3 kW), Cu Kα radiation with a wavelength λ = 1.540 Å. The presence of longitudinal and transverse phonon modes was analyzed using micro-Raman spectroscopy (make-HORIBA scientific, Labman HR-UV-open, 500 cm−1 to 3000 cm−1) using 532 nm laser, 1 mW power; the spectra were recorded at 50× objective. The surface morphology, lattice spacing, and EDS composition are studied by HRTEM analysis (Thermo Scientific, Talos F200X G2, 200 kV). The emission spectra of YGQDs were recorded using a benchtop spectrofluorometer (HORIBA, Fluromax-4; 300–650 nm). Time-correlated single photon counting was measured at εmax = 458 nm and 558 nm using (DeltaFlexTCSPC Lifetime Fluorimeter, 80–100 ps). XPS analysis was done using a PHI III Scanning XPS Microprobe. CHO analysis was performed using an Elementar Analysensysteme GmbH, Germany. TOF-mass was calculated using a Waters, USA XEVO G2-XS QTOF.Synthesis of GO and YGQDs from CW
The detailed procedure for synthesizing GO from CW is mentioned in the ESI.† For YGQDs synthesis, 0.1 g GO powder was added to a pre-stirred aqueous solution of H2O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH (3
NaOH (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mmol) to produce a final concentration of 1 mg L−1 at room temperature (RT). The contents were sonicated for 30 min and autoclaved at 121 °C in a polytetrafluoroethylene (PTFE)-lined bottle for 12 h. After filtering and cooling to RT, the mixture was neutralized with a 10% v/v 20 mL HCl solution. The dissolved salts were removed using a dialysis bag with a cut-off (MWCO) 3000 Da (Genaxy Pvt Ltd, Servapor dialysis membrane) for 48 h before drying at 60 °C to get pure YGQDs.
2 mmol) to produce a final concentration of 1 mg L−1 at room temperature (RT). The contents were sonicated for 30 min and autoclaved at 121 °C in a polytetrafluoroethylene (PTFE)-lined bottle for 12 h. After filtering and cooling to RT, the mixture was neutralized with a 10% v/v 20 mL HCl solution. The dissolved salts were removed using a dialysis bag with a cut-off (MWCO) 3000 Da (Genaxy Pvt Ltd, Servapor dialysis membrane) for 48 h before drying at 60 °C to get pure YGQDs.
Reactive oxygen species (ROS) scavenging activity and enzyme mimetic activity of YGQDs
The radical quenching ability of YGQDs was determined using an earlier established method reported by Brand-Williams et al.83 Briefly, 0.05 mL YGQDs solutions having concentrations in the range of 0–100 μM were mixed with 1.2 mL freshly prepared DPPH in 1 mM methanol solution and vortexed for 30 min. The same concentrations of AA (0–100 μM) were prepared similarly and taken as the positive control. Both solutions were then incubated in the dark for 30 min at RT. Using a UV-vis spectrophotometer, the absorbance was measured at 517 nm. The change in the color of the DPPH assay solution from purple to yellow with reduced absorbance at 517 nm indicates the scavenging activity by YGQDs and AA.ROS determination in C6-cells by 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) assay
C6-cells (1 × 106 cells per mL) were pre-mixed with YGQDs (2.5, 5, 10, and 25 μM) and then treated with H2O2 (200 μM) for 6 h. Untreated and H2O2-treated cells were taken as the negative and positive controls, respectively. These cells (2 × 106 cells per mL) were then incubated with H2DCFDA (20 μM) for 20 min at 37 °C. εmax was measured spectrofluorometrically at 530 nm at an excitation wavelength of 507 nm.Fluorescence-activated single-cell sorting (FACS) analysis
Annexin V-FITC (AV)/propidium iodide (PI) binding of C6-cells was studied using the AV apoptosis detection kit (BD Biosciences). Briefly, YGQDs (2.5, 5, and 10 μM) pre-mixed C6-cells (1 × 106 cells per mL) were treated with H2O2 (200 μM) for 6 h. The cells were harvested, washed, and dissolved in 1 mL PBS, followed by mixing with the sequential addition of 5 μL PI, 20 μL binding buffer, and 5 μL AV. Following 15 min of incubation without light, cells were analyzed by a BD FACS ARIA TM Fusion flow cytometer (Becton Dickinson, CA, USA).Data analysis was performed using BD FACSDiva 9.0.1 software.
Malondialdehyde (MDA) expression and protein carbonylation assay study
YGQDs (5, 10, and 25 μM) pre-treated C6-cells (4 × 106 cells per mL) were incubated with H2O2 (200 μM) for 6 h. After incubation, cells were collected and centrifuged at 7500g for 10 min at 4 °C. An enzyme-linked immunosorbent assay (ELISA) for MDA detection was performed using the MDA assay kit (Elabscience, Cat. no. EL-0060) as per the manufacturer's instructions. Due to redox imbalance, proteins, lipids, and nucleic acids are the major targets of ROS-mediated oxidation. The oxidative modification of the amino acids to carbonyl groups was derivatized with 2,4-dinitrophenylhydrazine (DNPH) (D199303 SigmaAldrich), followed by precipitation with 30% (w/v) trichloroacetic acid (TCA). Then, absorbance was measured at 360 nm after dissolving the DNPH-derivatized protein pellet in 1 mL 6 M guanidine HCl.Statistical statement
The stats were analyzed by one-way and two-way analysis of variance using GraphPad Prism (version 8.00; GraphPad Software, Inc., CA, USA). The results were measured as mean ± standard deviation of at least three independent experiments. The results were shown as approximate mean values. Differences between the group data were considered statistically significant and highly significant when p < 0.05 and p < 0.001, respectively.Note: *P < 0.05; **0.05 < P < 0.01; ***0.01 < P < 0.001.
All other methods are elaborated in the ESI.† Pretreatment of coal, Synthesis of GO from CW, HR-TEM sample preparation, UV-vis and pH study, Enzyme assay, RNA extraction, and RT-PCR, Cytotoxicity studies, H2O2 degradation, H2O2-induced damage control in Caco2 and HepG2 cells, H2O2 and LPS-induced damage control, ELISA (pro-inflammatory marker), YGQDs cellular uptake.
Abbreviations
Additional abbreviations
| EDS | Energy dispersive X-ray spectroscopy |
| SAED | Selected area electron diffraction |
| RNA | Ribonucleic acid |
| M.W | Molecular weight |
| cDNA | Complementary DNA |
| PL | Photoluminescence |
| p-XRD | Powder X-ray diffraction |
| EC50 | Effective concentration showing 50% of activity |
| DPPH & DPPHH | α,α-Diphenyl-β-picrylhydrazyl |
| RNS | Reactive nitrogen species |
| FACS | Fluorescence-activated cell sorting |
| IFN-γ | Interferon-gamma |
| TNF-α | Tumor necrosis factor-alpha |
| IL-6 & IL-β | Interleukin chemokine |
| PBMC | Peripheral blood mononuclear cells |
| LSM | Laser scanning microscopy |
| FITC | Fluorescein isothiocyanate |
| ROS | Reactive oxygen species |
| RSA | Radical scavenging activity |
| OS | Oxidative stress |
| TOF | Tandem time-of-flight |
| MTT | (3-[4,5-Dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay. |
Author contributions
TD: conceptualization, experiments, methodology, synthesis, design, data analysis, and validation. PK: methodology, validation, and data analysis. CB: manuscript review and correction, data validation, and supervision. DM: conceptualization, methodology, review, and manuscript editing. SD: supervision, manuscript review and editing, data validation, conceptualization.Conflicts of interest
All the authors declare that there is no competing financial interest involved.Acknowledgements
BRNS (54/14/15/2020-BRNS/35054). All the authors have contributed and acknowledged BRNS for providing funds for carrying out the work and the National Institute of Technology Patna and National Institute of Pharmaceutical Education and Research Hajipur for providing laboratory aid and other instrumentation facilities. All the authors thank the Central research facilities at IIT (ISM) Dhanbad and IIT Mumbai for providing advanced analysis. We also acknowledge Dr. S. K. Sahu, IIT (ISM) Dhanbad and Dr. C. Bora, Patna Science College, Patna for their suggestions. Finally, all the other anonymous reviewers are thanked for their valuable comments towards making this a successful contribution.References
- Y. Volkov, Biochem. Biophys. Res. Commun., 2015, 468, 419–427 CrossRef CAS PubMed.
- L. Lin and S. Zhang, ChemComm, 2012, 48, 10177 RSC.
- D. B. Shinde and V. K. Pillai, Chem. – Eur. J., 2012, 18, 12522–12528 CrossRef CAS PubMed.
- Y. Dong, H. Pang, S. Ren, C. Chen, Y. Chi and T. Yu, Carbon, 2013, 64, 245–251 CrossRef CAS.
- D. Torres, J. L. Pinilla, E. M. Gálvez and I. Suelves, RSC Adv., 2016, 6, 48504–48514 RSC.
- W. Wang, Z. Wang, J. Liu, Y. Peng, X. Yu, W. Wang, Z. Zhang and L. Sun, Ind. Eng. Chem. Res., 2018, 57, 9144–9150 CrossRef CAS.
- Z. Wang, J. Yu, X. Zhang, N. Li, B. Liu, Y. Li, Y. Wang, W. Wang, Y. Li, L. Zhang, S. Dissanayake, S. L. Suib and L. Sun, ACS Appl. Mater. Interfaces, 2016, 8, 1434–1439 CrossRef CAS PubMed.
- A. Abbas, L. T. Mariana and A. N. Phan, Carbon, 2018, 140, 77–99 CrossRef CAS.
- M. He, X. Guo, J. Huang, H. Shen, Q. Zeng and L. Wang, Carbon, 2018, 140, 508–520 CrossRef CAS.
- S. P. Sasikala, L. Henry, G. Yesilbag Tonga, K. Huang, R. Das, B. Giroire, S. Marre, V. M. Rotello, A. Penicaud, P. Poulin and C. Aymonier, ACS Nano, 2016, 10, 5293–5303 CrossRef CAS PubMed.
- S. Kang, K. M. Kim, K. Jung, Y. Son, S. Mhin, J. H. Ryu, K. B. Shim, B. Lee, H. S. Han and T. Song, Sci. Rep., 2019, 9, 4101 CrossRef PubMed.
- Y. Liu, B. Gao, Z. Qiao, Y. Hu, W. Zheng, L. Zhang, Y. Zhou, G. Ji and G. Yang, Chem. Mater., 2015, 27, 4319–4327 CrossRef CAS.
- A. Abbas, T. A. Tabish, S. J. Bull, T. M. Lim and A. N. Phan, Sci. Rep., 2020, 10, 21262 CrossRef CAS PubMed.
- R. Ye, C. Xiang, J. Lin, Z. Peng, K. Huang, Z. Yan, N. P. Cook, E. L. G. Samuel, C.-C. Hwang, G. Ruan, G. Ceriotti, A.-R. O. Raji, A. A. Martí and J. M. Tour, Nat. Commun., 2013, 4, 2943 CrossRef PubMed.
- Q. Liu, J. Zhang, H. He, G. Huang, B. Xing, J. Jia and C. Zhang, Nanomaterials, 2018, 8, 844 CrossRef PubMed.
- Y. Zhang, K. Li, S. Ren, Y. Dang, G. Liu, R. Zhang, K. Zhang, X. Long and K. Jia, ACS Sustainable Chem. Eng., 2019, 7, 9793–9799 CrossRef CAS.
- A. Kundu, B. Maity and S. Basu, New J. Chem., 2022, 46, 7545–7556 RSC.
- Y. Dong, J. Lin, Y. Chen, F. Fu, Y. Chi and G. Chen, Nanoscale, 2014, 6, 7410–7415 RSC.
- N. Morimoto, T. Kubo and Y. Nishina, Sci. Rep., 2016, 6, 21715 CrossRef CAS PubMed.
- A. D. Lemly, Hum Ecol. Risk. Assess., 2017, 24, 627–641 CrossRef.
- K. M. Zierold and C. Odoh, Rev. Environ. Health, 2020, 35, 401–418 CrossRef CAS PubMed.
- M. Whiteside and J. Herndon, J. Adv. Med. Med. Res., 2018, 25, 1–11 Search PubMed.
- S. Chung, R. A. Revia and M. Zhang, Adv. Mater., 2019, 33, 1904362 CrossRef PubMed.
- H. Wang, C. Liu, Z. Liu, J. Ren and X. Qu, Small, 2018, 14, 1703710 CrossRef PubMed.
- H. Sun, A. Zhao, N. Gao, K. Li, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2015, 54, 7176–7180 CrossRef CAS PubMed.
- Y. Lin, L. Wu, Y. Huang, J. Ren and X. Qu, Chem. Sci., 2015, 6, 1272–1276 RSC.
- H. Sun, Y. Zhou, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2018, 57, 9224–9237 CrossRef CAS PubMed.
- A. Sun, L. Mu and X. Hu, ACS Appl. Mater. Interfaces, 2017, 9, 12241–12252 CrossRef CAS PubMed.
- D. Kim, J. M. Yoo, H. Hwang, J. Lee, S. H. Lee, S. P. Yun, M. J. Park, M. J. Lee, S. Choi, S. H. Kwon, S. Lee, S.-H. Kwon, S. Kim, Y. J. Park, M. Kinoshita, Y.-H. Lee, S. Shin, S. R. Paik, S. J. Lee, S. Lee, B. H. Hong and H. S. Ko, Nat. Nanotechnol., 2018, 13, 812–818 CrossRef CAS PubMed.
- L. Hu, W. Sun, Y. Tang, S. Li, B. Zhang, X. Sun, W. Ji, L. Ma, H. Deng, S. Han and D. Zhu, Carbon, 2021, 176, 148–156 CrossRef CAS.
- Q. Xin, X. Jia, A. Nawaz, W. Xie, L. Li and J. R. Gong, Nano Res., 2020, 13, 1427–1433 CrossRef CAS.
- H. Sun, A. Zhao, N. Gao, K. Li, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2015, 54, 7176–7180 CrossRef CAS PubMed.
- N. R. Nirala, G. Khandelwal, B. Kumar, Vinita, R. Prakash and V. Kumar, Talanta, 2017, 173, 36–43 CrossRef CAS PubMed.
- Y. Wang, W. Kong, L. Wang, J. Z. Zhang, Y. Li, X. Liu and Y. Li, Phys. Chem. Chem. Phys., 2019, 21, 1336–1343 RSC.
- L. Nilewski, K. Mendoza, A. S. Jalilov, V. Berka, G. Wu, W. K. Sikkema, A. Metzger, R. Ye, R. Zhang, D. X. Luong, T. Wang, E. McHugh, P. J. Derry, E. L. Samuel, T. A. Kent, A.-L. Tsai and J. M. Tour, ACS Appl. Mater. Interfaces, 2019, 11, 16815–16821 CrossRef CAS PubMed.
- M. Krunić, B. Ristić, M. Bošnjak, V. Paunović, G. Tovilović-Kovačević, N. Zogović, A. Mirčić, Z. Marković, B. Todorović-Marković, S. Jovanović, D. Kleut, M. Mojović, Đ. Nakarada, O. Marković, I. Vuković, L. Harhaji-Trajković and V. Trajković, Free Radicals Biol. Med., 2021, 177, 167–180 CrossRef PubMed.
- L. Wang, Y. Li, Y. Wang, W. Kong, Q. Lu, X. Liu, D. Zhang and L. Qu, ACS Appl. Mater. Interfaces, 2019, 11, 21822–21829 CrossRef CAS PubMed.
- V. L. Feigin, T. Vos, E. Nichols, M. O. Owolabi, W. M. Carroll, M. Dichgans, G. Deuschl, P. Parmar, M. Brainin and C. Murray, Lancet Neurol., 2020, 19, 255–265 CrossRef PubMed.
- N. Khatri, M. Thakur, V. Pareek, S. Kumar, S. Sharma and A. K. Datusalia, CNS Neurol. Disord. Drug Targets, 2018, 17, 689–695 CrossRef CAS PubMed.
- J. P. Bolaños, M. A. Moro, I. Lizasoain and A. Almeida, Adv. Drug Delivery Rev., 2009, 61, 1299–1315 CrossRef PubMed.
- B. Uttara, A. Singh, P. Zamboni and R. Mahajan, Curr. Neuropharmacol., 2009, 7, 65–74 CrossRef CAS PubMed.
- Y. Gilgun-Sherki, E. Melamed and D. Offen, Neuropharmacology, 2001, 40, 959–975 CrossRef CAS PubMed.
- C. Ren, X. Hu and Q. Zhou, Adv. Sci., 2018, 5, 1700595 CrossRef PubMed.
- O. M. Ighodaro and O. A. Akinloye, Alexandria J. Med., 2018, 54, 287–293 CrossRef.
- (a) D. C. Marcano, D. V. Kosynkin, J. M. Berlin, A. Sinitskii, Z. Sun, A. Slesarev, L. B. Alemany, W. Lu and J. M. Tour, ACS Nano, 2010, 4, 4806–4814 CrossRef CAS PubMed; (b) P. Kumar, B. Yadav, A. Kumar, C. Bora, A. Kumar and S. Das, FlatChem, 2021, 27, 100245 CrossRef CAS.
- Y. Zhao, X. Wu, S. Sun, L. Ma, L. Zhang and H. Lin, Carbon, 2017, 124, 342–347 CrossRef CAS.
- U. Stoin, A. I. Shames, I. Malka, I. Bar and Y. Sasson, ChemPhysChem, 2013, 14, 4158–4164 CrossRef CAS PubMed.
- G.-L. Hong, H.-L. Zhao, H.-H. Deng, H.-J. Yang, H.-P. Peng, Y.-H. Liu and W. Chen, Int. J. Nanomed., 2018, 13, 4807–4815 CrossRef CAS PubMed.
- F. Liu, M.-H. Jang, H. D. Ha, J.-H. Kim, Y.-H. Cho and T. S. Seo, Adv. Mater., 2013, 25, 3657–3662 CrossRef CAS PubMed.
- G. Venugopal, M.-H. Jung, M. Suemitsu and S.-J. Kim, Carbon, 2011, 49, 2766–2772 CrossRef CAS.
- T. Livneh, T. L. Haslett and M. Moskovits, Phys. Rev. B, 2002, 66, 195110 CrossRef.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, Adv. Mater., 2010, 23, 776–780 CrossRef PubMed.
- (a) C. Hontoria-Lucas, A. J. López-Peinado, J. D. López-González, M. L. Rojas-Cervantes and R. M. Martín-Aranda, Carbon, 1995, 33, 1585–1592 CrossRef CAS; (b) P. Kumar, C. Bora, B. Kumar, P. Kr. Sukul and S. Das, Synth. Met., 2020, 264, 116381 CrossRef CAS.
- K. He, N. Chen, C. Wang, L. Wei and J. Chen, Cryst. Res. Technol., 2018, 53, 1700157 CrossRef.
- B. D. Cullity and J. W. Weymouth, Am. J. Phys., 1957, 25, 394–395 CrossRef.
- Y. Dong, H. Pang, H. B. Yang, C. Guo, J. Shao, Y. Chi, C. M. Li and T. Yu, Angew. Chem., Int. Ed., 2013, 52, 7800–7804 CrossRef CAS PubMed.
- S. Ahirwar, S. Mallick and D. Bahadur, ACS Omega, 2017, 2, 8343–8353 CrossRef CAS PubMed.
- Y. Chong, C. Ge, G. Fang, X. Tian, X. Ma, T. Wen, W. G. Wamer, C. Chen, Z. Chai and J.-J. Yin, ACS Nano, 2016, 10, 8690–8699 CrossRef CAS PubMed.
- B. Kong, T. Yang, F. Cheng, Y. Qian, C. Li, L. Zhan, Y. Li, H. Zou and C. Huang, J. Colloid Interface Sci., 2022, 611, 545–553 CrossRef CAS PubMed.
- B.-C. Lee, J. Y. Lee, J. Kim, J. M. Yoo, I. Kang, J.-J. Kim, N. Shin, D. J. Kim, S. W. Choi, D. Kim, B. H. Hong and K.-S. Kang, Sci. Adv., 2020, 6, eaaz2630 CrossRef CAS PubMed.
- L. Wang, Y. Li, L. Zhao, Z. Qi, J. Gou, S. Zhang and J. Z. Zhang, Nanoscale, 2020, 12, 19516–19535 RSC.
- P. Kumar, P. Shivam, S. Mandal, P. Prasanna, S. Kumar, S. R. Prasad, A. Kumar, P. Das, V. Ali, S. K. Singh and D. Mandal, Int. J. Nanomed., 2019, 14, 6073–6101 CrossRef CAS PubMed.
- J. Zhao, W. Gao, X. Cai, J. Xu, D. Zou, Z. Li, B. Hu and Y. Zheng, Theranostics, 2019, 9, 2843–2855 CrossRef CAS PubMed.
- X. Wang, H. Wang and S. Zhou, J. Phys. Chem. Lett., 2021, 12, 11751–11760 CrossRef CAS PubMed.
- M. Hassan, E. Haque, K. R. Reddy, A. I. Minett, J. Chen and V. G. Gomes, Nanoscale, 2014, 6, 11988–11994 RSC.
- S. Arunragsa, Y. Seekaew, W. Pon-On and C. Wongchoosuk, Diamond Relat. Mater., 2020, 105, 107790 CrossRef CAS.
- A. S. Jalilov, L. G. Nilewski, V. Berka, C. Zhang, A. A. Yakovenko, G. Wu, T. A. Kent, A.-L. Tsai and J. M. Tour, ACS Nano, 2017, 11, 2024–2032 CrossRef CAS PubMed.
- R. Yuan, Y. Li, S. Han, X. Chen, J. Chen, J. He, H. Gao, Y. Yang, S. Yang and Y. Yang, ACS Cent. Sci., 2021, 8, 10–21 CrossRef PubMed.
- A. Quincozes-Santos, L. D. Bobermin, A. Latini, M. Wajner, D. O. Souza, C.-A. Gonçalves and C. Gottfried, PLoS One, 2013, 8, 64372 CrossRef PubMed.
- H. Guo, H. Guo, Y. Xie, Y. Chen, C. Lu, Z. Yang, Y. Zhu, Y. Ouyang, Y. Zhang and X. Wang, Redox Biol., 2022, 56, 102441 CrossRef CAS PubMed.
- R. Omar, M. Smith, G. Perry and M. Pappolla, J. Neuropathol. Exp. Neurol., 1996, 55, 634 CrossRef.
- F. P. Zemlan, O. J. Thienhaus and H. B. Bosmann, Brain Res., 1989, 476, 160–162 CrossRef CAS PubMed.
- D. W. Cleveland and J. D. Rothstein, Nat. Rev. Neurosci., 2001, 2, 806–819 CrossRef CAS PubMed.
- J. Choi, H. D. Rees, S. T. Weintraub, A. I. Levey, L.-S. Chin and L. Li, J. Biol. Chem., 2005, 280, 11648–11655 CrossRef CAS PubMed.
- C. A. Massaad, T. M. Washington, R. G. Pautler and E. Klann, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13576–13581 CrossRef CAS PubMed.
- B. Mansuroğlu, S. Derman, A. Yaba and K. Kızılbey, Int. J. Biol. Macromol., 2015, 72, 79–87 CrossRef PubMed.
- A. Azari, R. Nabizadeh, S. Nasseri, A. H. Mahvi and A. R. Mesdaghinia, Chemosphere, 2020, 250, 126238 CrossRef CAS PubMed.
- S. Mandal, S. R. Prasad, D. Mandal and P. Das, ACS Appl. Mater. Interfaces, 2019, 11, 33273–33284 CrossRef CAS PubMed.
- S. Nayak, S. R. Prasad, D. Mandal and P. Das, J. Hazard. Mater., 2020, 392, 122287 CrossRef CAS PubMed.
- B. A. Al Jahdaly, M. F. Elsadek, B. M. Ahmed, M. F. Farahat, M. M. Taher and A. M. Khalil, Sustainability, 2021, 13, 2127 CrossRef.
- W. Gao, J. He, L. Chen, X. Meng, Y. Ma, L. Cheng, K. Tu, X. Gao, C. Liu, M. Zhang, K. Fan, D.-W. Pang and X. Yan, Nat. Commun., 2023, 14, 160 CrossRef CAS PubMed.
- P. Saxena, K. Selvaraj, S. K. Khare and N. Chaudhary, Biotechnol. Lett., 2021, 44, 1–22 CrossRef PubMed.
- W. Brand-Williams, M. E. Cuvelier and C. Berset, LWT–Food Sci. Technol., 1995, 28, 25–30 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Pretreatment of coal; synthesis of GO from CW; general instrumentation; HR-TEM sample preparation and EDS map of YGQDs and GO; UV-vis spectra of GO and pH-dependent photophysical studies of YGQDs; cytotoxic studies and hemolytic assay; H2O2 degradation activity; monitoring YGQDs internalization in C-6 cells using confocal microscopy; degradation of H2O2 free radical by YGQDs; MTT assay color changes for LPS + YGQDs; ELISA picture representing color intensities for MDA level estimation. See DOI: https://doi.org/10.1039/d3nr04259f |
| This journal is © The Royal Society of Chemistry 2023 |