 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Accurate biometal quantification per individual Caenorhabditis elegans†
Katherine
Ganio
a,
Simon A.
James
a,
Dominic J.
Hare
 ab,
Blaine R.
Roberts
a and
Gawain
McColl
*a
ab,
Blaine R.
Roberts
a and
Gawain
McColl
*a
aThe Florey Institute of Neuroscience and Mental Health, The University of Melbourne, 30 Royal Parade, Parkville, Victoria 3052, Australia. E-mail: gawain.mccoll@florey.edu.au; Tel: +61 3 903 56608
bElemental Bio-imaging Facility, University of Technology Sydney, Broadway, New South Wales, Australia
First published on 26th January 2016
Abstract
In the life sciences, small model-organisms are an established research platform. Due to the economy of culturing and maintenance animals such as the roundworm Caenorhabditis elegans, and the fly Drosophila melanogaster, have been instrumental for investigating key genetic pathways, early development, neuronal function, as well as disease pathogenesis and toxicology. Small model organisms have also found utility in the study of inorganic biochemistry, where the role of metal ion cofactors are investigated for numerous fundamental cellular processes. The metabolism and homeostasis of metal ions is also central to many aspects of biology and disease. Accurate quantification of endogenous metal ion content is an important determinant for many biological questions. There is currently no standardised method for quantifying biometal content in individual C. elegans or estimating the variation between individuals within clonal populations. Here, we have determined that ten or more adults are required to quantify physiologically important metals via inductively coupled plasma mass spectrometry (ICP-MS). The accuracy and precision of this method was then compared to synchrotron-based X-ray fluorescence microscopy (XFM) to determine the variation between isogenic, developmentally synchronous C. elegans adults.
Introduction
Accurate analytical tools for studying metals are important for a range of biological investigations, including fundamental inorganic biochemistry,1 toxicology2 and disease mechanisms.3 The roundworm Caenorhabditis elegans is a classic, versatile model organism that has been applied to the study of metals in biology. For example, the ecotoxicity of metal and metal oxide engineered nanoparticle exposure have been investigated in this organism.4–6C. elegans can be raised in tightly controlled experimental conditions, facilitating analysis of metal and metal–ligand complex uptake, localisation and metabolism, and the resulting effects on life history traits.7–9A growing number of diseases have been correlated with the loss of homeostasis or change in the metabolism of endogenous metal ions in cells and tissues.10,11 The underlying mechanisms by which metals may contribute to the onset of disease are areas of intense investigation due to better integration of contemporary analytical techniques into the biosciences. Metal cofactors are essential for the structural and catalytic activity of nearly one-third of the human proteome.12 For example, cytochromes use an iron (Fe)-containing heme group and numerous antioxidants rely upon a redox-active metal center. In addition, metal ions including manganese (Mn), Fe, copper (Cu) and zinc (Zn) can have specific deleterious effects when in overabundance or insufficiency.
To determine the concentration of metals per individual or between populations of small multicellular organisms, like C. elegans that weigh less than 1 μg, requires sensitive analytical techniques.13 Quantification of specific metals in C. elegans by inductively coupled plasma-mass spectrometry (ICP-MS) has been reported using forty to hundreds of thousands of individuals per assay.14–16 Collecting samples that consist of more than several hundred can present practical challenges, and include concerns that large populations may not be as developmentally synchronous as those cultured using standard smaller scale methods. These issues may confound the biological variation of metals and additionally prevent metal levels from being reported as concentration per individual. It is therefore adventitious to develop an analytical protocol that does not require culturing on an overly large scale. In addition, there is no standardised method of sample preparation for the purpose of elemental analysis per individual specimen. A validated approach that is capable of accurate and precise quantification of metals using smaller sample number would overcome these limitations, and may also be amenable for further development into high-throughput screens, for which C. elegans are well suited.
Experimental
Chemicals and reagents
Ultra-pure water (18.2 MΩ; Milli-Q H2O; Merk Millipore, Australia) was used for the dilution of all reagents and standards. Analytical grade 65% (v/v) nitric acid (HNO3; Merck) was used to digest samples and was diluted to 1% as the standard diluent. The instrument was calibrated for manganese (Mn), iron (Fe), copper (Cu) and zinc (Zn) using mixed 0, 0.5, 1, 3, 5, 10, 50, 100 and 500 parts per billion (ppb) standard calibration solutions in standard diluent from commercially-available ICP-MS-CAL2-1, ICP-MS-CAL3-1 and ICP-MS-CAL4-1 certified reference standards (AccuStandard, USA). A reference element solution containing 200 μg L−1 yttrium (Y) (ICP-MS-Internal Standard Solution-1, Accustandard) was introduced by a T-piece positioned after the peristaltic pump and was used to normalise all measurements. The tuning solution for instrument optimisation contained 1 μg L−1 of cerium (Ce), cobalt (Co), lithium (Li), thallium (Tl) and Y in 2% (v/v) HNO3 (Agilent Technologies, Australia). All standards were stored in polyethylene bottles washed using 1% HNO3 in MilliQ-H2O, prior to use. Seronorm™ Trace Elements Serum L-1 and L-2 (Sero, Norway, lot #0903106) were used to externally assess analytical performance.Caenorhabditis elegans strains
Wild type C. elegans (strain N2) were obtained from the Caenorhabditis Genetics Center and were maintained on standard nematode growth media (NGM) at 20 °C using established protocols.17Sample preparation for ICP-MS
Ten independent biological replicates of groups consisting of 10, 25, 50, 100 and 200 individual C. elegans were analysed. This was repeated and the data pooled to give a total of 20 independent replicates for each sample size. Gravid wild type hermaphrodites were allowed to lay eggs for 3 hours at 20 °C in order to obtain developmentally synchronous young adults (4 day old post egg lay). Young adults were counted and removed from NGM plates and then transferred into 1.5 mL polypropylene microfuge tubes (TechnoPlas, Australia) containing 200 μL S-basal (5.85 g L−1 NaCl; 1 g L−1 KH2PO4; 6 g L−1 K2HPO4). The microfuge tubes were gently inverted for 30 minutes to flush the gut of bacterial feed. Samples were then washed three times with 200 μL S-basal, followed by three washes in 200 μL ultra-pure water to remove any remaining bacteria. Aspiration between washes was performed using a stereomicroscope to avoid sample loss. Following the washes samples were then flash frozen in liquid N2 and lyophilized overnight. Following lyophilisation, samples were digested in 20 μL of 65% HNO3 for 12 hours at room temperature and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to a final volume of 200 μL using 1% HNO3. Ten digest blanks were prepared for each experiment, giving 20 blanks in total, to determine the limit of detection and limit of quantification for the elements measured, as well as, correcting for any contamination that may occur during the digestion process.
10 to a final volume of 200 μL using 1% HNO3. Ten digest blanks were prepared for each experiment, giving 20 blanks in total, to determine the limit of detection and limit of quantification for the elements measured, as well as, correcting for any contamination that may occur during the digestion process.
Inductively coupled plasma-mass spectrometry
All measurements were performed on an Agilent Technologies 7700× ICP-MS with a MicroMist nebulizer (Glass Expansion, Australia). Torch positioning, sample depth adjustment and lens optimization were set according to manufacturer recommendations while the other instrumental parameters (ESI Table 1†) were optimized during a batch-specific user tune prior to each experimental run. Helium collision gas at 3 mL min−1 was used to minimise polyatomic interferences. Samples were introduced directly from 1.5 mL polypropylene tubes via an integrated automation system (IAS) autosampler (Agilent) using a peristaltic pump. Tubing length consisted of 400 mm of 0.15 mm I.D. polypropylene tubing and 0.25 mm I.D. Tygon PeriPump tubing, with approximately 150 μL of sample needed for each measurement. Sample uptake time comprised of 52 s with a stabilisation time of 40 s and integration time of 0.1 s for each of the element isotopes monitored and internal standard.Sample preparation for hydrated XFM
A cohort of developmentally synchronous C. elegans adults (4 day old post egg lay; generated as described above) were washed four times in 500 μL S-basal17 to remove excess bacteria, anesthetised in S-basal kept at 4 °C with 0.2% (w/v) sodium azide (NaN3). Once completely immobilised, samples were transferred to an agarose (2%, w/v) pad approximately 10 μm thick with 0.2% (w/v) NaN3, affixed to a 2 μm thick silicon nitride window (Silson, UK), covered with 4 μm thick Ultralene film (Volga Instruments, India) to prevent evaporate loss and immediately mounted for XFM.X-ray fluorescence microscopy
The distribution of metals was mapped at the XFM beamline at the Australian Synchrotron. A beam of 15.6 keV was focused to a spot of 2 μm (full-width at half-maximum) using a Kirkpatrick-Baez mirror pair.18 The X-ray energy was chosen to induce K-shell ionisation of elements with atomic numbers below 37 as well as for separating the relatively intense elastic and inelastic scatter peaks from the fluorescence lines of lighter elements. Scanning of the specimen occurred continuously through the focus with a virtual pixel size of 0.8 μm in both the horizontal and vertical while X-ray fluorescence (XRF) was collected with the Maia detector system. An elemental map of 3200 × 1600 pixels (5.12 Mpixels) was collected using a dwell time of approximately 3.13 ms per pixel. Manganese and platinum (Micromatter, Canada) reference metal foils were used to calibrate XRF intensity to elemental areal density using a fundamental parameter approach. Analysis of the X-ray fluorescence spectra, including corrections for self-absorption, scatter from the air path and the efficiency of the detector were performed using GeoPIXE v7.2f (CSIRO, Australia).Statistical tests
The limit of detection (LOD) and limit of quantification (LOQ) were calculated according to Boumans19 (three times and ten times the standard deviation of the background signal, respectively). Variation of the mean metal concentration between sample groups was determined via a one-way ANOVA with Tukey's post hoc multiple comparisons test in Prism v6.0d (GraphPad, USA). Outliers were detected and removed using a method that combines robust regression with outlier detection, ROUT,20 with a false discovery rate, Q, set to 1% using Prism v6.0d. In total, four outliers were removed from the ICP-MS data set; two from the Cu analysis group containing 100 individuals per measure and one from the group containing 200 individuals per measure as well as one from the Zn analysis group containing 100 individuals per measure (Fig. 1).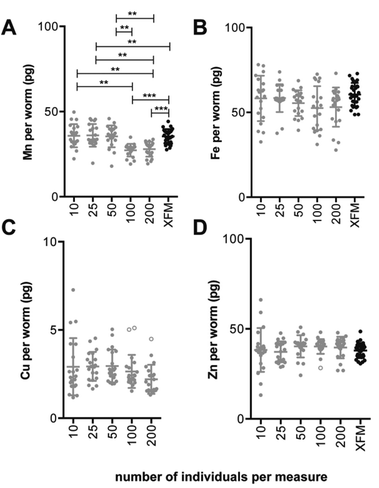 | ||
| Fig. 1 Comparison of ICP-MS and XFM estimates of Mn, Fe, Cu and Zn content per individual adult wild-type C. elegans (ICP-MS data: n = 20 replicates per measure, for XFM data: n = 29 replicates, Tukey's multiple comparisons test; **p < 0.01, ***p < 0.001). The ICP-MS data represents a combination of two independent analyses. (A) The average Mn content per worm measured by ICP-MS differed between sample groups. Sample groups containing 100 and 200 individuals had lower Mn per individual compared to other sample groups (p < 0.01) including XFM measurements (p < 0.001). (C) Note that Cu was below the limit of quantification for the XFM measurement parameters used in this study. (B–D) No significant difference for average Fe, Cu or Zn content per individual between groups measured via ICP-MS or between those and XFM measurements. Bar represents mean ± standard deviation. For mean, SD and coefficient of variation for each sample group refer to Table 2. Data excluded from analysis indicated as open circles. | ||
Results and discussion
Analytical sensitivity of ICP-MS
Here we describe a method that determines the minimum number of adult C. elegans per assay (i.e. minimum sample size) necessary to accurately quantify endogenous metals by ICP-MS, which can be easily adopted for use with other small model organisms. The accuracy and precision of this method was compared and contrasted with synchrotron-based X-ray fluorescence microscopy (XFM), an alternative analytical technique that we have previously used to produce quantitative micrometer resolution maps of metals in biological specimens.21,22 We observed good agreement between these two analytical techniques, supporting our approach for assessing low-level biometal content in small populations of C. elegans.To account for the small size of adult C. elegans and low abundance of Cu,23 samples were digested and subsequently diluted with minimal amounts of solvent to avoid diluting analyte concentrations beyond the ICP-MS detection limits (final volume = 200 μL). The experimental limits of detection and quantitation calculated from the standard deviation of the mean for each element in the digest blanks and the background equivalent concentration are presented in ESI Table 2.† The background equivalent concentrations are calculated using the background signal for a given element, using the analyte specific intensity of the background and as a result are usually greater than detection limits.24 Several factors can influence the detection limits of a given assay including elemental abundance in the 0 parts per billion (ppb) calibration standard, the linearity of the calibration curve, flow rate of the carrier gas, integration time for each element, as well as, polyatomic interference for certain elements. Some elements are more ubiquitous in a non-clean-room environment. Iron and zinc have a higher background intensity that is reflected in the background equivalent concentration for the given analyte.
ICP-MS data was pooled from two independent measurements. All samples included in the analysis yielded values above the limits of detection and the limits of quantification for all elements of interest and all outliers were removed. To verify the accuracy of the calibration standards triplicate measurements of two Seronorm standards were generated. All values were within acceptable range, as established by the standards manufacturer (Sero), with recoveries within 15% of the certified values for both standards (Table 1).
| Element | Seronorm standard | Expected value | Measured value | % Recovery |
|---|---|---|---|---|
| Mn | L-1 | 15.00 | 14.57 ± 0.38 | 94.3 ± 2.5 |
| L-2 | 19.90 | 19.07 ± 0.26 | 95.8 ± 1.3 | |
| Fe | L-1 | 1390 | 1383 ± 7 | 99.5 ± 0.5 |
| L-2 | 2030 | 2024 ± 13 | 99.7 ± 0.7 | |
| Cu | L-1 | 1691 | 1688 ± 16 | 99.9 ± 0.9 |
| L-2 | 2887 | 2826 ± 14 | 97.9 ± 0.5 | |
| Zn | L-1 | 1738 | 1597 ± 1 | 91.9 ± 0.1 |
| L-2 | 2520 | 2204 ± 4 | 87.5 ± 0.2 |
Quantification of metal content per individual
Our method suggests as few as 10 individuals per measure can be used to accurately quantify Mn, Fe, Cu and Zn via ICP-MS (Fig. 1). With the exception of Mn increasing the number of individuals sampled did not result in significant change in the average metal content. Across all elements, the coefficient of variation (as a percentage) ranged from 7.56 to 55.6% (Table 2), with the largest variance seen in Cu levels of measures using 10 adults. Copper is present in very low abundance in these nematodes (approximately 2 pg per individual) a level near the limit of detection of the ICP-MS. The difficulty of measuring such a low abundance is reflected by the high variation between estimates. Increased precision when measuring metals of relative low abundance may require increased number of biological replicates. For Mn and Zn the precision of estimates improved using samples comprising more than 10 individuals (Table 2). In contrast, the coefficient of variation for Fe and Cu estimates improved less with increasing sample size, and ranged from 22.92% and 55.61% (Fe and Cu, respectively, n = 10 individuals per measure) to 21.80% and 31.04% (n = 200 individuals per measure; Table 2). Overall similar levels of variation about the mean were comparable to estimates for the dagger nematode, Xiphinema vuittenezi, for Mn, Fe, Cu and Zn quantification via ICP-MS.25 Variation of element levels within sample groups may arise from biological, instrumental or procedural sources. Despite optimisation of sample preparation and instrumental settings, natural variation even between genetically identical and co-cultured individuals may still occur. The source of such variation may include small stochastic differences in development, reproductive status and nutritional status.| ICP-MS | ||||
|---|---|---|---|---|
| No. of individuals per measure | Mn | Fe | Cu | Zn |
| 10 | 35.83 ± 6.79 | 58.26 ± 13.36 | 2.92 ± 1.62 | 38.27 ± 12.17 |
| n = 20 | n = 20 | n = 20 | n = 20 | |
| 18.95 | 22.92 | 55.61 | 31.80 | |
| 25 | 35.42 ± 6.69 | 49.82 ± 11.22 | 2.93 ± 0.82 | 37.25 ± 5.83 |
| n = 20 | n = 20 | n = 20 | n = 20 | |
| 18.57 | 22.53 | 28.04 | 15.64 | |
| 50 | 36.05 ± 6.42 | 55.53 ± 7.61 | 2.95 ± 0.93 | 40.27 ± 6.16 |
| n = 20 | n = 20 | n = 20 | n = 20 | |
| 18.13 | 13.70 | 31.55 | 15.29 | |
| 100 | 27.31 ± 3.71 | 52.51 ± 12.95 | 2.38 ± 0.47 | 40.82 ± 3.09 |
| n = 20 | n = 20 | n = 18 | n = 19 | |
| 13.58 | 24.67 | 19.73 | 7.56 | |
| 200 | 27.92 ± 4.36 | 53.13 ± 11.58 | 2.09 ± 0.65 | 39.63 ± 5.99 |
| n = 20 | n = 20 | n = 19 | n = 20 | |
| 15.62 | 21.80 | 31.04 | 15.10 | |
| XFM | ||||
| 35.47 ± 4.22 | 60.56 ± 6.85 | No data | 37.95 ± 4.16 | |
| n = 29 | n = 29 | n = 29 | ||
| 11.90 | 11.30 | 10.95 | ||
The number of elements that can be analysed during each run is limited by the sample volume and abundance of the elements of interest, which may then require further optimisation of the quantitative parameters. However, our method can be applied to a wide range of elements restricted only by abundance and sensitivity of the instrument used. The background levels will also limit detection of less abundant elements. If the sample matrix contains significant background a higher number of individuals per sample or optimisation of the method parameters of the ICP-MS specific to that element may be required.
Comparison of analytical techniques
A complementary technique for accurate quantification of metals in biological systems is X-ray fluorescence microscopy (XFM), providing spatial maps of elements in situ, often with minimal disruption of the sample during perperation.13 A population of wild type adults was imaged using XFM (Fig. 2) to quantify Mn, Fe and Zn (Table 2, only whole imaged animals were used, see ESI Fig. 1† for masks used). The means were compared to those determined via ICP-MS. Estimates of Mn, Fe and Zn determined via ICP-MS were within 86.7 to 108.3% of those determined using XFM. However, significant disparity was observed for Mn in assays of 100 and 200 individuals per ICP-MS measure (27.31 ± 3.71 and 27.92 ± 4.36 pg per individual, respectively) that were lower than that for XFM (35.47 ± 4.22 pg per individual, p < 0.001). Previous comparison between XFM and inductively couple plasma-optical emission spectrometry (ICP-OES) showed variation of up to 11% in highly uniform, reference materials.26However, a direct comparison of XFM and laser ablation-ICPMS analysis of brain tissue, a complex biological matrix, showed a range of 70.8 to 164% variation for Fe, Cu and Zn content.27 Despite C. elegans also being a complex biological matrix, our methods provided good agreement between these different quantitative techniques.
The low abundance of Cu in C. elegans determined by ICP-MS could not be compared to XFM as a quantitative map for Cu distribution was not obtained within the available scan times. A measurement scheme with improved sensitivity, for example, achieved by increasing the dwell time, may be required to quantify the trace amounts of Cu fluorescence. Spatial distribution of Cu in C. elegans has been previously reported, but only following high Cu supplementation.28
Conclusions
In conclusion, the present work reports a comparison of biometals quantification using XFM and ICP-MS in order to determine minimum sample size necessary to accurately quantify these metals using ICP-MS as well as determining the variation between individuals within genetically identical and developmentally synchronous populations. To elucidate the role transition elements have in biology it is imperative to be able to accurately measure their abundance. Our method allows for the quantification of low abundance elements using the minimum sample size and can be adapted for the analysis of other trace elements. By comparing the values obtaining using ICP-MS with the most robust method available to quantify metals at the individual level, XFM, we have demonstrated that our method is a suitable alternative to XFM, when spatial resolution is not required. This method can be adapted to other small model organisms to understand the essential role metals have in biology.Acknowledgements
We gratefully acknowledge support from the Australian Research Council (DP130100357 and LP140100095) and the Victorian Government's Operational Infrastructure Support Program. We also thank Barbara Cardoso, Irene Volitakis, Neuroproteomics facility (Florey Institute) and the Australian Synchrotron for technical assistance. Part of this research was undertaken on the XFM beamline at the Australian Synchrotron (XFM9373). The nematode strain used in this study was provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).Notes and references
- S. J. Lippard, Nat. Chem. Biol., 2006, 2, 504–507 CrossRef CAS PubMed.
- N. Cedergreen, PLoS One, 2014, 9, e96580 Search PubMed.
- K. J. Barnham and A. I. Bush, Chem. Soc. Rev., 2014, 43, 6727–6749 RSC.
- J.-Y. Roh, S. J. Sim, J. Yi, K. Park, K. H. Chung, D.-Y. Ryu and J. Choi, Environ. Sci. Technol., 2009, 43, 3933–3940 CrossRef CAS PubMed.
- H. Ma, P. M. Bertsch, T. C. Glenn, N. J. Kabengi and P. L. Williams, Environ. Toxicol. Chem., 2009, 28, 1324–1330 CAS.
- H. Wang, R. L. Wick and B. Xing, Environ. Pollut., 2009, 157, 1171–1177 CrossRef CAS PubMed.
- D. Barsyte, D. A. Lovejoy and G. J. Lithgow, FASEB J., 2001, 15, 627–634 CrossRef CAS PubMed.
- A. N. Minniti, D. L. Rebolledo, P. M. Grez, R. Fadic, R. Aldunate, I. Volitakis, R. A. Cherny, C. Opazo, C. Masters and A. I. Bush, Mol. Neurodegener., 2009, 4, 2 CrossRef PubMed.
- B. Crone, M. Aschner, T. Schwerdtle, U. Karst and J. Bornhorst, Metallomics, 2015, 7, 1189–1195 RSC.
- R. McRae, P. Bagchi, S. Sumalekshmy and C. J. Fahrni, Chem. Rev., 2009, 109, 4780–4827 CrossRef CAS PubMed.
- S. J. Stohs and D. Bagchi, Free Radicals Biol. Med., 1995, 18, 321–336 CrossRef CAS PubMed.
- A. Lothian, D. J. Hare, R. Grimm, T. M. Ryan, C. L. Masters and B. R. Roberts, Front. Aging Neurosci., 2013, 5, 35 Search PubMed.
- D. J. Hare, E. J. New, M. D. de Jonge and G. McColl, Chem. Soc. Rev., 2015, 44, 5941–5958 RSC.
- G. McColl, D. W. Killilea, A. E. Hubbard, M. C. Vantipalli, S. Melov and G. J. Lithgow, J. Biol. Chem., 2008, 283, 350–357 CrossRef CAS PubMed.
- I. M. Klang, B. Schilling, D. J. Sorensen, A. K. Sahu, P. Kapahi, J. K. Andersen, P. Swoboda, D. W. Killilea, B. W. Gibson and G. J. Lithgow, Aging, 2014, 6, 975–991 Search PubMed.
- Y.-T. Lin, H. Hoang, S. I. Hsieh, N. Rangel, A. L. Foster, J. N. Sampayo, G. J. Lithgow and C. Srinivasan, Free Radicals Biol. Med., 2006, 40, 1185–1193 CrossRef CAS PubMed.
- S. Brenner, Genetics, 1974, 77, 71–94 CAS.
- D. Paterson, M. D. de Jonge, D. L. Howard, W. Lewis, J. McKinlay, A. Starritt, M. Kusel, C. G. Ryan, R. Kirkham, G. Moorhead and D. P. Siddons, AIP Conf. Proc., 2011, 1365, 219–222 CrossRef.
- P. W. J. M. Boumans, R. J. McKenna and M. Bosveld, Spectrochim. Acta, Part B, 1981, 36, 1031–1058 CrossRef.
- F. R. Hampel, E. M. Ronchetti, P. J. Rousseeuw and W. A. Stahel, Robust Statistics: the Approach Based on Influence Functions, John Wiley and Sons, New York, 1986 Search PubMed.
- D. J. Hare, M. W. M. Jones, V. C. Wimmer, N. L. Jenkins, M. D. de Jonge, A. I. Bush and G. McColl, Metallomics, 2016 10.1039/C5MT00288E.
- S. A. James, B. R. Roberts, D. J. Hare, M. D. de Jonge, I. E. Birchall, N. L. Jenkins, R. A. Cherny, A. I. Bush and G. McColl, Chem. Sci., 2015, 6, 2952–2962 RSC.
- S. A. James, M. D. de Jonge, D. L. Howard, A. I. Bush, D. Paterson and G. McColl, Metallomics, 2013, 5, 627 RSC.
- R. Thomas, Practical guide to ICP-MS: a tutorial for beginners, CRC press, 2nd edn, 2013 Search PubMed.
- Z. Sávoly, P. Nagy, K. Havancsák and G. Záray, Microchem. J., 2012, 105, 83–87 CrossRef.
- S. A. James, D. E. Myers, M. D. de Jonge, S. Vogt, C. G. Ryan, B. A. Sexton, P. Hoobin, D. Paterson, D. L. Howard, S. C. Mayo, M. Altissimo, G. F. Moorhead and S. W. Wilkins, Anal. Bioanal. Chem., 2011, 401, 853–864 CrossRef CAS PubMed.
- K. M. Davies, D. J. Hare, S. Bohic, S. A. James, J. L. Billings, D. I. Finkelstein, P. A. Doble and K. L. Double, Anal. Chem., 2015, 87, 6639–6645 CrossRef CAS PubMed.
- B. P. Jackson, P. L. Williams, A. Lanzirotti and P. M. Bertsch, Environ. Sci. Technol., 2005, 39, 5 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ESI table and figure. See DOI: 10.1039/c5an02544c |
| This journal is © The Royal Society of Chemistry 2016 |

