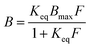Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
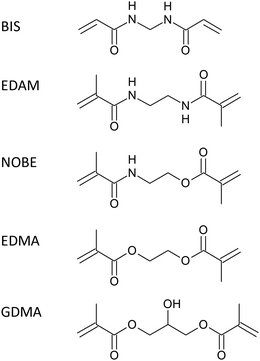
Rabbit IgG-imprinted nanoMIPs by solid phase synthesis: the effect of cross-linkers on their affinity and selectivity†
Matteo
Chiarello
 ,
Laura
Anfossi
,
Laura
Anfossi
 ,
Simone
Cavalera
,
Simone
Cavalera
 ,
Fabio
Di Nardo
,
Fabio
Di Nardo
 ,
Thea
Serra
,
Thea
Serra
 ,
Fabrizio
Sordello
,
Fabrizio
Sordello
 and
Claudio
Baggiani
and
Claudio
Baggiani
 *
*
Department of Chemistry, University of Torino, Via Giuria 7, 10125 - Torino, Italy. E-mail: claudio.baggiani@unito.it
First published on 16th March 2022
Abstract
Solid phase synthesis (SPS) of molecularly imprinted nanopolymers (nanoMIPs) represents an innovative method to prepare nanomaterials with tailor-made molecular recognition properties towards peptides and proteins. The synthesis of nanoMIPs by SPS usually involves a pre-polymerization formulation, where the cross-linker is invariably N,N′-methylen-bis-acrylamide (BIS). To date, the effect of cross-linkers on the binding properties of nanoMIPs prepared using cross-linkers other than BIS has never been reported. In this work, in order to investigate the effect of different cross-linkers in protein-imprinted nanoMIPs prepared by SPS, alongside BIS we considered other similar cross-linkers: N,N′-ethylene dimethacrylamide (EDAM), N,O-bis-methacryloylethanolamine (NOBE), ethylene glycol dimethacrilate (EDMA) and glycerol dimethacrylate (GDMA), replacing BIS with them in pre-polymerization mixtures. The synthetized nanoMIPs were homogeneous, with a polydispersity index of 0.24–0.30 and a mean diameter of 129–169 nm in water. The binding properties of the nanoMIPs were measured via equilibrium partition experiments with the template, rabbit IgG (RIgG), and the selectivity was evaluated with respect to bovine IgG (BIgG), bovine serum albumin (BSA) and hen egg lysozyme (LZM). The experimental results show that all the cross-linkers, with the exception of EDMA, endowed nanoMIPs with high binding affinities for the template (BIS: 16.0 × 106 mol−1 L, EDAM: 8.8 × 106 mol−1 L, NOBE: 15.8 × 106 mol−1 L, and GDMA: 12.8 × 106 mol−1 L), medium to high imprinting factors (BIS: 12.3, EDAM: 5.5, NOBE: 7.2, and GDMA: 11.6) and good selectivity towards other proteins but markedly dependent on the structure of the cross-linker, confirming the importance of the latter in the SPS of imprinted nanopolymers.
1. Introduction
Immunoglobulin G (IgG) is the most abundant protein showing immunological activity, accounting for 75–80% of all immunoglobulins. It strongly binds to the corresponding antigens – usually biomacromolecules foreign to the organism – with high specificity, playing a key role in the immune system of mammals.1 For this reason, IgG is extremely relevant not only in diagnostics,2,3 therapeutics4,5 and theragnostics6 but also in applications where very high selectivity towards a target molecule is mandatory, as (bio)sensoristics7,8 and affinity chromatography.9,10 IgG can be conveniently isolated from plasma by the classical Cohn's method based on the fractional precipitation of serum proteins by ethanol.11 Unfortunately, this method does not assure the complete separation of IgG from other serum proteins, and more efficient downstream purification strategies must be used to obtain pure IgG fractions. Several methods based on affinity ligands of natural or artificial origin have been proposed but, at present, affinity chromatography based on Protein A, a 42 kDa protein with high affinity at the Fc region of IgG, is the preferred method for preparative and industrial purposes.12–14 However, this method suffers from high costs and limited stability of Protein A, and harsh elution conditions which can sometimes lead to irreversible damage of IgG. Thus, man-made IgG-binding materials based on the molecular imprinting technology which could overcome these drawbacks are of significant interest. In the last 10 years, several papers describing different approaches to IgG imprinting have been published: cryogels,15,16 films,17,18 hydrogels,19 interpenetrating polymers,20 magnetic particles,21,22 membranes,23,24 microbeads25,26 and molecularly imprinted nanoparticles (nanoMIPs) prepared by solid phase synthesis (SPS).27This latter approach has proved particularly useful for obtaining high affinity protein-imprinted nanopolymers, characterized by high selectivity for the template and complete compatibility with aqueous environments.28–36 Moreover, as the template is covalently grafted onto the solid phase, the isolation and purification of nanoMIPs is an easy task, and no residual protein remains trapped in the nanoparticles, avoiding product contamination.
The synthesis of protein-imprinted nanoMIPs by SPS is normally performed in water, and the pre-polymerization formulations include a large excess – up to 98% by moles – of functional monomers.37 The selection of the functional monomer seems to be of lesser importance than in the case of templates made up of small molecules, because it has been shown that functional monomers that differ in their chemical properties are in any case able to interact with different functional groups present on the templates, invariably leading to nanoMIPs with good molecular recognition properties.34 About the cross-linker, it is added in a much more limited amount, and invariably is N,N′-methylen-bis-acrylamide (BIS).37 Its prevalent use may be justified by its good solubility in water and compatibility with proteins. It is nevertheless possible to consider using other cross-linkers, of which, however, the effect on the binding properties of nanoMIPs has never been reported in the literature to date, with the remarkable exception of the use of N,N′-ethylene dimethacrylamide for the solid phase synthesis of adenosine monophosphate-binding nanoMIPs.38
In this work, in order to investigate the effect of different cross-linkers in rabbit IgG-imprinted nanoMIPs prepared by SPS, alongside BIS, we have considered some other similar cross-linkers whose structural formulae are shown in Chart 1: N,N′-ethylene dimethacrylamide (EDAM), N,O-bis-methacryloylethanolamine (NOBE), ethylene glycol dimethacrylate (EDMA) and glycerol dimethacrylate (GDMA), replacing them for the BIS in pre-polymerization mixtures without changing the molar proportions with functional monomers. The binding properties of the nanoMIPs have been measured via equilibrium partition experiments with the template, rabbit IgG (RIgG), and the selectivity has been evaluated with respect to three other proteins of interest: bovine IgG (BIgG), bovine serum albumin (BSA) and hen egg lysozyme (LZM).
2. Materials and methods
2.1. Chemicals and materials
Glass beads, Spheriglass-2429, of 70–100 μm average particle size (Potters, UK) were aminated as previously reported.39N,O-Bis-methacryloyl ethanolamine (NOBE) was prepared in accordance with the literature.40 Acrylic acid (AA), 3-aminopropyltrimethoxysilane (APTMS), ammonium persulphate (APS), bovine IgG (BIgG), bovine serum albumin (BSA), N,N′-diisopropylcarbodiimide (DIC), 4-(N,N-dimethylamino)pyridine (DMAP), ethanolamine, N,N′-ethylenedimethacrylamide (EDAM), ethylene glycol dimethacrylate (EDMA), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), glycerol dimethacrylate (GDMA, a mixture of 1,2 and 1,3 isomers), N-hydroxysuccinimide (NHS), N-isopropylacrylamide (NIPAm), N,N′-methylen-bis-acrylamide (BIS), morpholineethanesulphonic acid (sodium salt, MES), rabbit IgG (RIgG), succinic anhydride, N-tertbutylacrylamide (TBAm), and N,N,N′,N′-tetramethylethylendiamine (TEMED) were purchased from Sigma-Merck (Milan, Italy). The hen egg lysozyme (LZM) was purchased from Boehringer Ingelheim (Milan, Italy). Solvents and all other chemicals were purchased from Sigma-Merck (Milan, Italy). All the solvents were of HPLC grade, whereas all chemicals were of analytical grade. The water used was ultra-purified using a Purelab Prima System from Elga (Marlow, UK). Protein stock solutions were prepared by dissolving 25 mg of proteins in 25 mL of phosphate buffer (20 mmol L−1, 0.13 mol L−1 NaCl, pH 7.4) and stored in the dark at −20 °C. The Coomassie Blue G250 protein assay reagent was purchased from VWR International (Milan, Italy).2.2. Rabbit IgG immobilization on glass beads
In a 100 mL round-bottom flask with a reflux condenser, 10 g of aminated glass beads (1.1 μmol g−1 of amino groups by the Kaiser method41), 5 mg (0.05 mmol) of succinic anhydride and about 1 mg of DMAP as a catalyst were suspended into 40 mL of anhydrous pyridine. The mixture was heated at 90 °C for six hours, cooled, filtered on a 0.22 μm nylon membrane, and washed with dimethylformamide.The hemisuccinated beads were transferred into a 100 mL flat-bottom flask containing 40 mL of dimethylformamide, 6 mg of NHS (0.050 mmol) and 8 μL of DIC (0.052 mmol). The suspension was incubated at 4 °C for 60 min onto a horizontal roller, filtered on a 0.22 μm nylon membrane, washed with cold dimethylformamide and dried under vacuum suction.
The activated glass beads were transferred into a 100 mL flat-bottom flask and 40 mL of 1 mg mL−1 of rabbit IgG dissolved in bicarbonate buffer (50 mmol L−1, pH 8.5) was added. The suspension was incubated at room temperature overnight onto a horizontal roller, filtered on a 0.22 μm nylon membrane, washed with water, dried under vacuum suction and stored in the dark at 4 °C.
2.3. Synthesis of nanoMIPs
Polymerization mixtures were prepared in accordance with the literature,34 with minor modifications and adjusting the dilution of monomers to avoid the formation of unwanted lumps of polymers. A pre-polymerization mixture (molar ratio BIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AA
AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NIPAM
NIPAM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TBAm = 2
TBAm = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48) was made by mixing 25 mL of ultrapure water with 0.0065 mmol cross-linkers (BIS: 1 mg, EDAM: 1.1 mg, NOBE: 1.3 mg, EDMA: 1.3 mg, and GDMA: 1.5 mg), 4.7 mg of AA (0.065 mmol), 11 mg of NIPAm (0.097 mmol) and 19.8 mg of TBAm (0.156 mmol, dissolved in 0.5 mL of ethanol) under sonication. Then, 5 mL of mixture was added to 50 mL polypropylene SPE cartridges containing 2.5 g of functionalized glass beads. The cartridges were purged with nitrogen for 5 min, 3 μL of TEMED and 100 μL of 30 mg mL−1 aqueous solution of APS were added and the polymerization was carried out at room temperature for 60 min in a roller-equipped incubator. The supernatant was drained by vacuum aspiration, the dry cartridges were cooled to 4 °C and polymerization by-products and low-affinity nanoMIPs were washed with 10 × 2 mL of ice-cold water. High affinity nanoMIPs were collected by eluting the cartridges at room temperature with 5 × 2 mL of 0.1 mol L−1 aqueous HCl. The eluates were immediately neutralized with aqueous ammonium hydroxide 1 mol L−1 and purified by gel-filtration in ultrapure water onto a 26 × 250 mm Sephadex G25 column. The nanoMIPs were isolated by centrifugation at 14
48) was made by mixing 25 mL of ultrapure water with 0.0065 mmol cross-linkers (BIS: 1 mg, EDAM: 1.1 mg, NOBE: 1.3 mg, EDMA: 1.3 mg, and GDMA: 1.5 mg), 4.7 mg of AA (0.065 mmol), 11 mg of NIPAm (0.097 mmol) and 19.8 mg of TBAm (0.156 mmol, dissolved in 0.5 mL of ethanol) under sonication. Then, 5 mL of mixture was added to 50 mL polypropylene SPE cartridges containing 2.5 g of functionalized glass beads. The cartridges were purged with nitrogen for 5 min, 3 μL of TEMED and 100 μL of 30 mg mL−1 aqueous solution of APS were added and the polymerization was carried out at room temperature for 60 min in a roller-equipped incubator. The supernatant was drained by vacuum aspiration, the dry cartridges were cooled to 4 °C and polymerization by-products and low-affinity nanoMIPs were washed with 10 × 2 mL of ice-cold water. High affinity nanoMIPs were collected by eluting the cartridges at room temperature with 5 × 2 mL of 0.1 mol L−1 aqueous HCl. The eluates were immediately neutralized with aqueous ammonium hydroxide 1 mol L−1 and purified by gel-filtration in ultrapure water onto a 26 × 250 mm Sephadex G25 column. The nanoMIPs were isolated by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, dried by lyophilisation and stored at 4 °C.
000 × g, dried by lyophilisation and stored at 4 °C.
Non-imprinted polymers (nanoNIPs) were prepared under the same experimental conditions in terms of the composition of the polymerization mixture and polymerization time, but using glass beads functionalized with diclofenac as a solid phase.42
2.5. Determination of the size and charge of nanoMIPs
The hydrodynamic particle size and zeta potential were measured with a ZetaView® Nanoparticle Tracking Analyzer PMX-120, (Analytik, Cambridge, UK) using a laser source at 488 nm. Solid samples of each of the nanoMIPs were dissolved in working dilution with ultrapure water under sonication, the pH was adjusted with HCl 0.1 mol L−1, and about 2 mL of the sample was immediately injected into the analyzer. The results are the average of three distinct measurements made at 25.5 ± 0.1 °C.2.6. Atomic force microscopy of nanoMIPs
Borosilicate glass slides, 10 × 10 mm, were washed with ‘piranha’ solution (98% sulphuric acid + 30% hydrogen peroxide, 3 + 1 v/v. Caution! It reacts violently with organic materials) for 10 min, rinsed with ultrapure water, dried under nitrogen and immersed overnight in a 1% v/v solution of APTMS in dry toluene. The aminated slides were washed with ethanol and ultrapure water and covered with an adequate volume of MES buffer (10 mmol L−1, pH 4.7) containing 1 mg mL−1 NHS-activated nanoMIPs (vide infra), incubated at room temperature overnight, rinsed with ultrapure water and dried under a nitrogen.Atomic force microscopy imaging was performed using a Park System XE-100 microscope (Park Systems Europe GmbH, Mannheim, Germany) in a non-contact mode (scan rate 0.4 Hz) using ACTA-10M cantilevers (Applied Nano Structures, Mountain View, USA).
2.7. Coupling of nanoMIPs to glass beads
In 4 mL vials, 1 mg of nanoMIPs were dissolved in 1 mL of MES buffer under sonication; 5 mg of NHS (44 nmol) and 7 mg of EDC (28 nmol) were added and the solutions were incubated at 4 °C for 60 min. Then, they were transferred into 3 mL vials containing 1 g of aminated glass beads. The suspensions were incubated at room temperature overnight, filtered on 0.22 μm nylon membranes, washed with ultrapure water, dried under vacuum at room temperature and stored at 4 °C.2.8. Protein determination
The protein determination was carried out using the Bradford assay method. Briefly, 50 μL of the protein sample was added to 200 μL of the protein assay reagent in polystyrene microplates (12 × 8 wells, flat bottom, VWR International, Milan, Italy). After shaking for 30 s, the absorbance values were read at 450 and 590 nm. Each experimental point was assessed as the average of four repeated measures. Concentrations were calculated from a calibration graph covering the 0.5–50 μg mL−1 range of the protein diluted in the same phosphate buffer plotting the ratio A590/A450vs. the concentration.432.9. Determination of binding properties
To measure binding isotherms, about 40 mg of glass bead supporting nanoMIPs were exactly weighed in 4 mL flat bottom amber glass vials. Then, 1.0 mL of phosphate buffer (20 mmol L−1, 0.13 mol L−1 NaCl, pH 7.4) containing increasing amounts of proteins ranging from 1 to 50 μg mL−1 was added. The vials were incubated overnight at room temperature under continuous agitation on a horizontal rocking table. Then, the solutions were filtered on 0.22 μm nylon membranes and the free amounts of proteins were measured by the Bradford assay. Each experimental point was assessed as the average of three repeated measures.Binding parameters were calculated using SigmaPlot 12 (Systat Software Inc., Richmond, CA, USA). Non-linear least square fitting was applied to the averaged experimental data. Binding isotherm parameters were calculated using a Langmuir binding isotherm model:
To assure robust results, weighted (1/y) Pearson VII limit minimization was chosen as the minimization method. To avoid being trapped in local minima, which would give incorrect results, minimizations were carried out several times by using different initial guess values for the binding parameters.
The imprinting factor, IF, was calculated as follows:
| IF = Keq(MIP)/Keq(NIP) |
The binding selectivity, α, was calculated as follows:
| α = Keq(protein)/Keq(RIgG) |
3. Results and discussion
In order to investigate the effect of different cross-linkers in IgG-imprinted nanoMIPs, in the pre-polymerization mixtures, BIS was replaced with other similar cross-linkers: N,N′-ethylene dimethacrylamide (EDAM), N,O-bis-methacryloylethanolamine (NOBE), ethylene glycol dimethacrylate (EDMA) and glycerol dimethacrylate (GDMA), without changing the molar proportions with functional monomers and using the same persulfate/TEMED-induced radical polymerization protocol in water at room temperature. After gel filtration, centrifugation and drying, nanoMIPs were collected as white solids, with yields calculated with respect to the amount of monomers in the polymerization mixtures of 15–18% (1–1.2 mg). When dissolved in water, nanoMIPs gave transparent and colourless solutions, without any perceivable turbidity. The composition of nanoparticles can be influenced by the different reactivities of the monomers, as well as the effective degree of crosslinking, but, because of the limited quantity of nanoparticles obtained, no attempts were made to establish the effective degree of crosslinking. Therefore, as a first approximation, we assume that it does not vary significantly between the different polymers.3.1. The size and charge of nanoMIPs
Acrylic acid was used as the charged functional monomer; thus, nanoMIPs can be seen as charged polyelectrolytes at neutral pH. This is confirmed by ζ potential measurements, reported in Table 1, where, at pH 7, all the nanoMIPs show a net negative potential, with ζ values between −7.4 mV (GDMA) and −23.9 mV (BIS), while, at pH 3, under more acidic conditions, where carboxyls are fully protonated, ζ turns positive, with values between +1.0 mV (EDMA) and +15.1 (NOBE).| d p (nm) | ΔV | PDI | ζ mV | Δζ | ||||
|---|---|---|---|---|---|---|---|---|
| pH 3 | pH 7 | pH 3 | pH 7 | pH 3 | pH 7 | |||
| BIS | 171 ± 83 | 129 ± 66 | 2.34 | 0.24 | 0.26 | +7.7 | −23.9 | 31.6 |
| EDAM | 189 ± 94 | 169 ± 84 | 1.40 | 0.25 | 0.25 | +10.4 | −8.3 | 18.7 |
| NOBE | 186 ± 87 | 148 ± 72 | 1.98 | 0.22 | 0.24 | +15.1 | −18.0 | 33.1 |
| EDMA | 158 ± 79 | 140 ± 73 | 1.44 | 0.25 | 0.27 | +1.0 | −22.2 | 23.2 |
| GDMA | 147 ± 77 | 129 ± 71 | 1.48 | 0.27 | 0.30 | +10.6 | −7.4 | 18.0 |
The hydrodynamic diameter, dp, measured by laser nanoparticle tracking at pH 7 shows nanoparticles with average diameters just over a hundred nm, ranging from 129 nm (BIS) to 169 nm (EDAM), and with a polydispersity index between 0.24 (NOBE) and 0.30 (GDMA), corresponding to moderately polydispersed nanoparticles. In a more acidic environment, at pH 3, the formation of aggregates larger than 1 μm (the instrumental limit of the particle tracker set-up), was indirectly observed, because the nanoparticle count fell by two orders of magnitude from 105 to 103. The fraction of nanoparticles remained in solution; the polydispersity index remains essentially constant, but diameters increase markedly, ranging from 147 nm (GDMA) to 189 nm (EDAM). These results show that in the solid phase synthesis the cross-linker structure marginally affects the dimensions of the resulting nanoparticles, which are probably mainly controlled by the formation of dangling long chains of monomers, some or most not cross-linked. Regardless, the cross-linker in some manner is yet capable of influencing the flexibility of nanoMIPs. In fact, while nanoparticles containing BIS or NOBE are able to double their volume from pH 7 to pH 3, nanoparticles containing EBIS, EDMA or GDMA swell significantly less. It must be noted that the swelling ability of the nanoparticles does not seem to be related to binding properties (vide infra, Section 3.3 for experimental results), as BIS- and GDMA-based nanopolymers show comparable binding constants but very different swelling abilities from pH 7 to pH 3. It is also noteworthy that the absolute difference in the ζ values measured between pH 7 and pH 3 is proportional to the swelling ability of the nanoparticles. This is not unexpected because as the volume changes, the surface charge density changes proportionally (whatever its sign), equally affecting the resulting ζ potential.
3.2. AFM imaging of nanoMIPs
Acrylic acid was used as the charged functional monomer; thus, the results obtained by laser nanoparticle tracking are confirmed by atomic force microscopy performed on nanoMIPs covalently grafted onto aminosilanized glass slides (see the ESI†). The imaging – an example of which is reported in Fig. 1 – performed on a relatively large area of 10 × 10 μm shows that the glass surface is randomly covered with what seems to be sparse clusters of nanoparticles. NanoMIPs were covalently grafted onto the glass slides at pH 4.7, in conditions within the pH increase covered by the tracking measurements (from pH 7 to pH 3); thus, the formation of these structures is likely to be due to the grafting of clustered nanoparticles stabilized by electrostatic interactions. The imaging of a cluster at a higher resolution (×25) on an area of 2 × 2 μm (Fig. 2) shows an overall shape rather irregular, with an approximate size of 1.2 × 1.2 × 0.3 μm, apparently composed of several tightly packed globular objects with a slightly wrinkled surface and with individual diameters comparable to those measured by nanoparticles laser tracking, therefore compatible with an aggregate of nanoparticles.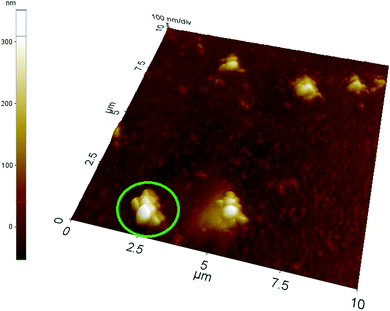 | ||
| Fig. 1 AFM image of a glass slide covalently grafted at low ionic strength with nanoMIPs prepared with GDMA as the cross-linker. The area delimited by green borders indicates the higher resolution image reported in Fig. 2. | ||
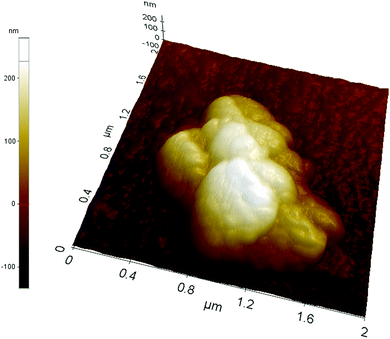 | ||
| Fig. 2 AFM image at a higher resolution of the cluster marked in green in Fig. 1. | ||
A further evidence of nanoparticles clustering induced by electrostatic interactions comes from the imaging of deposited nanoMIPs at a higher ionic strength (0.1 mol L−1 NaCl). In this case, clusters are significantly larger for all the nanoparticles examined (Fig. 3), often exceeding dimensions of 2 × 2 μm, even if their height with respect to the underlying glass surface does not seem to grow proportionally.
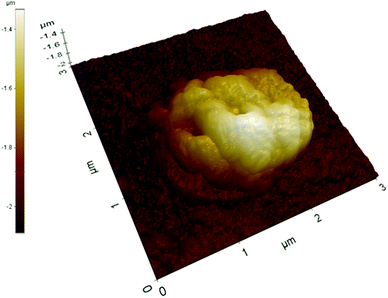 | ||
| Fig. 3 AFM image of a glass slide covalently grafted at high ionic strength with nanoMIPs prepared with GDMA as the cross-linker. | ||
3.3. The binding properties of nanoMIPs
In the traditional molecular imprinting techniques (bulk, suspension/emulsion, etc.) BIS is used very little and the cross-linker constitutes up to 80% molar of the polymerization mixture, thus exerting a deep effect not only on the morphology of the polymer and its bulk properties, but also on the binding properties.44–46 In contrast, in the SPS technique, BIS is the preferred cross-linker, and in the pre-polymerization mixture it is added in a much more limited amount, practically never more than 3% by moles. Consequently, it is to be expected that the effect on molecular recognition properties by the cross-linker is limited, and that the presence of structurally different cross-linkers is not able to affect these properties.Surprisingly, the determination of the equilibrium binding constant, Keq, by equilibrium partition experiments shows a distinctly different situation. In fact, as reported in Table 2, all the cross-linkers used for the imprinting of RIgG give nanoMIPs with a Keq value of around 107 mol−1 L, with the remarkable exception of EDMA, which gives a significantly lower value of 3.4 × 106 mol−1 L. In comparison, the corresponding nanoNIPs, prepared by SPS with diclofenac as the template, show Keq with values significantly lower than the values for the corresponding nanoMIPs and indistinguishable from each other (t-test: α = 0.05, n = 10, t < 2.101).
| RIgG | BIgG | BSA | LZM | ||
|---|---|---|---|---|---|
| BIS | nMIP | 16.0 ± 1.3 | 3.5 ± 0.4 | 2.2 ± 0.3 | 1.7 ± 0.3 |
| nNIP | 1.3 ± 0.3 | 3.4 ± 0.7 | 1.0 ± 0.2 | 0.6 ± 0.2 | |
| EDAM | nMIP | 8.8 ± 1.9 | 3.3 ± 0.8 | 2.1 ± 0.5 | 2.3 ± 0.6 |
| nNIP | 1.6 ± 0.3 | 2.8 ± 0.9 | 0.3 ± 0.2 | 1.3 ± 0.2 | |
| NOBE | nMIP | 15.9 ± 2.5 | 6.2 ± 1.2 | 0.6 ± 0.3 | 0.2 ± 0.2 |
| nNIP | 2.2 ± 0.5 | 1.7 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.1 | |
| EDMA | nMIP | 3.4 ± 0.7 | 2.4 ± 0.4 | 1.4 ± 0.4 | 0.3 ± 0.1 |
| nNIP | 1.2 ± 0.4 | 1.6 ± 0.4 | 0.9 ± 0.2 | 0.1 ± 0.0 | |
| GDMA | nMIP | 12.8 ± 1.5 | 4.3 ± 1.0 | 1.8 ± 0.3 | 1.4 ± 0.3 |
| nNIP | 1.1 ± 0.3 | 1.8 ± 0.5 | 0.6 ± 0.2 | 0.6 ± 0.0 |
The differing values of Keq obtained for each nanoMIP have an obvious influence on the imprinting factor, IF, that is an estimate of how much the binding affinity increases for an imprinted polymer with respect to a non-imprinted polymer of identical compositions. In Fig. 4, all the five nanoMIPs show IF values higher than unity, confirming the success of the SPS technique in the imprinting of RIgG. However, while nanoMIPs containing BIS and GDMA show IF values higher than 10 (BIS: 12.3 ± 3.0, GDMA 11.6 ± 3.5), corresponding to a very strong imprinting effect, others show markedly lower IF values (EDAM: 5.5 ± 1.6, NOBE: 7.2 ± 2.0), demonstrating that the choice of the right cross-linker is important to achieve an efficient SPS process, regardless of whether the cross-linker itself is present in the pre-polymerization mixture in very limited quantities compared to the other monomers. It should be noted that the GDMA-based nanoMIPs are to be considered structurally more complex than the other nanoMIPs, as they are composed of an almost equimolar mixture of two different cross-linkers, respectively, glycerol 1,2- and 1,3-dimethacrylate. However, the resulting nanoMIP does not appear to behave significantly differently from other nanoMIPs, except of course for those based on EDMA. It is also noteworthy that the polymer with the lowest IF value (2.8 ± 1.1) contains EDMA, which represents the predominant cross-linker used to prepare imprinted polymers with the traditional approaches. This fact is strong evidence of how the SPS technique differs from the other molecular imprinting approaches, and how it is necessary to pay attention to transfer pre-polymerization mixture formulations from one approach to another without a careful preliminary evaluation.
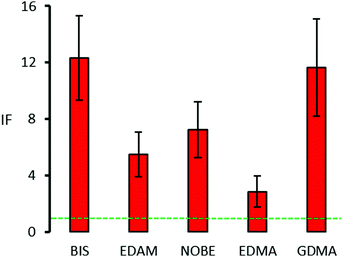 | ||
| Fig. 4 Imprinting factors (±1 s.e.) of the nanoMIPs prepared with different cross-linkers. The green dotted line indicates an IF of 1, i.e. no imprinting effect. | ||
In addition to the magnitude of the binding constant and imprinting factor, a third essential parameter for evaluating the molecular recognition properties of nanoMIPs is the binding selectivity, α. In this work, we evaluated the selectivity of RIgG-imprinted nanoMIPs towards a structurally very similar protein such as bovine IgG (BIgG), a protein structurally different but of similar isoelectric point, bovine serum albumin (BSA), and a protein of different structures and isoelectric points such as the hen egg lysozyme (LZM).
The α values in Fig. 5 show that all the nanopolymers characterized by high affinity (Keq ∼ 107 mol−1 L) are selective towards the template RIgG, with a limited but substantial recognition (0.2 ≤ α ≤ 0.4) towards BIgG, confirming the results reported in the literature for human IgG-imprinted nanoMIPs.27 This limited recognition can be explained on the basis of the shared presence in the IgG structure of the Fc fragment, which differs little between the proteins of different species.1 As a template RIgG is randomly grafted on the surface of the glass beads, nanoMIPs produced by SPS will have molecular recognition properties towards different parts of the template structure. Some will have binding sites recognizing the Fc fragment, common to IgG from different species, while others will recognize other portions of the protein, which are typical for IgG of a particular species. Therefore, during the rebinding of BIgG, some nanoMIPs will preferentially bind the Fc fragment, regardless of its origin (rabbit or bovine), while others, more selective, will not be able to bind the BIgG. Thus, the resulting binding will be an average between the full (Fc-binding nanoMIPs) and the weak (non-Fc-binding nanoMIPs) recognition of BIgG.
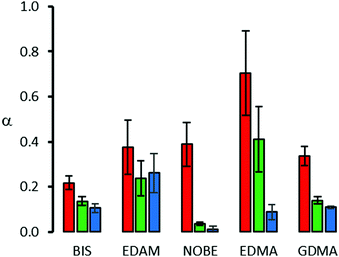 | ||
| Fig. 5 Binding selectivity (±1 s.e.) of the nanoMIPs prepared with different cross-linkers. Red bars: BIgG, green bars: BSA, and blue bars: LZM. | ||
In contrast, because of the low affinity resulting in a limited imprinting factor, with EDMA-based nanoMIPs, RIgG and BIgG are recognized almost in the same way, confirming the substantial absence of selectivity. It must be noted that the low values of Keq measured for EDMA-based nanoMIPs (2.4 ± 0.4 × 106 mol−1 L) and nanoNIPs (1.6 ± 0.4 × 106 mol−1 L) are statistically indistinguishable from each other (t-test: α = 0.05, n = 10, t = 1.414). Therefore, in this case, the binding to BIgG cannot be attributed for certain to the presence of imprinted binding sites.
Concerning BSA and LZM, as these proteins are very different from IgG, for all the nanopolymers, the molecular recognition results are very limited, lower than that observed for BIgG. However, it should be noted that the binding behaviour presents significant differences, since for BIS-, EDMA- and GDMA-based nanopolymers the recognition follows the order of similarity, i.e. BIgG > BSA > LZM, while in the case for EDAM- and NOBE-based nanopolymers it is different, as the first recognizes the three proteins in the same way, while, for the second, BSA and LZM show almost no recognition, confirming that small changes in the nature of the cross-linker – i.e. the replacement of an amide group with an ester group (NOBE vs. EDAM) – exert a significant effect on the binding properties of the nanoMIPs.
4. Conclusions
The results reported in this work confirm the relevance of the cross-linker structure in the SPS technique. Although present in minimal quantities compared to the other monomers in the pre-polymerization mixture, they are able to influence the binding affinity and selectivity of protein-imprinted nanopolymers through subtle differences in their structure, i.e. the replacement of an amide group with an ester group (NOBE vs. EDAM), the presence of a hydroxyl group (GDMA vs. EDMA) or the number of atoms in the molecular bridge (BIS vs. EDAM). The experimental results currently available are not sufficient to advance quantitative hypotheses on the relationship between the binding properties of nanoMIPs and structural properties of cross-linkers, but it is plausible that a further expansion of the number of cross-linkers tested could provide robust indications on the type of molecular structure optimal for obtaining nanoMIPs with high affinity and selectivity for the target molecule.Author contributions
Conceptualization: C. B.; methodology: M. C. and C. B.; investigation – laser particle tracking: T. S.; investigation – atomic force microscopy: F. S.; investigation – binding isotherms: M. C. and S. C.; resources: L. A.; data curation: F. D. N. and C. B.; writing – original draft preparation: C. B.; writing – review and editing: M. C.Conflicts of interest
There are no conflicts to declare.Notes and references
- G. Vidarsson, G. Dekkers and T. Rispens, Front. Immunol., 2014, 5, 520 CrossRef PubMed.
- S. K. Vashist, P. B. Luppa, L. Y. Yeo, A. Ozcan and J. H. T. Luong, Trends Biotechnol., 2015, 33, 692 CrossRef CAS PubMed.
- C. Dincer, R. Bruch, A. Kling, P. S. Dittrich and G. A. Urban, Trends Biotechnol., 2017, 35, 728 CrossRef CAS PubMed.
- P. Sharma and J. P. Allison, Science, 2015, 348, 56 CrossRef CAS PubMed.
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaia, Nat. Rev. Drug Discovery, 2017, 16, 315 CrossRef CAS PubMed.
- T. Lammers, S. Aime, W. E. Hennink, G. Storm and F. Kiessling, Acc. Chem. Res., 2011, 44, 1029 CrossRef CAS PubMed.
- F. S. Felix and L. Angnes, Biosens. Bioelectron., 2018, 102, 470 CrossRef CAS PubMed.
- A. Jones, L. Dhanapala, R. N. T. Kankanamage, C. V. Kumar and J. F. Rusling, Anal. Chem., 2020, 92, 345 CrossRef CAS PubMed.
- M. C. Hennion and V. Pichon, J. Chromatogr. A, 2003, 1000, 29 CrossRef CAS PubMed.
- E. L. Rodriguez, S. Poddar, S. Iftekhar, K. Suh, A. G. Woolfork, S. Ovbude, A. Pekarek, M. Walters, S. Lott and D. S. Hage, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2020, 1157, 122332 CrossRef CAS PubMed.
- E. J. Cohn, L. E. Strong, W. L. Hughes, D. J. Mulford, J. N. Ashworth, M. Melin and H. L. Taylor, J. Am. Chem. Soc., 1946, 68, 459 CrossRef CAS PubMed.
- K. Huse, H. J. Böhme and G. H. Scholz, J. Biochem. Biophys. Methods, 2002, 51, 217 CrossRef CAS PubMed.
- A. C. A. Roque, C. S. O. Silva and M. A. Taipa, J. Chromatogr. A, 2007, 1160, 44 CrossRef CAS PubMed.
- P. Gagnon, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 1221, 57 CrossRef CAS PubMed.
- S. Asliyuce, L. Uzun, R. Say and A. Denizli, React. Funct. Polym., 2013, 73, 813 CrossRef CAS.
- I. Perçin, N. Idil and A. Denizli, Proc. Biochem., 2019, 80, 13 CrossRef.
- A. Tretjakov, V. Syritski, J. Reut, R. Boroznjak, O. Volobujeva and A. Opik, Microchim. Acta, 2013, 180, 1433 CrossRef CAS.
- R. Bai, Y. Sun, M. Zhao, Z. Han, J. Zhang, Y. Sun, W. Dong and S. Li, Talanta, 2021, 226, 122160 CrossRef CAS PubMed.
- M. Cui, Y. Gong, M. Du, K. Wang, T. Li, X. Zhu, S. Wang and X. Luo, Sens. Actuators, B, 2021, 337, 129820 CrossRef CAS.
- A. Nematollahzadeh, P. Lindemann, W. Sun, J. Stute, D. Lütkemeyer and B. Sellergren, J. Chromatogr. A, 2014, 1345, 154 CrossRef CAS PubMed.
- J. Zhou, N. Gan, T. Li, F. Hu, X. Li, L. Wang and L. Zheng, Biosens. Bioelectron., 2014, 54, 199 CrossRef CAS PubMed.
- A.-A. Liang, B.-H. Hou, C.-S. Tang, D.-L. Sun and E.-A. Luo, Bioelectrochemistry, 2021, 137, 107671 CrossRef PubMed.
- D. X. Yin and M. Ulbricht, Biomacromolecules, 2013, 14, 4489 CrossRef CAS PubMed.
- S. Schwark, W. Sun, J. Stute, D. Lutkemeyer, M. Ulbricht and B. Sellergren, RSC Adv., 2016, 6, 53162 RSC.
- Y. Saylan, R. Üzek, L. Uzun and A. Denizli, J. Biomater. Sci., Polym. Ed., 2014, 25, 881 CrossRef CAS PubMed.
- W. Shi, K.-B. Li, S.-Q. Zhang, H.-L. Lu, R.-H. Shi, Y.-G. Zhan, Z.-L. Tong and D.-M. Han, J. Chem. Eng. Data, 2019, 64, 5385 CrossRef CAS.
- E. Moczko, A. Guerreiro, C. Cáceres, E. Piletska, B. Sellergren and S. A. Piletsky, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1124, 1 CrossRef CAS PubMed.
- S. Ambrosini, S. Beyazit, K. Haupt and B. Tse Sum Bui, Chem. Commun., 2013, 49, 6746 RSC.
- A. Poma, A. Guerreiro, S. Caygill, E. Moczko and S. Piletsky, RSC Adv., 2014, 4, 4203 RSC.
- J. Xu, S. Ambrosini, E. Tamahkar, C. Rossi, K. Haupt and B. Tse Sum Bui, Biomacromolecules, 2016, 17, 345 CrossRef CAS PubMed.
- J. Ashley, Y. Shukor, R. D’Aurelio, L. Trinh, T. L. Rodgers, J. Temblay, M. Pleasants and I. E. Tothill, ACS Sens., 2018, 3, 418 CrossRef CAS PubMed.
- X. Feng, J. Ashley, T. Zhou, A. Halder and Y. Sun, Analyst, 2018, 143, 2750 RSC.
- K. Smolinska-Kempisty, A. Guerreiro, J. Czulak and S. Piletsky, Sens. Actuators, B, 2019, 301, 126967 CrossRef CAS.
- T. S. Bedwell, N. Anjum, Y. Ma, J. Czulak, A. Poma, E. Piletska, M. J. Whitcombe and S. A. Piletsky, RSC Adv., 2019, 9, 27849 RSC.
- C. Cáceres, E. Moczko, I. Basozabal, A. Guerreiro and S. Piletsky, Polymers, 2021, 13, 314 CrossRef PubMed.
- S. S. Piletsky, E. Piletska, M. Poblock, S. Macip, D. J. L. Jones, M. Bragah, T. H. Cao, R. Singh, A. C. Spivey, E. O. Aboagye and S. A. Piletsky, Nano Today, 2021, 41, 101304 CrossRef CAS.
- F. Canfarotta, A. Poma, A. Guerreiro and S. A. Piletsky, Nat. Protoc., 2016, 11, 443 CrossRef CAS PubMed.
- C. A. Mourão, F. Bokeloh, J. Xu, E. Prost, L. Duma, F. Merlier, S. M. A. Bueno, K. Haupt and B. Tse Sum Bui, Macromolecules, 2017, 50, 7484 CrossRef.
- S. Cavalera, M. Chiarello, F. Di Nardo, L. Anfossi and C. Baggiani, React. Funct. Polym., 2021, 163, 104893 CrossRef CAS.
- D. Meador and D. A. Spivak, Org. Lett., 2014, 16, 1402 CrossRef CAS.
- E. Poli, V. Chaleix, C. Damia, Z. Hjezi, E. Champion and V. Sol, Anal. Methods, 2014, 6, 9622 RSC.
- A. Poma, A. Guerreiro, M. J. Whitcombe, E. C. Piletska, A. P. F. Turner and S. A. Piletsky, Adv. Funct. Mater., 2013, 23, 2821 CrossRef CAS.
- T. Zor and Z. Selinger, Anal. Biochem., 1996, 236, 302 CrossRef CAS PubMed.
- E. M. A. Dourado, C. Herdes, P. R. V. Tassel and L. Sarkisov, Int. J. Mol. Sci., 2011, 12, 4781 CrossRef CAS PubMed.
- G. D. Olsson, B. C. G. Karlsson, E. Schillinger, B. Sellergren and I. A. Nicholls, Ind. Eng. Chem. Res., 2013, 52, 13965 CrossRef CAS.
- K. Golker, B. C. G. Karlsson, G. D. Olsson, A. M. Rosengren and I. A. Nicholls, Macromolecules, 2013, 46, 1408 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Binding isotherms for nanoMIPs and nanoNIPs, and AFM imaging of nanoMIPs grafted onto glass surfaces. See DOI: 10.1039/d2tb00245k |
| This journal is © The Royal Society of Chemistry 2022 |