A benzothiazolium-based fluorescent probe with ideal pKa for mitochondrial pH imaging and cancer cell differentiation†
Bo
Lin
a,
Li
Fan
 a,
Ying
Zhou
a,
Jinyin
Ge
a,
Xueli
Wang
b,
Chuan
Dong
a,
Ying
Zhou
a,
Jinyin
Ge
a,
Xueli
Wang
b,
Chuan
Dong
 a,
Shaomin
Shuang
a,
Shaomin
Shuang
 *a and
Man Shing
Wong
*a and
Man Shing
Wong
 *b
*b
aCollege of Chemistry and Chemical Engineering, Shanxi University, Taiyuan, 030006, China. E-mail: smshuang@sxu.edu.cn; Fax: +86-351-7018842; Tel: +86-351-7018842
bDepartment of Chemistry and Institute of Advanced Materials, Hong Kong Baptist University, Hong Kong Special Administrative Region, China. E-mail: mswong@hkbu.edu.hk
First published on 28th September 2020
Abstract
A mitochondrial pH sensing fluorescent probe namely 2-(2-(6-hydroxynaphthalen-2-yl)vinyl)-3-(6-(triphenyl-phosphonio)hexyl)benzothiazol-3-ium bromide (HTBT2) was designed and facilely synthesized via the Knoevenagel condensation reaction. HTBT2 displayed a linear fluorescence enhancement at 612 nm in response to pH changes between 8.70 and 7.20. The pKa value was determined to be 8.04 ± 0.02, which might be ideal for mitochondrial pH (pHmito ∼8.0) detection. HTBT2 also exhibited a remarkable large Stokes shift of 176 nm, which could diminish the interference of excitation light. The results of live cell imaging studies suggested that HTBT2 showed excellent targeting ability for mitochondria. Importantly, it was successfully applied to visualize mitochondrial pH changes in live cells and differentiate the pHmito difference between cancer cell lines and normal cell lines. Our results consistently supported that HTBT2 held practical promise for the investigation of physiological processes related to pHmito changes and clinical potential for cancer cell differentiation.
Introduction
Various complex physiological processes in eukaryotic cells can be implemented through a high level of compartmentalization via independent division and/or close cooperation between subcellular organelles. Compartmentalization is regarded as the result of evolving to provide distinct environmental conditions for matching the requirements from the optimal individual metabolic process, and to store energy via electromotive force between inside and outside the membrane.1 Each organelle with different function requires a specific physiological pH condition to facilitate its metabolic process. For instance, lysosome with pH 4.5–5.0, Golgi with pH 6.0–6.7 and endosome with pH 5.5–6.5 are typical acidic organelles; the pH of the cytoplasm (6.8–7.2) and endoplasmic reticulum (∼7.2) is nearly neutral.2–5 However, the mitochondria pH (pHmito) environment is markedly alkaline (∼8.0) in resting cells resulting from proton (H+) extrusion in the process of the respiratory electron transport chain reaction.2–4,6 It is noted that information transmission, Na+/K+/Ca2+ exchange and homeostasis, reactive oxygen species (ROS) production and bioenergy are closely related to pHmito.4,7 In particular, mitochondrial autophagy (mitophagy) and apoptosis are often associated with the acidification of mitochondria.6,8,9 Moreover, some reports indicated that various pathological conditions are closely related to abnormal levels of mitophagy, for instance, neurodegenerative diseases, cardiovascular diseases, Parkinson's disease, Reye's syndrome and Alzheimer's disease.8,10,11 Consequently, a reliable and accurate method for monitoring pHmito fluctuation in situ holds promise to elucidate mitochondria related physiology and diseases.Fluorescent probes, particularly small molecular probes, have recently attracted great attention in bioanalysis and real-time bioimaging technologies in the imaging of live cells and organisms due to their excellent sensitivity and high spatial and temporal resolution. Cationic molecular probes have been found to be desirable for targeting mitochondria because the mitochondrial inner membrane potential is negative.2–4,6,12 Tang et al.13 reported the first mitochondrial pH molecular probe (spring red, pKa = 6.3) based on the quaternary ammonium ion as a targeting group and tertiary amines as a pH responsive site, which provided a new strategy for the mitochondrial pH measurement. Subsequently, various mitochondrial pH fluorescent probes using tertiary amines to respond to the pH alterations have been developed successively.10,14–17 However, the pKa values of these probes are far from ideal for pHmito (∼8.0) measurement because tertiary amines are only sensitive to pH variations in an acidic environment, which poses a sensitivity issue. Song et al.18 synthesized a fluorescein probe with a carboxyl group as a sensing site to respond to the pH changes for pHmito detection; nevertheless, the pKa (5.77 ± 0.03) of this probe is far from satisfactory. To improve sensitivity, Qi et al.19–23 prepared fluorescein or rhodamine-based probes by introducing cycloimide as a pH responsive group, which raised the pKa of the probe close to the neutral range. By employing the phenolic hydroxyl groups as a pHmito-sensing group, several pHmito probes with higher fluorescence yield and wider monitoring range have been reported.6,24–29 The pKa values of these probes respectively were 5.88,24 6.79,26 8.85,25 7.33,27 7.33,28 and 7.25,29 which were far away from mitochondrial pH. Very recently, Zhang's group30 and Wu's group31 reported two probes utilizing tertiary amines and phenolic hydroxyl groups as pH-sensitive sites, respectively. However, these pKa values were still much lower than mitochondrial pH. Fortunately, Sarkar et al.6 reported a 2-naphthol-derived probe, namely CMP1, exhibiting a pKa value of 7.86 ± 0.05 in living cells. Furthermore, pHmito changes in living cells introduced by carbonyl cyanide m-chlorophenyl hydrazone treatment were successfully observed with CMP1. However, to date, very few pH fluorescent probes with an ideal pKa value (∼8.0) for mitochondrial imaging in live cells have been reported.
Benzothiazole dyes have been extensively used as various fluorescent probes due to their excellent optical performance, such as excellent photostability, large molar extinction coefficients and Stokes shift. Recently, we have constructed a ratiometric pH fluorescent probe (BTNO) by conjugated double bond bridged benzothiazole and binaphthol.32BTNO displays a pKa (7.91 ± 0.03) nearing mitochondrial pH, which was attributed to the use of phenolic hydroxyl groups as the pH sensitive group. Unfortunately, BTNO cannot be used to specifically detect mitochondrial pH because its structure did not contain the mitochondrial targeting group. Herein, we designed and synthesized a novel biocompatible fluorescent probe HTBT2 (Scheme 1) by introducing triphenylphosphonium (TPP) as a mitochondrial targeting unit into compound BTNO. Besides, owing to the enhanced intramolecular charge transfer (ICT) process from the naphthol functionality to benzothiazolium, HTBT2 exhibits a larger Stokes shift and longer emission wavelength compared with BTNO. Under neutral and acidic conditions, HTBT2 emits a strong orange-red fluorescence, while under alkaline conditions, naphthoxide (TBT) was formed via the deprotonation of the hydroxyl group, then the ICT process and the fluorescence quenched due to the conversion of naphthoxide to naphthoquinone (OBT), in which charge separation in the conjugate structure is hardly stable. The fluorescence intensities of HTBT2 at 612 nm and pH values exhibited a linear relationship in the range from pH 8.70 to 7.20. Its pKa value was found to be 8.04 ± 0.02, which matched well with mitochondrial pH (pHmito ∼8.0). Importantly, HTBT2 showed potential to detect the pHmito fluctuations in live cells and differentiate the pHmito differences between cancer cells and normal cells. These results consistently suggested that HTBT2 could be practically effective to investigate physiological processes associated with pHmito alterations.
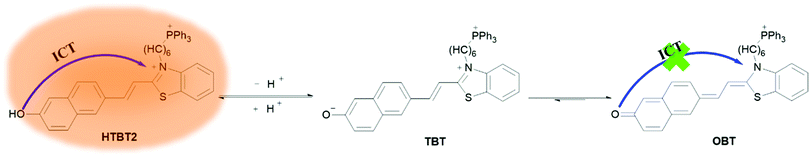 | ||
| Scheme 1 Molecular structure of HTBT2 in acidic and alkaline environments as well as its sensing mechanism for pH changes. | ||
Results and discussion
Effect of pH on spectroscopic properties of HTBT2
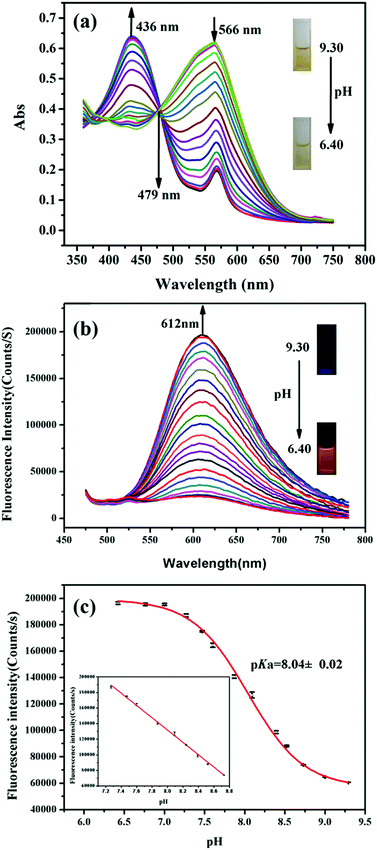
The pH-dependent absorption properties of HTBT2 were investigated in the DMSO/Tris–HCl buffer (1/2, v/v) system under different pH conditions. As depicted in Fig. 1a, HTBT2 exhibited a strong absorption band around 566 nm (ε1 = 2583 L mol−1 cm−1) at pH 9.30. Upon decreasing pH from 9.30 to 6.40, the absorption band blue-shifted progressively from 566 nm to 436 nm (ε2 = 2647 L mol−1 cm−1), giving rise to a well-defined isosbestic point at 479 nm. Concomitantly, the colour of the solution changed from yellow to yellow-green. Such a blue-shift phenomenon was attributed to the weaker π-electron delocalization in the protonated state of HTBT2. It should be noted that the peak at 556 nm still existed when the pH value was 3.0, which could be attributed to the ionization of HTBT2 (Fig. S2, ESI†).The fluorescence spectra of HTBT2 at pH 6.40 (Fig. S1, ESI†) indicated that the excitation and emission maxima of HTBT2 are 436 and 612 nm, respectively. The fluorescence spectra of HTBT2 at various pH (Fig. 1b) showed that almost no fluorescence was observed at pH 9.30. However, a new emission band at ∼612 nm appeared with a gradual increase in intensity upon the decrease of pH from 9.30 to 6.40. Meanwhile, the orange-red fluorescence of solution enhanced gradually upon decreasing pH. Moreover, the lifetimes of HTBT2 at various pH values neared 7.01 μs (Table S1, ESI†). The fluorescence enhancement as a result of the formation of the protonated HTBT2 improves the ICT process under acidic conditions. The fluorescence quantum yield of HTBT2 was found to be 0.112 (DMSO) relative to Rhodamine 6G (Φ = 0.95 in water). It is worth noting that HTBT2 showed a remarkably large Stokes shift of 176 nm, which could effectively reduce the excitation interference and then drastically improve the sensitivity of measurements.
To determine the pKa of HTBT2, Boltzmann function fitting of the pH-dependent fluorescence intensity at 612 nm was adapted. As shown in Fig. 1c, the pKa was found to be 8.04 ± 0.02, which was very close to the pH of the mitochondrial matrix (∼8.0). There was a good linearity in the pH range of 8.70 to 7.20, based on the regression equation: F = 764314.81192 − 79080.76184 × pH with R2 of 0.9991. These results indicated that HTBT2 could be potentially suitable for detecting mitochondrial pH variations.
Mass spectra and 1H NMR spectra of HTBT2 assays with pH changes
To confirm the sensing mechanism of HTBT2, mass and 1H NMR titration experiments were implemented. The results of the mass spectra titration assays were displayed in Fig. S3 (ESI†), and the strong characteristic peak of HTBT2 at 324.6273 and the negligible characteristic peak of OBT at 648.2699 were measured when the pH of the aqueous solution containing the probe was 6.40. The intensities of the peak at 314.6173 progressively decreased and the intensities of the peak at 648.2699 gradually increased as pH increased from 6.40 to 8.30, which was attributed to the conversion of HTBT2 to OBT in the process. As shown in Fig. S4 (ESI†), the chemical shifts of 1H in HTBT2 (in d6-DMSO) at 10.02 ppm disappear step by step with pH increased from 6.40 to 9.30 in the 1H NMR spectra, which indicated that the mechanism of the probe sensing pH changes was achieved by the protonation and deprotonation of phenolic hydroxyl groups. Meanwhile, some of the proton resonances were clearly shifted upfield. Particularly, 2H vinyl proton significantly shifted upfield to 4.93 ppm. The upfield shift of those protons suggested that the generated naphthoxide via phenolic hydroxyl deprotonation was quickly transformed to naphthoquinone leading to the increase of electron density around those protons. The transformation was attributed to the fact that charge separation in the conjugate structure is extremely unstable. Thence, the above results indicated that deprotonation and protonation occurred on the phenolic hydroxyl of HTBT2 and the generated TBT was quickly converted to OBT, providing a sensing mechanism for pH changes.Selectivity, photostability and reversibility of HTBT2
Considering the complexity of measured environments in live cells, the accuracy of pH measurement could be affected by various ions and biomolecules in live cells. Thus, the selectivity of HTBT2 to OH− in the presence of other potential interfering species was studied at pH 6.40. The result depicted in Fig. S5 (ESI†) indicated that physiologically common metal cations, such as K+, Li+ and Mg2+, along with other transition and heavy metal ions including Co2+, Hg2+, Ni2+, Ba2+ and Cd2+, hardly caused noticeable emission intensity changes in the measurements. Furthermore, some familiar anions (such as Cl−, Br−, I−, SO42−, S2O32−, SO32−, HS−, NO3−, NO2−, Ac−, HCO3− and ClO4−) and some other important substances (e.g. H2O2, HClO, O2−, NO, ONOO−, L-glutathione, homocysteine and cysteine) exhibited negligible effect. These facts clearly indicated that HTBT2 showed outstanding selectivity toward pH changes in the presence of background ions and biologically important species.Subsequently, HTBT2's stability was investigated via monitoring the fluorescence intensity change for 2 h in continuous light of 436 nm at room temperature. The fluorescence responses of HTBT2 against time at pH 6.40 and 9.30 are shown in Fig. S6 (ESI†). As seen, the photostability HTBT2 was very good at the measured pH range for 2 h.
The reversibility was one of the important prerequisites for real-time monitoring pH variations in actual biological samples. Subsequently, the pH value was modulated repeatedly between 6.40 and 9.30 by adding trace volume of HCl (1 M) and NaOH (1 M), and HTBT2's fluorescence intensities were recorded at every moment. As clearly shown in Fig. S7 (ESI†), the response of HTBT2 was fully reversible. Therefore, HTBT2 is practically effective for use in tracking pH alterations in the real-time scale.
Cytotoxicity assessment
To assess the cytotoxicity of the probe to live cells, the IC50 value was determined by using the MTT assay.33 The IC50 value of HTBT2 was obtained as 64.48 μM (Fig. S8, ESI†). This result clearly demonstrated that the probe showed low toxicity and is suitable for cell imaging experiments under the concentrations of 10 μM.Fluorescence imaging in live cells
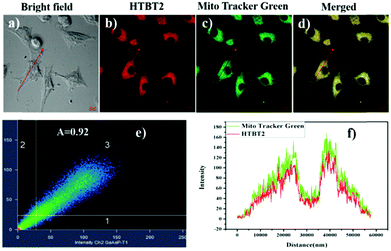
Since mitochondria are the typical weakly alkaline organelles in cells, it is practically useful for HTBT2 to sense the alkalinity if it can show subcellular localization of mitochondria. To assess the mitochondria staining ability of HTBT2, we conducted co-localization studies on SMMC-7721 cells with MitoTracker Green, a commercial mitochondria-specific green fluorescent dye. The bright red fluorescence images from HTBT2 (Fig. 2a) merged well with those from MitoTracker Green (Fig. 2b–d), with a large mean Pearson's co-localization coefficient (A) of 0.92. Meanwhile, the changes in the intensity profile of HTBT2 and MitoTracker Green in the linear regions tended toward synchronization (Fig. 2f). These results suggested that HTBT2 selectively accumulated in mitochondria of SMMC-7721 cells, in addition to showing good cell membrane permeability (Fig. S9–S11, ESI†).To explore the capability of HTBT2 for imaging mitochondrial pH variations, the pH-dependent fluorescence images of HTBT2 in live cells were investigated. As demonstrated in Fig. 3, the red fluorescence intensity in the SMMC-7721 cells gradually decreases upon increasing the pH from 7.0 to 9.0 and there is only little fluorescence detected at pH 9.00. These results meant that the HTBT2 probe could effectively measure the mitochondrial pH in live cells. Moreover, the optical stability of the probe in the living cells at these pH values was also measured. As shown in Fig S12 (ESI†), the fluorescence intensities in cells decreases slightly (<5%) at various pH. The phenomenon revealed that HTBT2 possesses the potential to monitor mitochondrial pH changes in living cells for a long time.
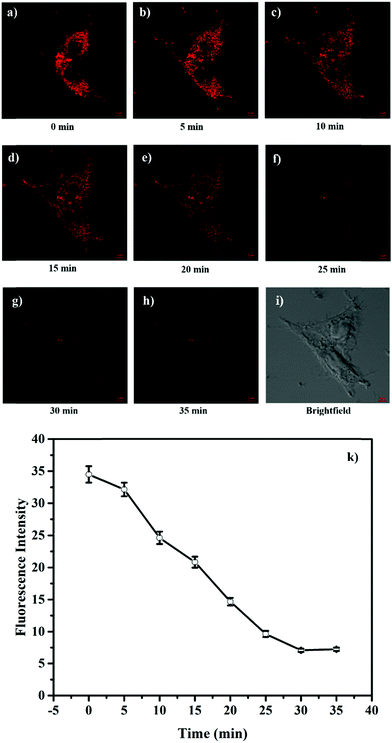
The attractive mitochondria specificity and pH responsive ability of HTBT2 urged us to further investigate the monitoring capability of pHmito changes in real time upon different stimulation. After incubation with 10 μM HTBT2 for 10 min and washing three times with PBS, SMMC-7721 cells were further treated with 5 mM NH4Cl, which could induce a dramatic increase in pHmitovia neutralizing acid in the cells.34 As depicted in Fig. 4 and Fig. S13 (ESI†), the fluorescence intensities in NH4Cl-treated cells faded gradually within 25 min, and the changes of the fluorescence intensities from these cells after 30 min were negligible. This fact suggested that HTBT2 exhibited a fast response to the alkalization in mitochondria.
It was reported that reactive oxygen species (ROS) as signalling molecules of apoptosis could be generated when the homeostasis of mitochondrial redox was destroyed, concomitant with variations in mitochondria pH.35 In order to examine the relationship between mitochondrial pH and oxidative stress, SMMC-7721 cells stained with HTBT2 were treated with 0.1 mM H2O2 and 1 mM NAC for 1 h, respectively. As depicted in Fig. S14 (ESI†), compared with non-treated cells (the second row in Fig. S14d–f, ESI†), the mean fluorescence intensity (Fig. S14a–c, ESI†) in H2O2-treated cells obviously enhanced, unambiguously implying that the mitochondrial pH decreased. On the other hand, cells treated with NAC resulted in a slightly increased mitochondrial pH (Fig. S14g–i, ESI†). All of the above results clearly demonstrated that HTBT2 could be employed for real-time detection of mitochondrial pH fluctuations in live cells.
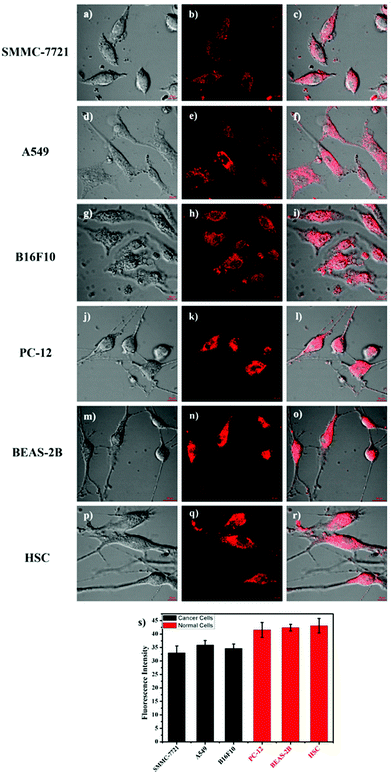
It is well known that some diseases are closely related to mitochondrial pH abnormality, such as cancer.8,10,11,36,37 To investigate the application and capability of HTBT2 in differentiating the pHmito of cancer and normal cell lines by fluorescence imaging, three types of cancer cell lines including SMMC-7721 (human liver cancer cells); A549 (human alveolar cancer cells); and B16F10 (mouse melanoma cells) and three types of normal cell lines including PC-12 (rat neural cells); BEAS-2B (normal human alveolar cells); and HSC cells (human hematopoietic stem cells) were selected for the studies. Fig. 5 showed that the fluorescence intensities in the three normal cells lines were stronger than those of three cancer cells lines, consistently confirming that the pHmito in the cancer cells was higher than that in the normal cells.38 Interesting, the fluorescence in cancer cells can be neglected when the thresholds were raised from 0.4 to 82 (Fig. S15, ESI†), while the fluorescence in normal cells can be observed easily. Furthermore, the results of dynamic assays shown that the time required for HTBT2 to stain BEAS-2B cells (∼12 min, Fig. S10, ESI†) and pc-12 cells (∼13 min, Fig. S11, ESI†) was slightly shorter than that of SMMC-7721 cells (∼18 min, Fig. S9, ESI†). Our results strongly supported that HTBT2 showed a great clinical potential for differentiating cancer cells from normal cells.
Conclusion
In summary, a benzothiazolium-based mitochondrial pH fluorescent probe HTBT2 was facilely synthesized via Knoevenagel condensation reaction, for real-time visualization of mitochondrial pH fluctuations in live cells. HTBT2 exhibited not only a significant Stokes shift of 176 nm, but also excellent selectivity toward slight pH changes, good reversibility and photostability, and low cytotoxicity. It is noted that the pKa value of the probe (8.04 ± 0.02) matched pHmito (∼8.0) well with a linear response range between pH 8.70 and 7.20. In addition to the excellent mitochondrial targeting ability, HTBT2 showed an ability to detect and monitor mitochondrial pH fluctuations in live SMMC-7721 cells. Impressively, it was also successfully applied to differentiate the pHmito between cancer cell lines and normal cell lines. Therefore, HTBT2 as the pH imaging probe offers practical potential to investigate physiological processes inside mitochondria.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The work was supported by the National Natural Science Foundation of China (No. 91843301, 21874087 and 81901814), the Key R&D project of Shanxi Province (201803D421031), Talents Project and General Research Fund (GRF) (HKBU 12301317) from the Research Grant Council of Hong Kong. We also appreciate Lanqi Huang, Di Xu, Tongxin Zhang and Wu Chun from Hong Kong Baptist University, Hong Kong for assisting with experiments.Notes and references
- E. Guzel, S. Arlier, O. Guzeloglu-Kayisli, S. M. Tabak, T. Ekiz, N. Semerci, K. Larsen, F. Schatz, J. C. Lockwood and A. U. Kayisli, Int. J. Mol. Sci., 2017, 18, 792 CrossRef.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS.
- J.-T. Hou, W. X. Ren, K. Li, J. Seo, A. Sharma, X.-Q. Yu and J. S. Kim, Chem. Soc. Rev., 2017, 46, 2076–2090 RSC.
- P. Paroutis, N. Touret and S. Grinstein, Physiology, 2004, 19, 207–215 CrossRef CAS.
- X. Wang, L. Fan, Y. Wang, C. Zhang, W. Liang, S. Shuang and C. Dong, J. Mater. Chem. B, 2020, 8, 1466–1471 RSC.
- A. R. Sarkar, C. H. Heo, L. Xu, H. W. Lee, H. Y. Si, J. W. Byun and H. M. Kim, Chem. Soc., 2016, 7, 766–773 CAS.
- L. P. Roma, J. Duprez, Hilton K. Takahashi, P. Gilon, A. Wiederkehr and J.-C. Jonas, Biochem. J., 2012, 441, 971 CrossRef CAS.
- A. H. V. Schapira, Lancet, 2006, 368, 70–82 CrossRef CAS.
- H. Iwashita, S. Torii, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, S. Shimizu and K. Okuma, ACS Chem. Biol., 2017, 12, 2546–2551 CrossRef CAS.
- M. H. Lee, N. Park, C. Yi, J. H. Han, J. H. Hong, K. P. Kim, D. H. Kang, J. L. Sessler, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2014, 136, 14136–14142 CrossRef CAS.
- D. C. Wallace, Nat. Rev. Cancer, 2012, 12, 685–698 CrossRef CAS.
- Y. Yue, F. Huo, S. Lee, C. Yin and J. Yoon, Analyst, 2017, 142, 30–41 RSC.
- P. Li, H. Xiao, Y. Cheng, W. Zhang, F. Huang, W. Zhang, H. Wang and B. Tang, Chem. Commun., 2014, 50, 7184–7187 RSC.
- L. Gui, Z. Yuan, H. Kassaye, J. Zheng, Y. Yao, F. Wang, Q. He, Y. Shen, L. Liang and H. Chen, Chem. Commun., 2018, 54, 9675–9678 RSC.
- M. J. Chang, K. Kim, K. S. Park, J. S. Kang, C. S. Lim, H. M. Kim, C. Kang and M. H. Lee, Chem. Commun., 2018, 54, 13531–13534 RSC.
- X. Han, Z. Wang, Q. Cheng, X. Meng, D. Wei, Y. Zheng, J. Ding and H. Hou, Dyes Pigm., 2017, 145, 576–583 CrossRef CAS.
- S. Siriwibool, N. Kaekratoke, K. Chansaenpak, K. Siwawannapong, P. Panajapo, K. Sagarik, P. Noisa, R.-Y. Lai and A. Kamkaew, Sci. Rep., 2020, 10, 1283 CrossRef CAS.
- G. Li, B. Zhang, X. Song, Y. Xia, H. Yu, X. Zhang, Y. Xiao and Y. Song, Sens. Actuators, B, 2017, 253, 58–68 CrossRef CAS.
- S. Qi, Q. Li, W. Liu, H. Ren, H. Zhang, J. Wu, J. Ge and P. Wang, Spectrochim. Acta, Part A, 2018, 204, 590–597 CrossRef CAS.
- H. Yu, G. Li, B. Zhang, X. Zhang, Y. Xiao, J. Wang and Y. Song, Dyes Pigm., 2016, 133, 93–99 CrossRef CAS.
- D. Wang, Z. Wang, Y. Li, Y. Song, Y. Song, M. Zhang and H. Yu, New J. Chem., 2018, 42, 11102–11108 RSC.
- X. Bi, Y. Wang, D. Wang, L. Liu, W. Zhu, J. Zhang and X. Zha, RSC Adv., 2020, 10, 26874–26879 RSC.
- S. Xia, J. Wang, Y. Zhang, N. Whisman, J. Bi, T. E. Steenwinkel, S. Wan, J. Medford, M. Tajiri, R. L. Luck, T. Werner and H. Liu, J. Mater. Chem. B, 2020, 8, 1603–1615 RSC.
- M.-Y. Wu, K. Li, Y.-H. Liu, K.-K. Yu, Y.-M. Xie, X.-D. Zhou and X.-Q. Yu, Biomaterials, 2015, 53, 669–678 CrossRef CAS.
- B. Lin, L. Fan, J. Ge, W. Zhang, C. Zhang, C. Dong and S. Shuang, Analyst, 2018, 143, 5054–5060 RSC.
- P. Wang, J. Huang and Y. Gu, RSC Adv., 2016, 6, 95708–95714 RSC.
- L. Cao, Z. Zhao, T. Zhang, X. Guo, S. Wang, S. Li, Y. Li and G. Yang, Chem. Commun., 2015, 51, 17324–17327 RSC.
- Y. Chen, C. Zhu, J. Cen, Y. Bai, W. He and Z. Guo, Chem. Sci., 2015, 6, 3187–3194 RSC.
- L.-Q. Niu, J. Huang, Z.-J. Yan, Y.-H. Men, Y. Luo, X.-M. Zhou, J.-M. Wang and J.-H. Wang, Spectrochim. Acta, Part A, 2019, 207, 123–131 CrossRef CAS.
- H. Wang, J. Hu, G. Yang, X. Zhang, R. Zhang, K. Uvdal, Z. Zhang, X. Wu and Z. Hu, Sens. Actuators, B, 2020, 320, 128418 CrossRef CAS.
- X. Jiang, Z. Liu, Y. Yang, H. Li, X. Qi, W. X. Ren, M. Deng, M. Lü, J. Wu and S. Liang, Spectrochim. Acta, Part A, 2020, 224, 117435 CrossRef CAS.
- B. Lin, L. Fan, Z. Ying, J. Ge, X. Wang, T. Zhang, C. Dong, S. Shuang and M. S. Wong, Talanta, 2020, 208, 120279 CrossRef CAS.
- M. Li, J. Chou, K. W. King, J. Jing, D. Wei and L. Yang, J. Lab. Autom., 2014, 20, 32–45 CrossRef.
- A. K. Ho, A. Ling and C. L. Chik, J. Neurochem., 2000, 75, 1845–1851 CrossRef CAS.
- K. Nomura, H. Imai, T. Koumura, M. Arai and Y. Nakagawa, J. Biol. Chem., 1999, 274, 29294–29302 CrossRef CAS.
- E. Carafoli and I. Roman, Mol. Aspects Med., 1980, 3, 295–429 CrossRef CAS.
- L. Galluzzi, N. Larochette, N. Zamzami and G. Kroemer, Oncogene, 2006, 25, 4812–4830 CrossRef CAS.
- Y. Shi, S. K. Lim, Q. Liang, S. V. Iyer, H.-Y. Wang, Z. Wang, X. Xie, D. Sun, Y.-J. Chen, V. Tabar, P. Gutin, N. Williams, J. K. De Brabander and L. F. Parada, Nature, 2019, 567, 341–346 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: These data include experimental procedures; synthetic scheme, excitation and emission spectrum; mass and 1H NMR spectra assays, fluorescence lifetime, selectivity, photostability, reversibility and MTT assay of HTBT2; dynamic experiments of HTBT2 in SMMC-7721 cells, BEAS-2B cells and pc-12 cells; and fluorescence images of HTBT2 stained SMMC-7721 cells after being treated with H2O2, PBS and NAC. See DOI: 10.1039/d0tb01253j |
| This journal is © The Royal Society of Chemistry 2020 |