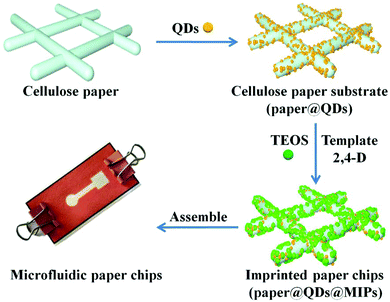
Deposition of CdTe quantum dots on microfluidic paper chips for rapid fluorescence detection of pesticide 2,4-D†
Zhong
Zhang
ab,
Xin
Ma
a,
Mengfan
Jia
a,
Bowei
Li
 c,
Jianhui
Rong
*b and
Xingbin
Yang
c,
Jianhui
Rong
*b and
Xingbin
Yang
 *a
*a
aShaanxi Engineering Laboratory for Food Green Processing and Safety Control, and Shaanxi Key Laboratory for Hazard Factors Assessment in Processing and Storage of Agricultural Products, College of Food Engineering and Nutritional Science, Shaanxi Normal University, Xi'an 710119, China. E-mail: xbyang@snnu.edu.cn; Fax: +86 29-85310580; Tel: +86 29-85310580
bSchool of Chinese Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, 10 Sassoon Road, Pokfulam, Hong Kong, China. E-mail: jrong@hku.hk; Fax: +852-39176537; Tel: +852-39176537
cKey Laboratory of Coastal Environmental Processes and Ecological Remediation, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai 264003, China
First published on 3rd December 2018
Abstract
Rapid detection of pesticides in fruits is an ongoing challenge. The objective of the present study was to develop novel fluorescent microfluidic paper chips for specific recognition and sensitive detection of the pesticide 2,4-D through the electron-transfer-induced fluorescence quenching mechanism. CdTe quantum dots (QDs) were deposited onto cellulose paper (base material) to yield imprinted paper chips (paper@QDs@MIPs). This method allows the transferability of the molecularly imprinted fluorescence sensor from the liquid phase to the solid phase (paper base) for rapid and portable analysis. The resultant imprinted paper chips were effectively characterized, and they exhibited ideal ordered spatial network structure, chemical stability, and fluorescence property. The paper@QDs@MIPs showed that 2,4-D binding significantly reduced the fluorescence intensity within less than 18 min, and it achieved satisfactory linearity in the range of 0.83–100 μM and high detectability of 90 nM. The recognition specificity for 2,4-D relative to its analogues was shown, and the imprinting factor was 2.13. In addition, the recoveries of the spiked bean sprouts at three concentration levels ranged within 94.2–107.0%, with a relative standard deviation of less than 5.9%. Collectively, the device provided an effective platform for rapid recognition, convenience, and detection of trace food pollutants in complex matrices, thereby ensuring food safety and further promoting surface imprinting studies.
Introduction
2,4-Dichlorophenoxyacetic acid (2,4-D) is a well-known plant growth regulator that facilitates the effective control of broadleaf weeds in an extensive range of crops.1 2,4-D is listed as one of the top 10 bestselling pesticides because of its promising endocrine disrupting activities and plant hormone activity.2 The accumulation of 2,4-D in agricultural products and natural environment not only causes serious pollution to the environment and ecosystem, but also jeopardizes human health, public safety, and economic development.3 Upon accumulation in food chain, indeed, 2,4-D is associated with the occurrence of human cancer, endocrine disruption, acute congestion, and neurodegeneration.42,4-D concentration has already been evaluated by different analytical methods, such as LC-MS,1,5 chromatography,2,6 electrochemistry,7 and surface-enhanced Raman spectroscopy.8 These technologies are powerful analytical techniques that provide high sensitivity and specificity. However, these detection methods generally require expensive instruments, complicated sample preparation, and time-consuming labor; further, the requirements for rapid detection hinder their further applications. Alternatively, enzyme-based biosensors appear to be a rapid, cost-effective, and simple strategy, although these methods require the separation of enzymes from the detected pesticides.9 Therefore, the convenient, rapid, and selective detection of trace amounts of 2,4-D in the environment remains a formidable challenge.
Quantum dots (QDs) have been often used as fluorescent markers in fluorescence detection techniques for good photostability, high quantum yield, narrow emission, long fluorescence lifetime, large extinction coefficient, and large Stokes shift.10,11 QDs are widely noted as an effective and attractive fluorescence sensor for the analysis of metal ions,12 small molecules,13 and even biological macromolecules.10,14 Chen's group constructed a new ratiometric fluorescence sensor for visually identifying and detecting 2,4-D in water samples, combining green nitrobenzoxadiazole (NBD) fluorescent dye and red QDs embedded in silica nanoparticles as signal sources. The sensor can be used for the selective and sensitive determination of 2,4-D.15
Molecular imprinting technology (MIT) appears to be able to enhance the selectivity of QD-based fluorescence sensors in the detection of specific target substances in a highly sensitive and selective manner.16–18 Molecularly imprinted polymers (MIPs) were tailored by selectively forming a 3D polymer network structure with complementary size, shape, and functional recognition sites for specific target analytes.19 Such an advantage of MIT leads to the development of a novel surface imprinting technology for overcoming the shortcomings of traditional MIPs, such as incomplete template removal, small binding capabilities, low affinity, and irregular material shapes.18,20,21 Moreover, surface imprinting techniques have been extensively studied based on core materials such as silica nanoparticles,22 magnetic Fe3O4 particles,23 nanotubes,24 and polystyrene beads.25 Therefore, surface imprinted polymers exhibit better onsite accessibility, faster mass transfer rate, and complete template removal.26–28
MIP-based fluorescence sensors were constructed on cellulose paper for its relative low cost, convenience, and availability.29,30 Microfluidic paper-based analytical devices (μPADs) have recently attracted increased attention owing to their several benefits, such as miniaturization, automation, and integration. A microfluidic paper chip is made of paper as the base material, which has several advantages.31 Firstly, a paper chip is rich in source, low in cost, and easy to store. Secondly, paper is porous and is a hydrophilic fiber material with many hydroxyl groups on the surface, is easy to modify, has a white background with low detection background, and can initiate flow in the sample by capillary force without necessitating pumping. Finally, cellulose paper has good biocompatibility for DNA and protein, and the post-treatment is very simple, environmentally friendly, and non-polluting.32–34 Ngo et al.35 studied the effects of gold, silver, and titanium dioxide nanoparticle properties on nanoparticle-functionalized paper. Dou et al.36 performed a Raman test on pig hair extract using a paper chip. The use of MIPs employing 2,4-D as analysts has been reported.2,8,37 Such methods generally require complicated chemical synthesis or modification, and the analysis time is longer. It is still very necessary to develop a rapid detection method in monitoring this field. However, there are a few reports on the combination of MIT with microfluidic paper chips.
Inspired by these studies, we developed novel CdTe QDs embedded with MIPs on 3D origami microfluidic paper chips that can directly monitor 2,4-D based on the electron-transfer-induced fluorescence quenching mechanism. CdTe-QDs-embedded cellulose paper acted as the support material, and the imprinted polymer layer was deposited onto the paper to yield an extraordinary MIP-based fluorescence sensor named paper@QDs@MIPs. Paper@QDs@MIPs was characterized in detail, and its preparation process, detection sensitivity, response time, and selectivity were systematically studied. Moreover, microfluidic paper chips were successfully applied to complex bean sprout samples with satisfactory results, proving their extraordinary potential toward the fast detection, low cost, and accurate quantification of 2,4-D in complex matrices.
Experimental
Reagents and chemicals
2,4-D, 3-aminopropyltriethoxysilane (APTES), 2-N-morpholinoethanesulfonic acid (MES), tellurium powder, 3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), cadmium nitrate (Cd(NO3)2), carbendazol, and sodium borohydride (NaBH4) were obtained from Sigma-Aldrich (Shanghai, China). Whatman No. 1 filter cellulose paper was acquired from GE Company (Shanghai, China). Thioglycolic acid, toluene, phenol, ethanol, and other chemical reagents were of at least the analytical grade and used directly without further purification, unless otherwise specified. All the aqueous solutions used in this work were prepared with ultrapure water (18.2 MΩ), which was obtained using a Millipore Milli-Q Plus system (Millipore, Bedford, MA, USA).Synthesis of carboxylated water-soluble CdTe QDs
The carboxylated water-soluble CdTe QDs were prepared according to a previously reported method.38 Finally, carboxylated CdTe QDs were obtained and stored for use in the dark.Preparation of fluorescence-sensing cellulose paper substrate (paper@QDs)
Cellulose paper was cut into 1 cm × 1 cm pieces (10 sheets per batch). Each cellulose paper was dipped in a culture dish and about 40 mL of 0.2 mol L−1 HCl solution was added. After activating treatment for 20 min, rinsing was carried out three times with distilled water to remove the excess HCl solution. Then, these papers were put in 40 mL of 50% (v/v) ethanol/water solution with 400 μL APTES for 2.5 h. Finally, the excess ethanol was removed with distilled water to obtain cellulose paper with amino groups. During the operation, care was taken to maintain each piece of cellulose paper on the same side.Next, 18 mL of 20 mg mL−1 EDC solution dissolved with MES solution (pH = 5.2) was added to 30 mL CdTe QDs solution. After mixing for 10 min, 18 mL of 10 mg L−1 NHS solution dissolved with MES (pH = 5.2) was added. Further, the CdTe QDs solution was poured into a culture dish with a cellulose paper sheet and soaked for 1 h, followed by shaking at room temperature for 12 h under light protection. Finally, the fluorescence-sensing cellulose paper substrate was washed 3 times in 0.01 mol L−1 phosphate buffer solution (pH = 7.0).
Preparation of imprinted paper chips (paper@QDs@MIPs)
Here, 10 mg of 2,4-D was taken in a culture dish and 20 mL phosphate buffer was added and mixed well. Then, 40 μL APTES solution was added and prepolymerization was initiated by shaking for 20 min. Then, the cellulose paper substrate grafted with CdTe QDs (2 to 3 sheets per culture dish) was immersed, taking care to maintain the paper sheet to be in the upward position. Then, 50 μL TEOS solution was added and 50 μL ammonia solution was shaken for 30 min. The grafted 2,4-D imprinted paper chips (paper@QDs@MIPs) were shaken for 4 h.Finally, the imprinted paper was rinsed upon removal. The resulting fluorescent imprinted paper chips were washed with distilled water to remove the template and impurities on the surface, maintaining the same side up, immersed in a mixture solution of methanol and 0.01 M acetic acid (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v), and shaken for 2 h; the eluent solution was changed every hour. Then, the paper chip was immersed in distilled water, shaken for 10 min, and the excess methanol and acetic acid solution were repeatedly removed 3 times. The prepared imprinted paper chips were placed in a refrigerator at 4 °C.
2, v/v), and shaken for 2 h; the eluent solution was changed every hour. Then, the paper chip was immersed in distilled water, shaken for 10 min, and the excess methanol and acetic acid solution were repeatedly removed 3 times. The prepared imprinted paper chips were placed in a refrigerator at 4 °C.
For comparison, a control group of paper@QDs@NIPs was prepared using the same procedure and under the same conditions without the addition of the 2,4-D molecule.
Preparation of microfluidic paper chips
The microfluidic chip was designed using Adobe Illustrator software and then directly printed on cellulose paper at a resolution of 2400 dpi through a wax printer. The printed paper was placed in an oven at 150 °C for 20 s to allow the wax to completely penetrate into the paper to form a hydrophobic barrier. The 3D origami microfluidic paper chips were composed of three layers, as shown in Fig. S1.† The size of the chips were designed to be 5.5 cm × 2.7 cm, diameter of the sample inlet was 0.6 cm, and distance between the sample inlet and detection zone (8 × 8 mm2) was 1.2 cm. The central sample cell was attached to the fluid channel in the uppermost layer, and the wax hydrophobic barrier could draw the fluid sample from the sample cell to the detection zone by capillary force without requiring any external force. The middle layer was patterned and an 8 × 8 mm2 cavity was cut with a knife to fit the paper@QDs@MIPs assembly. The third layer was completely printed and then assembled with paper@QDs@MIPs.33Characterization of imprinted paper chips
The prepared imprinted paper chips were treated with a gold spray, and the morphology was observed with an environmental scanning electron microscope. Infrared spectrum scanning was performed to observe changes in the groups at each step of the modification. To examine the thermal stability, thermogravimetric analysis was performed to study the relationship between the quality of imprinted paper chips and temperature changes. In addition, the fluorescence properties of the prepared imprinted paper chips were examined. Fluorescence spectrophotometer determination conditions: excitation wavelength, 350 nm; emission wavelength, 450–650 nm; excitation and emission wavelength slit width, 5 nm. Fluorescence images of the microfluidic paper chips were observed using a fluorescence confocal microscope (FV1200, Olympus) equipped with a CCD camera.Applications of microfluidic paper chips
Bean sprout samples were used to determine the actual application of the microfluidic paper chips on the detection of 2,4-D. After removing the bean sprouts skin and washing for 2–3 times, yellow bean sprouts and mung bean sprouts were rinsed with fresh water and then squeezed into a juice with a juicer, passed through a 0.45 μm aqueous phase filter, and finally diluted with water to 100 times and then spiked. Bean sprout juice and green bean sprout juice were spiked with 2,4-D at three concentration levels, namely, 5, 10, and 20 μmol L−1. Then, 50 μL solution was added dropwise to the detection zone of the microfluidic paper chip, and the measurement was carried out with a fluorescence spectrophotometer to determine and evaluate the practicality of the method in the actual complex samples. At the same time, the same samples were checked using HPLC as the control test.Results and discussion
Preparation of microfluidic paper chips
The key to preparing 3D microfluidic paper chips with good fluorescence recognition performance is to ensure that the basic component of the imprinted paper chip has a good structure and function. The main preparation processes (Fig. 1) were divided into three parts: (1) CdTe QDs grafting on the surface of modified cellulose paper, (2) formation of imprinted shell on the surface of paper@QDs, and (3) assembly of the imprinted paper chips into microfluidic channels. In order to prepare an imprinted paper chip with good fluorescence performance, it was necessary to investigate the major influence factors including the ratio of the template molecule to the functional monomer, amount of crosslinking agent and QDs, catalytic grafting method of CdTe QDs, and elution reagent. These influence factors were closely related to the fluorescence properties of the paper chips. For example, if the amount of crosslinking agent is too small, CdTe QDs cannot be fixed on the surface of the paper chip and the QDs can easily peel off from the paper. Moreover, the number of imprinting sites formed is limited. If there are too many crosslinking agents, the imprinting layer becomes too thick to block the fluorescence of the CdTe QDs, which will weaken the fluorescence of the paper chip; it is also disadvantageous to allow the interaction between the CdTe QDs and template molecules, which lowers the sensor's sensitivity. Through a few steps of processing, an imprinted paper chip with excellent fluorescence performance can be achieved.Conventional paper chips are usually based on enzyme-linked immunosorbent assays, and recent paper chips based on colloidal gold color change have been developing rapidly.39–41 The design of 3D origami microfluidic paper chips is shown in Fig. S1.† Briefly, a 3D origami microfluidic paper chip mainly consists of three layers of paper. The top layer comprises a central sample inlet and detection zone connected by a fluid channel that facilitates the inhalation of the fluid from the sample inlet to the detection zone by capillary forces. The middle layer is pre-patterned and a square cavity is measured and cut to fit the assembly of the paper@QDs@MIPs component. The bottom layer is completely printed with a hydrophobic material and assembled with the paper@QDs@MIPs component to maintain good contact with the square area of the top layer, as shown in Fig. S1A and B.† Finally, the 3D microfluidic paper chip was assembled using clamps (Fig. S1C†). The prepared imprinted paper chip can be assembled in a prefabricated microfluidic chip to form a cellulose paper chip with a 3D structure.
Characterization of imprinted paper chips
The morphological structures of the original cellulose paper (paper), cellulose paper substrate (paper@QDs), and imprinted paper chips (paper@QDs@MIPs) with an imprinted layer were observed by SEM, as shown in Fig. 2. From the image shown in Fig. 2A, it was found that the original bare paper had a large number of cellulose bundles that intersect to form a 3D ordered spatial network structure, while the cellulose bundle surface was smooth. However, many nanosized spherical particles appeared on the cellulose bundle of paper@QDs (Fig. 2B). The distribution of nanoparticles on the cellulose bundle was relatively uniform. By ultraviolet light irradiation, the surface of paper@QDs had obvious yellow fluorescence. The fluorescence emission wavelength was also about 566 nm, which was consistent with the emission wavelength of CdTe QDs. These results indicate that QDs can be successfully grafted onto the surface of cellulose paper by catalyzing the reaction of carboxyl group and amino group to form an amide bond. As evident from paper@QDs@MIPs (Fig. 2C), many large agglomerates appeared on the fiber bundle, indicating that the imprinted shell layer was successfully synthesized on the fiber bundle. The fiber structure of the cellulose paper was effectively preserved and exhibited stable properties during each polymerization process. The inset in Fig. 2C shows the photographs of the prepared MIPs under UV light. Evidently, the yellow fluorescence of paper@QDs@MIPs was very obvious, which can be used for the fluorescence detection of target molecules. This result indicated that paper@QDs@MIPs with a hybrid structure was successfully synthesized. Cellulose paper was used as a support platform, CdTe QDs were used as fluorescence signal units, and the outermost molecularly imprinted thin layer exhibited specific recognition performance.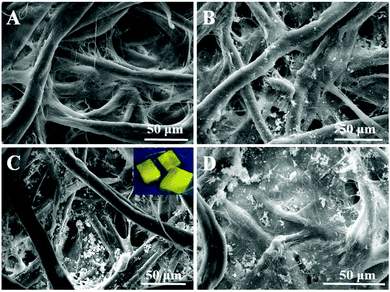 | ||
| Fig. 2 SEM images of (A) cellulose paper, (B) paper@QDs, (C) paper@QDs@MIPs, and (D) paper@QDs@NIPs. Inset photographs show the paper@QDs@MIPs under a UV lamp. | ||
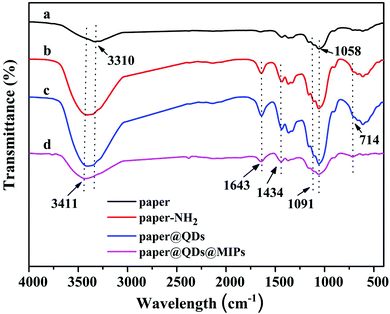
FT-IR spectroscopy was used to determine whether each step of the reaction carried out on the paper was successfully grafted or not. As shown in Fig. 3a, the peaks at 1110 cm−1 and 1058 cm−1 were strong, which was a stretching vibration peak caused by –C–O–C, and they correspond to the characteristic peak of the cellulose paper itself. The Si–O–Si at 1091 cm−1 had a wide antisymmetric stretching vibration, and 714 cm−1 was the peak of the Si–O symmetric stretching vibration peak. These characteristic peaks were very obvious in Fig. 3b and c, indicating that there were a large number of Si–O bonds on cellulose paper, which proved that the modification of the amino group on the paper by the silylating agent APTES was successful. At the same time, except for Fig. 3a, in Fig. 3b–d, the peaks of 3411 cm−1 and 1643 cm−1 appeared simultaneously, which correspond to the bending vibrations of the N–H bond, instead of the hydroxyl group of 3310 cm−1. This is because the hydroxyl group has no peak at 1643 cm−1. This symbol shows that a large amount of N–H was contained, except for bare paper. The C–N vibration absorption peak was 1434 cm−1 owing to the fact that the amide bond formed by the carboxyl group reacts with the amino group. Since only CdTe QDs have a carboxyl group, the presence of the characteristic peaks of the amide bond confirmed the successful grafting of CdTe QDs. In the paper@QDs@MIPs spectrum, because the imprinted material synthesized onto the surface of the paper@QDs, the Si–O characteristic peak was found further enhanced, and the vibration absorption peak of the CdTe QDs was weakened. These FT-IR results proved that the grafting reaction of each step had reached the ideal result.
EDX mapping analysis was carried out to further confirm whether the modification was successful. As shown in Fig. S2,† the paper sample had only three peaks. One peak was the gold sprayed during the sample prepared to facilitate conduction, so the paper sample had only two characteristic peaks. The results are listed in Table S1.† It was proven that there were only carbon and oxygen elements in the original paper sample, and Si and Cd elements were added to the paper@QDs@MIPs sample. The presence of oxygen (45.71%) and carbon (51.13%) came from the base material of the paper, Si (0.34%) was derived from the imprinted layer material, and Cd (2.82%) was derived from CdTe QDs, demonstrating that both these materials were grafted onto the paper. Therefore, the currently used grafting method can conveniently modify the CdTe QDs and the imprinted shell layer on paper.
In addition, thermogravimetric curves have been applied to investigate the thermal stability of imprinted paper chips. Thermal stability is also a very significant property of paper chips in actual use. The results are shown in Fig. S3† for the TG and DTG curves for paper@QDs@MIPs and paper@QDs@NIPs. As the temperature increased from 30 °C to 100 °C, the weight losses of paper@QDs@MIPs and paper@QDs@NIPs were mainly attributed to the volatility loss of moisture absorption, and the mass loss was 4.74% and 5.69% for MIP and NIP, respectively. In the range of 100–300 °C, there was almost no weight loss in both the paper chips, which indicated that the imprinted and nonimprinted paper chips have effective thermal stability at temperatures less than 300 °C. As the temperature continues to increase, the quality of the paper chip rapidly decreased in the temperature range of 300–400 °C; this may be due to the decomposition of the organic material in the paper chips. The fastest degradation rate of the imprinted paper chip is at 361.7 °C. The mass was stable at 400 °C. The residual amounts for the imprinted and nonimprinted paper chips were 29.1% and 9.84%, respectively, and the detailed transition temperature can also be obtained from the peak temperature in the DTG curve. After the temperature continued to increase to 800 °C, the quality of the two remains basically unchanged. Therefore, it can be concluded that the obtained paper@QDs@MIPs and paper@QDs@NIPs possessed favorable thermal stability at temperatures below 300 °C.
Fluorescence properties of imprinted paper chip
Fluorescence CdTe QDs (green) were grafted onto the paper chip and covered with a surface synthetic imprinting layer. The ability of the imprinted paper chip to continue to emit strong fluorescence is a critical issue, which was closely related to the recognition and sensitivity of the paper chip sensor. The CdTe QDs and the imprinted paper chip had similar fluorescence spectra, and the fluorescence intensity of the imprinted paper chip was slightly lower than that of the CdTe QDs fluorescence. However, the position of the largest emission peak shifted slightly. This was because the covalent attachment of the carboxyl group on the surface of the CdTe QDs to the amino group of the silica considerably reduces the surface charge number of CdTe QDs, thereby lowering the directional polarization of the molecule next to the CdTe QDs and reducing the Stokes shift. The simultaneously formed imprinted shell layer increased the particle radius, resulting in a red-shift in its emission spectrum relative to the fluorescence emission spectrum of CdTe QDs.In addition, the pH of the test solution played an important role in the fluorescence properties of the imprinted paper chips because it had a significant effect on the spatial configuration of 2,4-D, surface charge, and fluorescence intensity of paper@QDs@MIPs.42,43 Therefore, the binding ability of the sensor toward 2,4-D was changed at different pH values. Fig. S4† shows the effect of pH on the fluorescence intensity of paper@QDs@MIPs. As the pH increased from 5.0 to 7.0, the fluorescence intensity gradually increased; however, for pH values above 7.0, the fluorescence intensity decreased with increasing pH. The fluorescence intensity is the highest at pH 7.0. In order to achieve better fluorescence detection, we selected pH 7.0 as the ideal condition for the sample determination of the experiment samples.
Meanwhile, the response time of the imprinted paper chips to 2,4-D has been tested. As shown in Fig. S5A,† when the template molecule 2,4-D was added at a concentration of 50 μM in paper@QDs@MIPs, the fluorescence intensity rapidly decreased within 18 min; thereafter, the curve basically became balanced, indicating that the paper chip can achieve a fluorescence balance within 18 min. As shown in Fig. S5B,† the fluorescence intensity of paper@QDs@MIPs showed a marginal change over the measurement period of 60 min, indicating that the sensor has good physical stability and chemical inertness under the protection of the imprinted layer. Therefore, the corresponding fluorescence was determined after reaction for 18 min as a condition for further study.
Cellulose paper was a relatively uniform material that was intended to be a commercial paper chip of the future. The reproducibility and stability of paper chips have been discussed. At the same time, the stability of paper chips prepared in separate batches was determined, as shown in Fig. S6A.† The relative standard deviation (RSD) of the fluorescence change was 4.8%, indicating that the paper chip prepared by the method was relatively stable and had good repeatability. Meanwhile, the fluorescence intensity of the same paper in different parts was measured, as shown in Fig. S6B.† Five points on the paper chip, namely, upper, lower, left, right, and center, were measured at least three times. The RSD of the fluorescence was found to be 3.7%, which prove that CdTe QDs were evenly grafted onto the surface of the paper chip. The fluorescence at the center was comparatively stronger than that in the surrounding area, and the fluorescence around the center was weaker than that at the center. In subsequent measurements, it was the best to use a paper chip in the central area for measurements, which exhibits stronger fluorescence and sensitivity than the surrounding area.
Fluorescence sensing mechanism
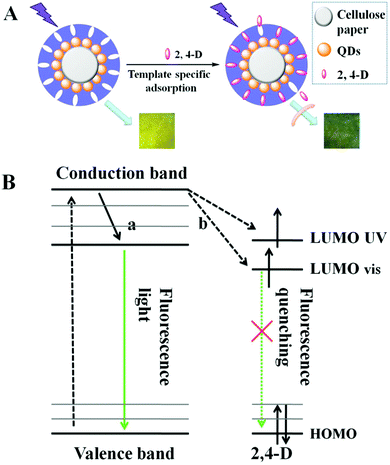
The prepared imprinted paper chips can be used for 2,4-D recognition and fluorescence detection, and its schematic diagram is shown in Fig. 4A. When the 2,4-D solution was dropped on paper@QDs@MIPs, the Meisenheimer complex was induced between the 2,4-D and amino groups in the QD surface imprinting site. A strong charge transfer interaction occurred between the 2,4-D aromatic ring and amino-grafted paper, and the charge transfers from the carboxyl group to the amino group. At the same time, the energy received by the CdTe QDs can be transferred to the complex and gets consumed in a nonfluorescent manner, which results in fluorescence quenching of the CdTe QDs. A change in the fluorescence intensity of paper@QDs@MIPs can be recorded using a spectrophotometer. Therefore, it can be observed that the fluorescence intensity of the CdTe QDs also changed as 2,4-D changed. The quenching mechanism can also be explained by the molecular orbital theory, as shown in Fig. 4B. In general, the ground-state electrons of CdTe QDs were capable of accepting the energy of ultraviolet light and then get excited from a stable valence band to an unstable conduction band. Subsequently, the excited electrons return to the ground state due to instability. The CdTe QDs would emit green fluorescence during this return process. In addition, after the addition of 2,4-D, a hydrogen bond was formed between the 2,4-D and the amino group on the surface of the CdTe QDs, and a strong interaction force causes electron transfer between CdTe QDs and 2,4-D. The UV absorption of 2,4-D was mainly at 285 nm, which was very close to the conduction band of CdTe QDs. Therefore, as indicated by the arrows in Fig. 4B, the excited electrons can jump directly to the LUMO UV and the visible energy level of 2,4-D. Therefore, the excited electrons return to the ground state in the manner shown by the dashed line, causing the QDs to undergo fluorescence quenching.According to the previous illustration of the detection mechanism, when the template molecules were adsorbed onto the surface of the paper chips, the quenching constant would remain constant within a certain concentration range, and the concentration of 2,4-D was positively correlated with fluorescence quenching. Therefore, the detection of 2,4-D concentration can be achieved by the change in fluorescence. Meanwhile, fluorescence quenching during this test followed the Stern–Volmer eqn (1) as follows:
| F0/F = 1 + KSVCq | (1) |
Sensitivity and selectivity of imprinted paper chips
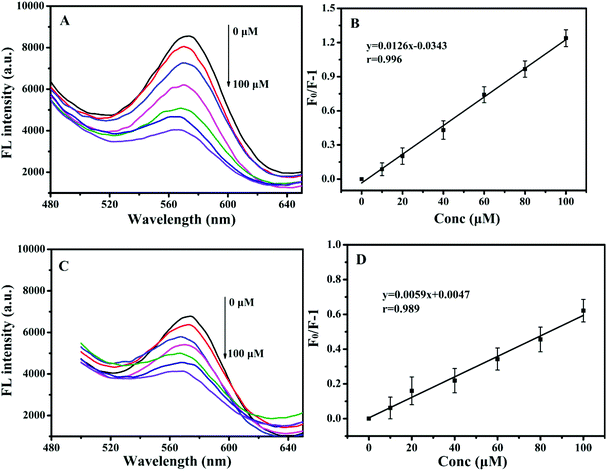
To further evaluate the sensitivity of paper@QDs@MIPs fluorescence sensors, 2,4-D as a template was examined. Under optimized conditions, the imprinted paper chips detected the fluorescence spectra with a series of concentrations of 2,4-D for its linearity range and sensitivity. As shown in Fig. 5A, as the 2,4-D concentration increased, the fluorescence intensity was significantly reduced within a linear range of 0.83–100 μM and correlation coefficient of 0.996 (Fig. 5B). The limit of detection (LOD, S/N = 3) was 90 nM, suggesting the potential for trace analysis. In addition, as shown in Fig. 5C, under the same 2,4-D concentration, the fluorescence intensity of the corresponding paper@QDs@NIPs was not significantly reduced, and the linear range was very narrow, as shown in Fig. 5D. The difference in this phenomenon was caused by the lack of specific recognition sites in paper@QDs@NIPs; further, most of the 2,4-D molecules cannot be identified by paper@QDs@NIPs. Therefore, this imprinted paper chip sensor has excellent sensitivity and high selectivity for 2,4-D, and the calculation of the imprinting factor was 2.13, indicating that the sensor has the feasibility of determining the residuals of 2,4-D. Moreover, fluorescence confocal microscopy can also verify the corresponding fluorescence changes. From Fig. 6, it is evident that a large amount of red fluorescence appeared in the cellulose bundle of the paper@QDs@MIPs, which was red fluorescence emitted by the QDs grafted onto the cellulose bundle under the UV lamp. When there was no template molecule, the imprinted paper chips exhibited the strongest red fluorescence. As the 2,4-D concentration increased, the imprinted paper chips exhibited increasingly weak red fluorescence. The correlated fluorescence confocal images also further demonstrated the fluorescence quenching of 2,4-D for QDs. This method proved to be a facile detection strategy without involving complex sample preparation and purification requirements.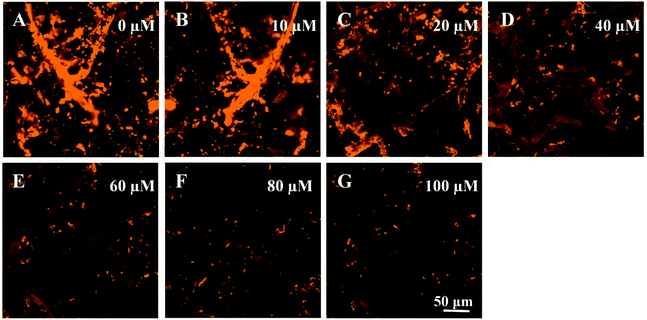 | ||
| Fig. 6 Fluorescence microscope images of paper@QDs@MIPs with addition of the (A) 0, (B), 10, (C) 20, (D) 40 (E) 60, (F) 80, and (G) 100 μM of 2,4-D. | ||
In addition, the selectivity for 2,4-D was the main indicator of paper@QDs@MIPs in the actual sample analysis. In contrast, the selectivity of the imprinted paper chip was further examined. Phenol, carbendazim, and toluene were selected for comparison, because these substances are similar to the 2,4-D structure. The results are shown in Fig. 7. It is evident that the imprinted paper chip has a certain recognition ability for the template molecule 2,4-D, and the fluorescence quenching intensity of the imprinted paper chip was the largest, indicating that the imprinted paper chip has better selectivity for 2,4-D, followed by carbendazim and phenol, with toluene being the worst. Although the imprinted paper chip had a certain degree of fluorescence quenching on the structural analogues of 2,4-D, the quenching intensity on 2,4-D was the highest. The nonimprinted paper chip shows a marginal difference in the fluorescence quenching of the four substances, indicating that it has no specific selectivity for 2,4-D and the recognition performance is not worthwhile. This difference may be explained by the difference in the molecular weights and spatial structures between 2,4-D and the analogues.
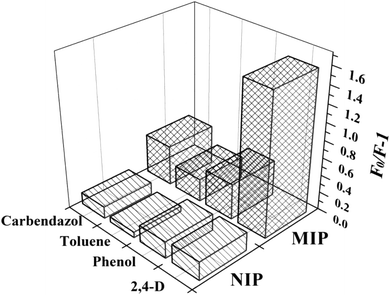 | ||
| Fig. 7 Selectivity of paper@QDs@MIPs and paper@QDs@NIPs to other template analogues (2,4-D, phenol, toluene, and carbendazol) at the same concentration. | ||
Practical applications of fluorescent microfluidic paper chips
To further investigate the applications of paper@QDs@MIPs in actual sample testing, the prepared imprinted paper chips were used to detect 2,4-D in bean sprout samples. The samples were spiked at three concentration levels to obtain the average recovery and RSD values. The results are listed in Table 1. The recovery of 2,4-D is similar to the addition of the standard, revealing that the prepared imprinted paper chips could be used to detect actual food samples. The recovery rate of the spiked bean sprout sample was 94.5–105.5% and its RSD was 4.7–5.9%. The recovery rate of the mung bean sprout sample was 94.2–107.0%, and the RSD was 4.4–5.6%. Moreover, the experiments have shown that the results of this method were in good consistency with the results obtained by the HPLC method (Table 1). In other words, the paper@QDs@MIPs sensor had broader application potential and can be used to actually detect 2,4-D in actual food samples.| Sample | Spiked (μmol L−1) | paper@QDs@MIPs | HPLC | |
|---|---|---|---|---|
| Found (μmol L−1) | Recovery ± RSD (%) | Determined (μmol L−1) | ||
| Soybean sprouts | 0 | 0 | — | — |
| 5.00 | 5.14 | 102.8 ± 4.7 | 4.96 | |
| 10.00 | 9.45 | 94.5 ± 5.9 | 9.89 | |
| 20.00 | 21.10 | 105.5 ± 5.1 | 20.91 | |
| Mung bean sprout | 0 | 0 | — | — |
| 5.00 | 4.71 | 94.2 ± 4.4 | 4.93 | |
| 10.00 | 10.33 | 103.3 ± 5.6 | 10.12 | |
| 20.00 | 21.40 | 107.0 ± 4.8 | 19.62 | |
Conclusions
The present study used cellulose paper as the base material and CdTe QDs as the fluorescence signal material to construct CdTe QDs microfluidic paper chips. The recognition units were imprinted on the surface of CdTe QDs for selective detection. The paper@QDs@MIPs sensor exhibited a fast recognition rate, low detection limit of 0.12 μM, and imprinted factor of 2.13 within a linear range of 0.83–100 μM. The imprinted paper chip has obvious selective recognition performance for 2,4-D, and it is stable, safe, environmentally friendly, and can be used for detecting 2,4-D in complex food substrates. Owing to the unique properties of cellulose paper, this method provides many advantages such as disposability, portability, low cost, and simple post-processing, which are not available in generic methods. Moreover, 3D origami microfluidic paper chips display the potential and expansion of the use of appropriate fluorescent materials to detect different types of food contaminants.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (31701705, 31671823), the Hong Kong Scholars Program (XJ2017060), the Key Laboratory of Coastal Environmental Process and Ecological Restoration (2018KFJJ01), the Open Project Program of State Key Laboratory of Food Science and Technology, Nanchang University (No. SKLF-KF-201813) and the Fundamental Research Fund of the Central University in China (GK201803084).References
- C. Xie, S. Gao, Q. Guo and K. Xu, Electrochemical sensor for 2, 4-dichlorophenoxy acetic acid using molecularly imprinted polypyrrole membrane as recognition element, Microchim. Acta, 2018, 169, 145–152 CrossRef.
- S. M. Wang, L. Ge, L. Li, M. Yan, S. G. Ge and J. H. Yu, Molecularly imprinted polymer grafted paper-based multi-disk micro-disk plate for chemiluminescence detection of pesticide, Biosens. Bioelectron., 2013, 50, 262–268 CrossRef PubMed.
- S. M. Hays, L. L. Aylward, J. Driver, J. Rose and C. Kiman, 2, 4-D exposure and risk assessment: comparison of external dose and biomonitoring based approaches, Regul. Toxicol. Pharmacol., 2012, 64, 481–489 CrossRef PubMed.
- A. M. Smith, M. T. Smith and M. A. La Merrill, 2, 4-Dichlorophenoxyacetic acid (2, 4-D) and risk of non-Hodgkin lymphoma: a meta-analysis accounting for exposure levels, Ann. Epidemiol., 2017, 27, 281–289 CrossRef PubMed.
- A. J. A. Charlton, V. Stuckey and M. D. Sykes, Determination of the phenoxyacid herbicides MCPA, mecoprop and 2, 4-D in kidney tissue using liquid chromatography with electrospray tandem mass spectrometry, Bull. Environ. Contam. Toxicol., 2009, 82, 711–715 CrossRef PubMed.
- M. Shamsipur, N. Fattahi, M. Pirsaheb and K. Sharafi, Simultaneous preconcentration and determination of 2, 4-D, alachlor and atrazine in aqueous samples using dispersive liquid–liquid microextraction followed by high-performance liquid chromatography ultraviolet detection, J. Sep. Sci., 2012, 35, 2718–2724 CrossRef PubMed.
- S. Viswanathan, H. Radecka and J. Radecki, Electrochemical biosensor for pesticides based on acetylcholinesterase immobilized on polyaniline deposited on vertically assembled carbon nanotubes wrapped with ssDNA, Biosens. Bioelectron., 2009, 24, 2772–2777 CrossRef PubMed.
- M. Z. Hua, S. Feng, S. Wang and X. Lu, Rapid detection and quantification of 2, 4-dichlorophenoxyacetic acid in milk using molecularly imprinted polymers–surface-enhanced Raman spectroscopy, Food Chem., 2018, 258, 254–259 CrossRef PubMed.
- I. B. Tahirbegi, J. Ehgartner, P. Sulzer, S. Zieger, A. Kasjanow and M. Paradiso, Fast pesticide detection inside microfluidic device with integrated optical pH, oxygen sensors and algal fluorescence, Biosens. Bioelectron., 2017, 88, 188–195 CrossRef PubMed.
- L. Zhu, X. Cui, J. Wu, Z. Wang, P. Wang, Y. Hou and M. Yang, Fluorescence immunoassay based on carbon dots as labels for the detection of human immunoglobulin G, Anal. Methods, 2014, 6, 4430–4436 RSC.
- A. Valizadeh, H. Mikaeili, M. Samiei, S. M. Farkhani, N. Zarghami and M. Kouhi, Quantum dots: synthesis, bioapplications, and toxicity, Nanoscale Res. Lett., 2012, 7, 480–494 CrossRef PubMed.
- J. L. Chen, A. F. Zheng, Y. C. Gao, C. Y. He, G. H. Wu and Y. C. Chen, Functionalized CdS quantum dots-based luminescence probe for detection of heavy and transition metal ions in aqueous solution, Spectrochim. Acta, Part A, 2008, 69, 1044–1052 CrossRef.
- H. D. Chen and Y. S. Xia, Compact hybrid (gold nanodendrite-quantum dots) assembly: plasmon enhanced fluorescence-based platform for small molecule sensing in solution, Anal. Chem., 2014, 86, 11062–11069 CrossRef.
- Y. X. Ma, S. Y. Xu, S. G. Wang and L. Y. Wang, Luminescent molecularly-imprinted polymer nanocomposites for sensitive detection, TrAC, Trends Anal. Chem., 2015, 67, 209–216 CrossRef.
- X. Wang, J. Yu, X. Wu, J. Q. Fu, Q. Kang and D. Z. Shen, A molecular imprinting-based turn-on ratiometric fluorescence sensor for highly selective and sensitive detection of 2, 4-dichlorophenoxyacetic acid (2, 4-D), Biosens. Bioelectron., 2016, 81, 438–444 CrossRef.
- S. F. Xu, H. Z. Lu, J. H. Li, X. L. Song, A. X. Wang and L. X. Chen, Dummy molecularly imprinted polymers-capped CdTe quantum dots for the fluorescent sensing of 2, 4, 6-trinitrotoluene, ACS Appl. Mater. Interfaces, 2013, 5, 8146–8154 CrossRef.
- K. Zhang, H. Zhou, Q. Mei, S. Wang, G. Guan and R. Liu, Instant visual detection of trinitrotoluene particulates on various surfaces by ratiometric fluorescence of dual-emission quantum dots hybrid, J. Am. Chem. Soc., 2011, 133, 8424–8427 CrossRef.
- L. X. Chen, X. Y. Wang, W. H. Lu, X. Q. Wu and J. H. Li, Molecular imprinting: perspectives and applications, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- M. F. Jia, Z. Zhang, X. B. Yang, J. H. Li and L. X. Chen, Design and application of novel molecular imprinting fluorescent sensors, Sci. Sin.: Chim., 2017, 47, 300–314 Search PubMed.
- X. L. Hu, X. Wu, F. F. Yang, Q. Wang, C. Y. He and S. R. Liu, Novel surface dummy molecularly imprinted silica as sorbent for solid-phase extraction of bisphenol A from water samples, Talanta, 2016, 148, 29–36 CrossRef.
- Z. Zhang, J. H. Li, L. W. Fu, D. Y. Liu and L. X. Chen, Magnetic molecularly imprinted microsensor for selective recognition and transport of fluorescent phycocyanin in seawater, J. Mater. Chem. A, 2015, 3, 7437–7444 RSC.
- X. D. Zheng, J. M. Pan, L. Gao, X. Wei, J. D. Dai, W. D. Shi and Y. S. Yan, Silica nanoparticles doped with a europium(III) complex and coated with an ion imprinted polymer for rapid determination of copper(II), Microchim. Acta, 2015, 182, 753–761 CrossRef.
- R. X. Gao, X. Kong, X. Wang, X. W. He, L. X. Chen and Y. K. Zhang, Preparation and characterization of uniformly sized molecularly imprinted polymers functionalized with core-shell magnetic nanoparticles for the recognition and enrichment of protein, Mater. Chem., 2011, 21, 17863–17871 RSC.
- M. S. Zhang, J. R. Huang, P. Yu and X. Chen, Preparation and characteristics of protein molecularly imprinted membranes on the surface of multiwalled carbon nanotubes, Talanta, 2010, 81, 162–166 CrossRef.
- L. Qin, X. W. He, W. Zhang, W. Y. Li and Y. K. Zhang, Surface-modified polystyrene beads as photografting imprinted polymer matrix for chromatographic separation of proteins, J. Chromatogr. A, 2009, 1216, 807–814 CrossRef.
- H. C. Chen, D. Y. Yuan, Y. Y. Li, M. J. Dong, Z. H. Chai and J. Kong, Silica nanoparticle supported molecularly imprinted polymer layers with varied degrees of crosslinking for lysozyme recognition, Anal. Chim. Acta, 2013, 779, 82–89 CrossRef.
- B. Deiminiat, I. Razavipanah, G. H. Rounaghi and M. H. A. Zavar, A novel electrochemical imprinted sensor for acetylsalicylic acid based on polypyrrole, sol-gel and SiO2@Au core-shell nanoparticles, Sens. Actuators, B, 2017, 244, 785–795 CrossRef.
- J. H. Li, J. Q. Fu, Q. Yang, X. Y. Wang and L. X. Chen, Thermosensitive molecularly imprinted core-shell CdTe quantum dots as a ratiometric fluorescence nanosensor for phycocyanin recognition and detection in seawater, Analyst, 2018, 143, 3570–3578 RSC.
- A. W. Martinez, S. T. Phillips and G. M. Whitesides, Patterned paper as a platform for in expensive, low-volume, portable bioassays, Angew. Chem., Int. Ed., 2017, 46, 1318–1320 CrossRef.
- Q. K. Kong, Y. H. Wang, L. N. Zhang, S. G. Ge and J. H. Yu, A novel microfluidic paper-based colorimetric sensor based on molecularly imprinted polymer membranes for highly selective and sensitive detection of bisphenol A, Sens. Actuators, B, 2017, 243, 130–136 CrossRef.
- X. F. Wei, T. Tian, S. S. Jia, Z. Zhu and Y. L. Ma, Microfluidic Distance Readout Sweet Hydrogel Integrated Paper Based Analytical Device (mu DiSH-PAD) for Visual Quantitative Point-of-Care Testing, Anal. Chem., 2016, 88, 2345–2352 CrossRef CAS.
- S. Choi, S. K. Kim, G. J. Lee and H. K. Park, Paper-based 3D microfluidic device for multiple bioassays, Sens. Actuators, B, 2015, 219, 245–250 CrossRef CAS.
- B. Li, Z. Zhang, J. Qi, N. Zhou, S. Qin and J. Choo, Quantum dot-based molecularly imprinted polymers on three-dimensional origami paper microfluidic chip for fluorescence detection of phycocyanin, ACS Sens., 2017, 2, 243–250 CrossRef CAS.
- J. Qi, B. W. Li, X. Y. Wang, L. Fu, L. Luo and L. X. Chen, Rotational paper-based microfluidic-chip device for multiplexed and simultaneous fluorescence detection of phenolic pollutants based on a molecular-imprinting technique, Anal. Chem., 2018, 90, 11827–11834 CrossRef CAS.
- Y. G. Ngo, D. Li, G. P. Simon and G. Garnier, Paper surfaces functionalized by nanoparticles, Adv. Colloid Interface Sci., 2011, 163, 23–38 CrossRef CAS.
- B. Dou, Y. Luo, X. Chen, B. Shi, Y. Du and Z. Gao, Direct measurement of beta-agonists in swine hair extract in multiplexed mode by surface-enhanced Raman spectroscopy and microfluidic paper, Electrophoresis, 2015, 36, 485–487 CrossRef CAS.
- I. Pereira, M. F. Rodrigues, A. R. Chaves and B. G. Vaz, Molecularly imprinted polymer (MIP) membrane assisted direct spray ionization mass spectrometry for agrochemicals screening in foodstuffs, Talanta, 2018, 178, 507–514 CrossRef CAS.
- Z. Zhang, J. H. Li, X. Y. Wang, D. Z. Shen and L. X. Chen, Quantum dots based mesoporous structured imprinting microspheres for the sensitive fluorescent detection of phycocyanin, ACS Appl. Mater. Interfaces, 2015, 7, 9118–9127 CrossRef CAS.
- S. Ma, Y. Tang, J. Liu and J. Wu, Visible paper chip immunoassay for rapid determination of bacteria in water distribution system, Talanta, 2014, 120, 135–140 CrossRef CAS.
- L. Ma, A. Nilghaz, J. R. Choi, X. Liu and X. Lu, Rapid detection of clenbuterol in milk using microfluidic paper-based ELISA, Food Chem., 2018, 246, 437–441 CrossRef CAS.
- D. Wu, J. Zhang, F. Xu, X. Wen, P. Li and X. Zhang, A paper-based microfluidic Dot-ELISA system with smartphone for the detection of influenza A, Microfluid. Nanofluid., 2017, 21–43 Search PubMed.
- S. Gamderouich, C. Bourdillon, W. D. De Marcillac, L. Coolen and A. Maitre, Correction: Quantum dot-imprinted polymers with size and shell-selective recognition properties, Chem. Commun., 2015, 51, 14933–14936 RSC.
- S. Gamderouich, C. Bourdillon, C. S. Lakhdar, L. Coolen and A. Maitre, Imprinted photonic hydrogels for the size and shell-selective recognition of nanoparticles, Angew. Chem., Int. Ed., 2017, 56, 9710–9714 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an02051e |
| This journal is © The Royal Society of Chemistry 2019 |