Advancements in DNA computing: exploring DNA logic systems and their biomedical applications
Yuewei
Zhao
a,
Xvelian
Li
b,
Yan
Zhou
b,
Xiaoting
Tian
b,
Yayou
Miao
b,
Jiayi
Wang
*ab,
Lin
Huang
 *ab and
Fanyu
Meng
*ab and
Fanyu
Meng
 *ab
*ab
aDepartment of Clinical Laboratory, Shanghai Chest Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200030, P. R. China. E-mail: fanyumeng@shsmu.edu.cn; linhuang@shsmu.edu.cn; jiayi.wang@sjtu.edu.cn
bInstitute of Thoracic Oncology, Shanghai Chest Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200030, P. R. China
First published on 6th September 2024
Abstract
DNA computing is regarded as one of the most promising candidates for the next generation of molecular computers, utilizing DNA to execute Boolean logic operations. In recent decades, DNA computing has garnered widespread attention due to its powerful programmable and parallel computing capabilities, demonstrating significant potential in intelligent biological analysis. This review summarizes the latest advancements in DNA logic systems and their biomedical applications. Firstly, it introduces recent DNA logic systems based on various materials such as functional DNA sequences, nanomaterials, and three-dimensional DNA nanostructures. The material innovations driving DNA computing have been summarized, highlighting novel molecular reactions and analytical performance metrics like efficiency, sensitivity, and selectivity. Subsequently, it outlines the biomedical applications of DNA computing-based multi-biomarker analysis in cellular imaging, clinical diagnosis, and disease treatment. Additionally, it discusses the existing challenges and future research directions for the development of DNA computing.
1. Introduction
DNA computing represents a novel paradigm in molecular computation, integrating principles from information science and biology. The fundamental concept involves leveraging the information processing capabilities of organic biological molecules to replace traditional digital physical switch components.1–3 The duplex helical configuration and base complementarity of DNA molecules enable the encoding of problems into specific DNA sequences. When paired with a specified DNA sequence acting as a switch, an output DNA sequence with a concentration proportional to the input DNA concentration is generated, resembling a simple analog circuit.4,5 DNA circuits utilize the concentration of specific DNA strands as signals, achieving addition, subtraction, and multiplication operations through the synthesis or cleavage of chemical bonds.6–8 In contrast, these DNA molecule reaction strategies are relatively simple, with few reports focusing on their application capabilities in analytical science. With the swift progress of synthetic biology technologies, DNA computing has garnered widespread research interest due to its powerful programmable and parallel computing capabilities.9–11Currently, although bioelectronics based on DNA computing is still in its developmental stage, the concept of assembling microscale biological component circuits to construct parallel biological computing systems has been preliminarily established.12 In 2004, Okamoto and colleagues first merged digital circuits with DNA computing, cascading DNA logic gates to form complex circuits, ultimately achieving universal DNA computing.13–15 By integrating the structural and functional characteristics of DNA, various DNA logic systems can be designed and integrated into autonomous computing devices.16,17 These systems employ binary-coded DNA or other triggers as inputs (with “0” representing absence and “1” representing presence), with DNA strands or optical/electrochemical signals serving as outputs (with “0” indicating low and “1” indicating high).18–20 Leveraging the inherent advantages of DNA, along with its strict Boolean logic and biological compatibility, DNA computing finds extensive applications in fields such as biological analysis, diagnostics, imaging, and drug delivery.21–23 However, DNA computing remains limited in its clinical applications, owing to insufficient clarification of the relationship between biological computing and analytical performance.
Previous reviews mainly focused on the basic concepts and functional principles of DNA in data storage,2,24,25 logic programming,20,26 and molecular computing.13,14 For comparison, the novelty and significance of this work lie in the development of novel DNA logic gates designed to enhance sensing performance (such as sensitivity and specificity), and to expand the scope of computational applications especially for biomedical use. In the past three years, significant progress has been made in DNA logic gates, particularly through the use of DNA motifs, nanomaterials, and DNA nanostructures. Therefore, we concentrated on exploring improvements in the efficiency, sensitivity, and selectivity of cascading molecular reactions, providing insights that may guide the future development of analytical science. Additionally, we comprehensively reviewed and analyzed the latest advancements in DNA computation for biomedical applications, including but not limited to cellular imaging, clinical diagnosis, and disease treatment. Finally, we discussed the challenges faced by DNA computing and proposed future prospects for this field. Scheme 1 illustrates DNA logic gates constructed using various materials, along with their associated biomedical applications, including cellular imaging, clinical diagnosis, and disease treatment. We anticipate that the review will contribute to advancing the field of DNA computing and provide insights for intelligent biomedical diagnostic applications.
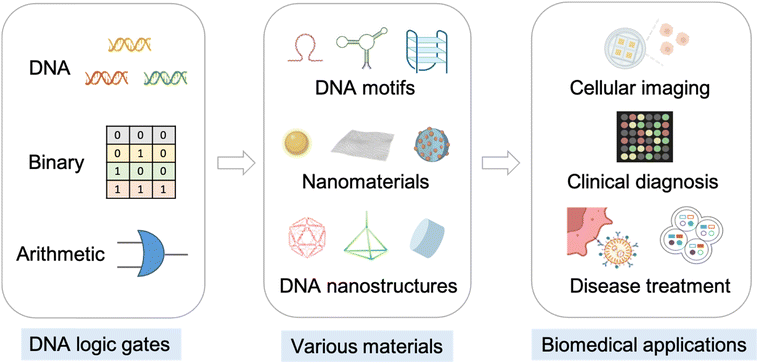 | ||
| Scheme 1 DNA logic gates, various materials for constructing DNA logic gates, and related biomedical applications. | ||
2. DNA logic systems
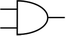
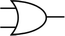
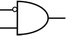
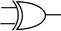
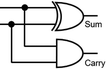
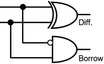
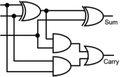
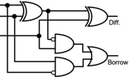
To construct a DNA-based computer, it is indispensable to initially build DNA logic gates, which function as devices that transform sets of inputs into observable outputs through logical processes. From fundamental logic gates (AND, OR, YES, NOT, XOR, and INHIBIT) to complex logical operations (addition, subtraction, and multiplication), these are essential computing components for constructing DNA logic systems.27,28 DNA molecules are designed to perform logical operations on binary inputs. In a DNA logic system, “inputs” are typically DNA strands with specific sequences, and “outputs” are other DNA strands produced as a result of biochemical reactions.29,30 The presence or absence of specific DNA strands in the system can represent binary values (0 or 1). Logic gates can be represented in terms of equivalent logic circuits and truth tables (Table 1). For example, the “AND” system has two input DNA strands, A and B, each representing a binary “1”. If both strands are present, they bind to a complementary “template” strand, triggering the production of a new output strand, C, which represents the binary “1”. If either A or B is missing, the reaction doesn't proceed, and no output is produced (binary “0”). Additionally, fundamental arithmetic logic operations (the half-adder/subtractor) include addition, which combines two numbers to produce a sum, and subtraction, which determines the difference between two numbers (Table 2).14,31 In summary, a DNA logic system is a molecular computing approach where DNA molecules and biochemical reactions are engineered to perform logical operations.DNA computing combines the analytical and computational capabilities of Boolean logic with the functionality of DNA as a material.32,33 DNA logic systems possess programmability, parallel processing capability, and biocompatibility, allowing for the design and adaptation of logic circuits while facilitating data processing and real-time monitoring. In biological analysis, integrating various materials into DNA logic systems can significantly enhance detection efficiency, sensitivity, and specificity. Here, we classify DNA logic systems based on various materials, including functional DNAs (motifs, DNAzymes, G-quadruplexes, etc.), nanomaterials (metal nanoparticles, metal nanoclusters, graphene oxides, magnetic beads, etc.), and DNA nanostructures (DNA tetrahedrons, DNA origamis, DNA tile assembly, etc.).34
2.1. Functional DNA-based DNA logic systems
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FSD = 20
FSD = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8). What's more, Lapteva et al. have advanced DNA strand-displacement circuits by combining temporal memory and logic computation (AND gate), allowing these circuits to make decisions based on specific input combinations and timing (Fig. 1B).41 The approach preserved temporal information within memory strands, with each one encoding the relative timing. The incorporation of temporal information into Boolean logic computation, holds promise for diverse circuit structures, including DNA-based neural networks.
8). What's more, Lapteva et al. have advanced DNA strand-displacement circuits by combining temporal memory and logic computation (AND gate), allowing these circuits to make decisions based on specific input combinations and timing (Fig. 1B).41 The approach preserved temporal information within memory strands, with each one encoding the relative timing. The incorporation of temporal information into Boolean logic computation, holds promise for diverse circuit structures, including DNA-based neural networks.
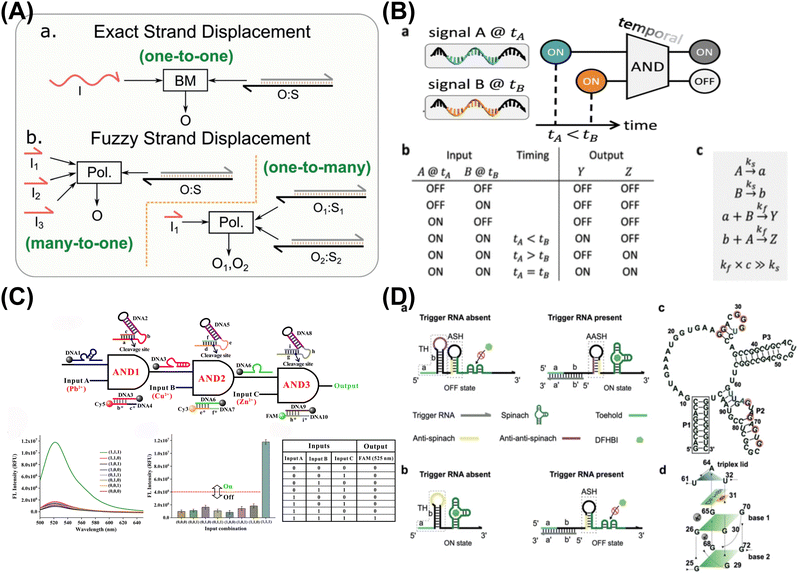 | ||
| Fig. 1 Functional DNA-based DNA logic systems. (A) Fuzzy DNA strand displacement to decrease the complexity of the DNA network. Reproduced with permission.40 Copyright 2020, Wiley-VCH. (B) DNA strand-displacement temporal logic circuits. Reproduced with permission.41 Copyright 2022, American Chemical Society. (C) DNAzyme-based scalable logic circuits with multiple outputs and automatic reset functions. Reproduced with permission.42 Copyright 2021, American Chemical Society. (D) G-quadruplex-based riboswitch architectures for the switchable fluorescent logic gate platform. Reproduced with permission.43 Copyright 2023, Wiley-VCH. | ||
2.2. Nanomaterial-based DNA logic systems
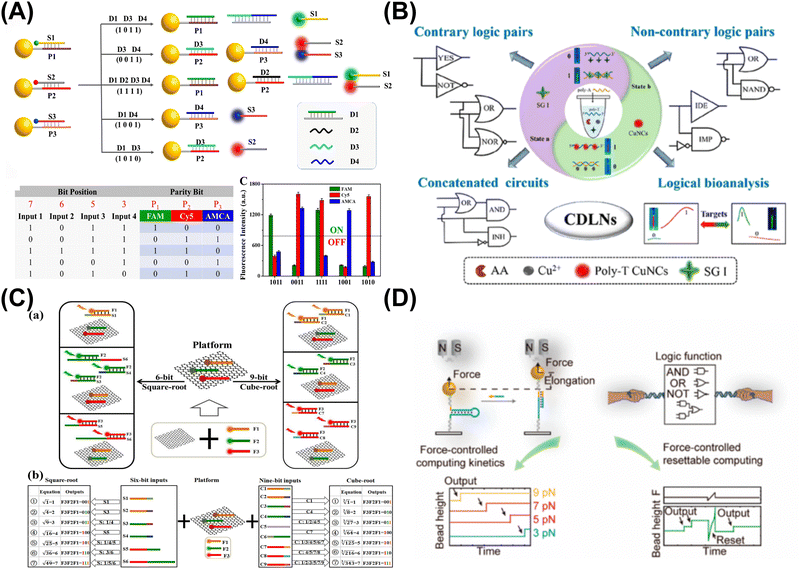 | ||
| Fig. 2 Nanomaterial-based DNA logic systems. (A) Metal nanomaterials and leveraging DNA-based nanostructures for data communication. Reproduced with permission.51 Copyright 2023, American Chemical Society. (B) Metal nanoclusters-based DNA logic nanodevices for minute-level DNA computing. Reproduced with permission.53 Copyright 2023, American Chemical Society. (C) Graphene oxide-based computing system for multiplexing logic operations. Reproduced with permission.54 Copyright 2020, Elsevier. (D) Magnetic tweezers-based single-molecule resettable DNA computing. Reproduced with permission.55 Copyright 2022, American Chemical Society. | ||
2.3. DNA nanostructure-based DNA logic systems
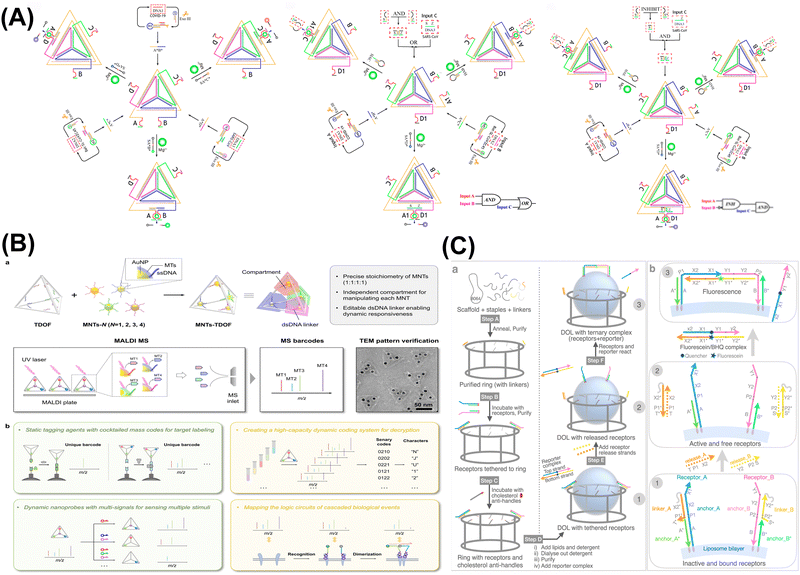 | ||
| Fig. 3 DNA nanostructure-based DNA logic systems. (A) Tetrahedron-based constitutional dynamic network. Reproduced with permission.69 Copyright 2022, American Chemical Society. (B) Compartmented DNA origami frames for precision information coding and logic mapping. Reproduced with permission.70 Copyright 2024, Wiley-VCH. (C) Three-dimensional digital nanoreactors to control absolute stoichiometry and spatiotemporal behavior of DNA receptors. Reproduced with permission.71 Copyright 2023, Springer Nature. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 homodimers and heterodimers (which refer to the ratios of DNA strands involved in interactions and are crucial for determining the binding affinities and functional properties of DNA constructs), was facilitated through SDR and monitored using a DNA logic gate and fluorescence measurements. Understanding these stoichiometries advances the design of more efficient DNA logic systems with predictable behaviors and enhanced performance in various applications. This tethering allowed the determination of the native stoichiometry and kinetics of membrane protein complexes, advancing the understanding of signaling pathways and drug discovery. The approach provided a versatile platform for studying membrane protein behavior and interaction dynamics in a variety of ratios and contexts. Lv et al. developed a programmable DNA integrated circuit system by integration of multilayer DNA-based programmable gate arrays (DPGAs) for multifunctional logic computing.75 They utilized DNA origami to address issues like transient binding and signal attenuation, thereby overcoming limitations on circuit size and depth by controlling molecular interactions in solution. The integration of DNA origami registers was critical to achieving higher-order integration of DPGAs, providing directionality for asynchronous computational processing in cascaded DPGAs. This work introduced a highly organized and reconfigurable DNA computing platform, representing a significant advancement in the progress of general-purpose DNA computing.
2 homodimers and heterodimers (which refer to the ratios of DNA strands involved in interactions and are crucial for determining the binding affinities and functional properties of DNA constructs), was facilitated through SDR and monitored using a DNA logic gate and fluorescence measurements. Understanding these stoichiometries advances the design of more efficient DNA logic systems with predictable behaviors and enhanced performance in various applications. This tethering allowed the determination of the native stoichiometry and kinetics of membrane protein complexes, advancing the understanding of signaling pathways and drug discovery. The approach provided a versatile platform for studying membrane protein behavior and interaction dynamics in a variety of ratios and contexts. Lv et al. developed a programmable DNA integrated circuit system by integration of multilayer DNA-based programmable gate arrays (DPGAs) for multifunctional logic computing.75 They utilized DNA origami to address issues like transient binding and signal attenuation, thereby overcoming limitations on circuit size and depth by controlling molecular interactions in solution. The integration of DNA origami registers was critical to achieving higher-order integration of DPGAs, providing directionality for asynchronous computational processing in cascaded DPGAs. This work introduced a highly organized and reconfigurable DNA computing platform, representing a significant advancement in the progress of general-purpose DNA computing.
3. Applications in biomedical diagnostics
Through the development and implementation of DNA logic systems, DNA computing-based technologies and platforms find applications in biomedical diagnosis.76,77 These include cell imaging, disease monitoring, biomarker analysis, drug therapy, and personalized healthcare.78–80 DNA computing in biomedical diagnosis offers advantages such as high sensitivity, specificity, and multiplexing ability, making it a promising approach for advancing diagnostic methodologies and improving patient care.81,823.1. DNA computing for cellular imaging
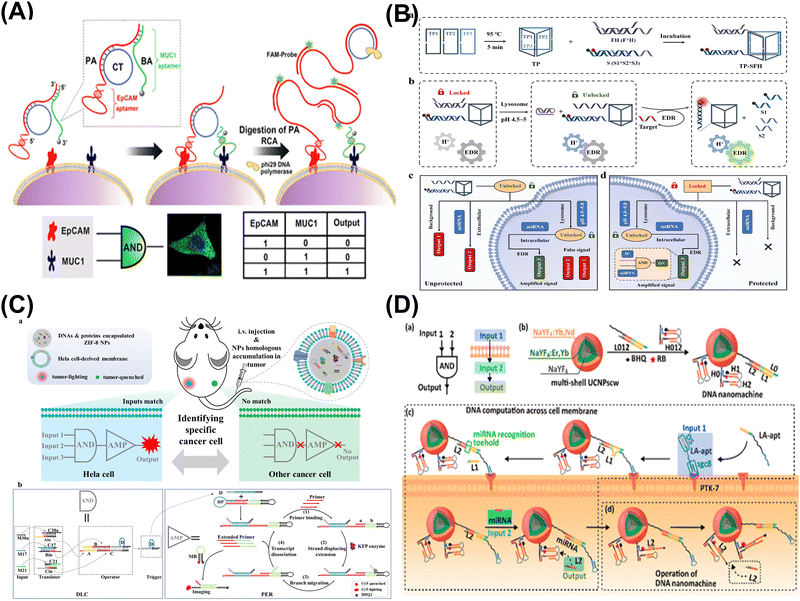 | ||
| Fig. 4 DNA computing for cellular imaging. (A) DNA logic nanomachine for imaging analysis of proteins on cell membranes. Reproduced with permission.84 Copyright 2020, American Chemical Society. (B) Intracellular activated logic nanomachines for detection of miRNAs in living cells. Reproduced with permission.86 Copyright 2023, Royal Society of Chemistry. (C) DNA logic circuit equipped with biomimetic ZIF-8 nanoparticles for the identification of specific cancers in vivo. Reproduced with permission.87 Copyright 2023, Wiley-VCH. (D) DNA nanomachine activated via computation across cancer cell membranes for therapy of solid tumors. Reproduced with permission.88 Copyright 2021, American Chemical Society. | ||
In summary, DNA computing shows promise in cellular imaging, enabling intelligent analysis of cell membranes and intracellular biomarkers. Through logic computation, it not only facilitates precise analysis of different cell subtypes in clinical samples but can also be utilized for targeted therapy of tumors in vivo, providing more efficient and applicable strategies for future intelligent diagnosis and treatment.
3.2. DNA computing for clinical diagnosis
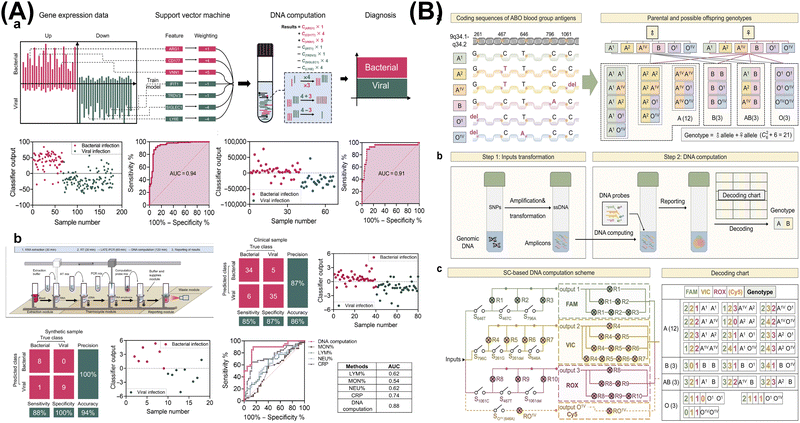 | ||
| Fig. 5 DNA computing for clinical diagnosis. (A) An automated DNA computing platform for rapid etiological diagnostics. Reproduced with permission.93 Copyright 2022, American Association for the Advancement of Science. (B) Programmable DNA molecular computation based logical analysis for clinical diagnostics. Reproduced with permission.94 Copyright 2022, Wiley-VCH. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) fM for miRNAs, 1
fM for miRNAs, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pM for mRNA, 0.05
pM for mRNA, 0.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ng mL−1 for prostate-specific antigen, and 10
ng mL−1 for prostate-specific antigen, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nM for sarcosine. By analyzing 32 patients with PCa and 50 healthy controls, they accurately distinguished between the cancer groups and the healthy groups (P value
nM for sarcosine. By analyzing 32 patients with PCa and 50 healthy controls, they accurately distinguished between the cancer groups and the healthy groups (P value![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) <
<![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01), with an AUC of 100%, a specificity of 100%, and a sensitivity of 100%. The utilization of multidimensional molecular classifiers for the analysis of diverse molecular biomarkers illuminates the path towards precision diagnosis and therapy.
0.01), with an AUC of 100%, a specificity of 100%, and a sensitivity of 100%. The utilization of multidimensional molecular classifiers for the analysis of diverse molecular biomarkers illuminates the path towards precision diagnosis and therapy.
In total, DNA arithmetic computation enables precise execution of mathematical models at the molecular level, facilitating complex assay of diagnostic biomarkers. By intelligently designing systems to cascade different logic gates and generate corresponding computational outputs, more precise disease diagnosis and analysis can be achieved. This will inspire further clinical applications, thereby advancing intelligent biomedical diagnosis and treatment.
3.3. DNA computing for disease treatment
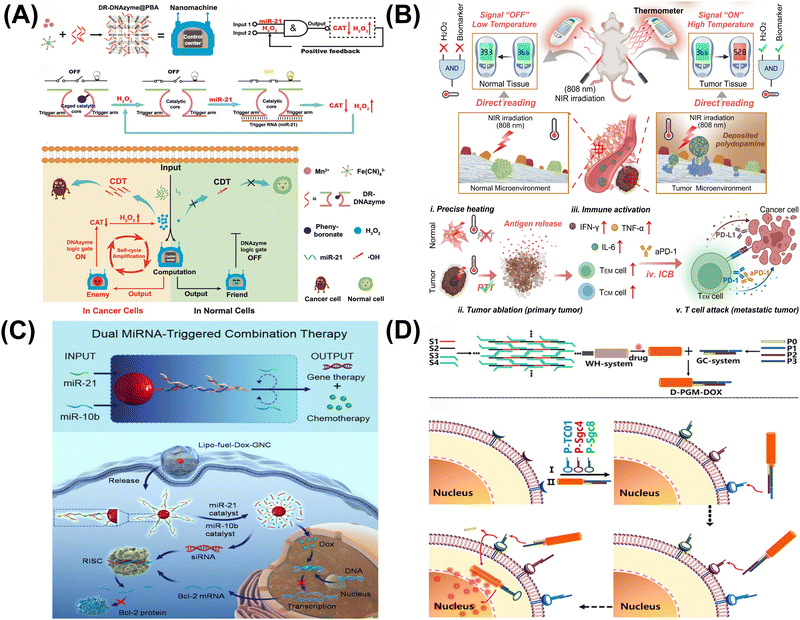 | ||
| Fig. 6 DNA computing for disease treatment. (A) DNAzyme logic-guided nanomachine for precise therapy.96 Copyright 2022, Wiley-VCH. (B) Intelligent DNA nanoreactor for tumor imaging and therapy.97 Copyright 2024, Wiley-VCH. (C) Dual microRNA logic-triggered drug release system.98 Copyright 2020, American Chemical Society. (D) Logic-guided missile-like DNA structure for targeted drug delivery.99 Copyright 2020, American Chemical Society. | ||
In conclusion, DNA logic gates offer targeted cancer treatment by precisely recognizing and acting within cancer cells, reducing systemic toxicity and improving outcomes. However, their complexity and reliance on specific conditions present challenges to widespread application, though they show promise in overcoming multidrug resistance.
4. Conclusions
This review aims to summarize the latest developments in the field of DNA computing, including recent advances in constructing logic systems based on various materials and the progress made in intelligent biomedical diagnostic applications. However, DNA computing is still in the exploratory stage and faces several challenges: (1) DNA biochemical reactions can be readily constrained by implementation conditions, and as computational complexity increases, enhancing reaction efficiency becomes a crucial factor in determining computational accuracy. Factors such as temperature, pH, ionic strength, and reactant concentration can affect the efficiency and accuracy of DNA computing. (2) Most DNA logic platforms can only perform limited logic operations and address specific types of problems, hindering the widespread adoption of DNA computing due to the lack of universal computing systems. Current DNA logic platforms can only perform simple operations (like AND, OR, NOT) and cannot handle complex data processing or multi-step operations, limiting their potential to become general-purpose computing systems. (3) The majority of DNA computing systems are still in the concept validation or lab-scale stages, presenting significant challenges for normal operation in complex biological samples. Applying DNA computing in complex biological environments requires overcoming challenges related to scalability, stability, and integration with biological processes.Therefore, key directions to address these challenges include finding novel DNA hybridization strategies, integrated nanomaterials, and designing multifunctional DNA nanostructures. Firstly, developing more robust DNA hybridization strategies and optimizing implementation conditions (such as temperature, ionic strength, and reactant concentration) to operate effectively in diverse and less controlled environments, while enhancing reaction efficiency and maintaining accuracy, is crucial for transitioning DNA computing from the laboratory to clinical applications. Secondly, it is necessary to design general-purpose computing systems capable of handling more complex and diverse tasks. In addition to new CRISPR–Cas editing tools, DNAzymes, and DNA barcoding technologies, integrating advanced nanomaterials (such as hydrogel, metal–organic frameworks, MXenes, two-dimensional transition metal dichalcogenides, and novel quantum dots) can increase the versatility of DNA logic gates.101,102 Finally, designing multifunctional DNA nanostructures, such as DNA nanorobots, DNA origami, and dynamic nanostructures, that can operate reliably, in combination with new DNA computers and molecular logic circuits, can pave the way for more powerful and versatile DNA computing systems in clinical settings.103,104 As these systems evolve, their enhanced analytical performance and diagnostic capabilities are expected to revolutionize precision medicine, providing more accurate and personalized diagnostic tools.
It is worth noting that the integration of artificial intelligence (AI) and machine learning (ML) with DNA computing can significantly optimize the design of DNA logic operations, improve data processing efficiency, and support personalized precision medical decisions.105 AI and ML can enhance the design of DNA logic operations by using predictive and optimization algorithms to achieve higher computational accuracy and efficiency. They also enable the rapid analysis and processing of complex biological data, extracting key biological information to accelerate data processing. Based on DNA computing results, AI and ML can assist in developing personalized treatment plans for more precise medical interventions.106,107 The integration of DNA computing with other emerging technologies, such as AI and ML, could further amplify its impact, enabling faster, more reliable, and more targeted precision medicine solutions, thereby significantly improving patient outcomes in the future.
Author contributions
Conceptualization: Jiayi Wang, Lin Huang, and Fanyu Meng; writing – original draft: Yuewei Zhao, Xvelian Li, and Yan Zhou; writing – review and editing: Jiayi Wang, Lin Huang, and Fanyu Meng; visualization: Xiaoting Tian and Yayou Miao.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no competing financial interests among the authors.Acknowledgements
This work was supported by the National Natural Science Funds (Grant No. 82372148) and the National Key R&D Program of China (No. 2021YFF0703500 and 2022YFE0103500).Notes and references
- L. M. Adleman, Science, 1994, 266, 1021–1024 CrossRef CAS.
- S. Yang, B. W. A. Bögels, F. Wang, C. Xu, H. Dou, S. Mann, C. Fan and T. F. A. de Greef, Nat. Rev. Chem., 2024, 8, 179–194 CrossRef.
- P. G. Moerman and R. Schulman, Nat. Nanotechnol., 2020, 15, 626–627 CrossRef CAS PubMed.
- A. Okamoto, K. Tanaka and I. Saito, J. Am. Chem. Soc., 2004, 126, 9458–9463 CrossRef CAS PubMed.
- F. Meng, W. Yu, C. Chen, S. Guo, X. Tian, Y. Miao, L. Ma, X. Zhang, Y. Yu, L. Huang, K. Qian and J. Wang, Small, 2022, 18, 2200784 CrossRef CAS PubMed.
- J. A. N. Brophy and C. A. Voigt, Nat. Methods, 2014, 11, 508–520 CrossRef CAS PubMed.
- L. Qian, E. Winfree and J. Bruck, Nature, 2011, 475, 368–372 CrossRef CAS PubMed.
- M. Manikandan, P. Nicolini and P. Hapala, ACS Nano, 2024, 18, 9969–9979 CrossRef CAS PubMed.
- J. Li, A. A. Green, H. Yan and C. Fan, Nat. Chem., 2017, 9, 1056–1067 CrossRef CAS.
- L. Huang, L. Wang, X. Hu, S. Chen, Y. Tao, H. Su, J. Yang, W. Xu, V. Vedarethinam, S. Wu, B. Liu, X. Wan, J. Lou, Q. Wang and K. Qian, Nat. Commun., 2020, 11, 3556 CrossRef CAS PubMed.
- Y. Wu, L. Zhang, S. Berretti and S. Wan, IEEE Trans. Ind. Inform., 2023, 19, 2089–2098 Search PubMed.
- L. Huang, Y. Zhou, Y. Zhu, H. Su, S. Yang, L. Feng, L. Zhao, S. Liu and K. Qian, Biosens. Bioelectron., 2022, 210, 114254 CrossRef CAS PubMed.
- D. Fan, J. Wang, E. Wang and S. Dong, Adv. Sci., 2020, 7, 2001766 CrossRef CAS.
- Q. Ma, C. Zhang, M. Zhang, D. Han and W. Tan, Small Struct., 2021, 2, 2100051 CrossRef CAS.
- W. Xu, J. Lin, M. Gao, Y. Chen, J. Cao, J. Pu, L. Huang, J. Zhao and K. Qian, Adv. Sci., 2020, 7, 2002021 CrossRef CAS.
- Y. Zhu, X. Xiong, M. Cao, L. Li, C. Fan and H. Pei, Sci. Adv., 2023, 9, eaax7983 CrossRef CAS PubMed.
- R. Wang, Z. Gu, Y. Wang, X. Yin, W. Liu, W. Chen, Y. Huang, J. Wu, S. Yang, L. Feng, L. Zhou, L. Li, W. Di, X. Pu, L. Huang and K. Qian, Adv. Funct. Mater., 2022, 32, 2206670 CrossRef CAS.
- F. Wang, H. Lv, Q. Li, J. Li, X. Zhang, J. Shi, L. Wang and C. Fan, Nat. Commun., 2020, 11, 121 CrossRef CAS PubMed.
- S. Piranej, A. Bazrafshan and K. Salaita, Nat. Nanotechnol., 2022, 17, 514–523 CrossRef CAS PubMed.
- Y. Zhang, N. Hu, J. Xu and Z. Wang, View, 2024, 5, 20230062 CrossRef CAS.
- M. Cao, X. Xiong, Y. Zhu, M. Xiao, L. Li and H. Pei, TrAC, Trends Anal. Chem., 2023, 159, 116911 CrossRef CAS.
- A. A. Green, J. Kim, D. Ma, P. A. Silver, J. J. Collins and P. Yin, Nature, 2017, 548, 117–121 CrossRef CAS.
- T. Kitada, B. DiAndreth, B. Teague and R. Weiss, Science, 2018, 359, eaad1067 CrossRef.
- Q. Liu, K. Yang, J. Xie and Y. Sun, IEEE Internet Things J., 2022, 9, 897–915 Search PubMed.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS.
- F. Yin, F. Wang, C. Fan, X. Zuo and Q. Li, View, 2021, 2, 20200038 CrossRef CAS.
- X. Yin, D. Yao, M. H. W. Lam and H. Liang, J. Mater. Chem. B, 2022, 10, 3055–3063 RSC.
- H. Yan, G. Cao, J. Wang, X. Zhu, S. Dong, Y. Huang, M. Chao, Y. Li, F. Gao and L. Hua, Biosens. Bioelectron., 2024, 256, 116278 CrossRef CAS.
- X. Chang, C. Zhang, C. Lv, Y. Sun, M. Zhang, Y. Zhao, L. Yang, D. Han and W. Tan, J. Am. Chem. Soc., 2019, 141, 12738–12743 CrossRef CAS.
- M. Zhang, C. Yancey, C. Zhang, J. Wang, Q. Ma, L. Yang, R. Schulman, D. Han and W. Tan, Sci. Adv., 2024, 10, eadn3329 CrossRef CAS.
- Y. Liu, Y. Zhai, H. Hu, Y. Liao, H. Liu, X. Liu, J. He, L. Wang, H. Wang, L. Li, X. Zhou and X. Xiao, Adv. Sci., 2024, 11, 2400011 CrossRef CAS PubMed.
- K. N. Lin, K. Volkel, C. Cao, P. W. Hook, R. E. Polak, A. S. Clark, A. San Miguel, W. Timp, J. M. Tuck, O. D. Velev and A. J. Keung, Nat. Nanotechnol., 2024, 1–11 Search PubMed.
- N. Diao, J. Hou, X. Peng, Y. Wang, A. He, H. Gao, L. Yang, P. Guo, J. Wang and D. Han, Angew. Chem. Int. Ed., 2024, e202406330 Search PubMed.
- B. Wang, S. S. Wang, C. Chalk, A. D. Ellington and D. Soloveichik, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2217330120 CrossRef CAS PubMed.
- N. Chen, J. Yu, Z. Liu, L. Meng, X. Li and K. C. Wong, Nucleic Acids Res., 2024, 52, 4137–4150 CrossRef PubMed.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, X. Liu, Q. Ling, R. Wu, J. Yang and C. Zhang, ACS Nano, 2024, 18, 5089–5100 CrossRef CAS.
- P. Miao and Y. Tang, ACS Cent. Sci., 2021, 7, 1036–1044 CrossRef CAS.
- Q. Chai, J. Chen, S. Zeng, T. Zhu, J. Chen, C. Qi, G. Mao and Y. Liu, Small, 2023, 19, 2207736 CrossRef CAS.
- Z. Wang, Y. Hu and L. Pan, Angew. Chem. Int. Ed., 2020, 59, 14979–14985 CrossRef CAS PubMed.
- A. P. Lapteva, N. Sarraf and L. Qian, J. Am. Chem. Soc., 2022, 144, 12443–12449 CrossRef CAS.
- G. Shi, C. Yan and J. Chen, Anal. Chem., 2021, 93, 3273–3279 CrossRef CAS.
- T. Wang and F. C. Simmel, Angew. Chem. Int. Ed., 2023, 62, e202302858 CrossRef CAS PubMed.
- M. Hu, X. Li, J. Wu, M. Yang and T. Wu, ACS Nano, 2024, 18, 2184–2194 CrossRef CAS.
- W. J. Zeng, X. R. Li, W. Liu, R. Yuan, W. B. Liang and Y. Zhuo, Anal. Chem., 2024, 96, 2117–2123 CrossRef CAS.
- Z. Li, M. Yang, Y. Shu and J. Wang, Sens. Actuators, B, 2023, 391, 134063 CrossRef CAS.
- M. Li, B. Yao, C. Jing, H. Chen, Y. Zhang and N. Zhou, Anal. Chim. Acta, 2022, 1198, 339559 CrossRef CAS.
- J. Wang, J. Han, J. Wang, X. Lv, D. Fan and S. Dong, Biosens. Bioelectron., 2024, 244, 115801 CrossRef CAS PubMed.
- W. Y. Lv, C. H. Li, F. F. Yang, Y. F. Li, S. J. Zhen and C. Z. Huang, Angew. Chem. Int. Ed., 2022, 61, e202115561 CrossRef CAS PubMed.
- E. Bethur, R. Guha, Z. Zhao, B. B. Katz, P. D. Ashby, H. Zeng and S. M. Copp, ACS Nano, 2024, 18, 3002–3010 CrossRef CAS PubMed.
- R. Gao, X.-S. Wei, Z. Chen, A. Xie and W. Dong, ACS Nano, 2023, 17, 18055–18061 CrossRef CAS PubMed.
- J. Yu, Q. Liu, L. Qi, Q. Fang, X. Shang, X. Zhang and Y. Du, Biosens. Bioelectron., 2024, 252, 116137 CrossRef CAS PubMed.
- J. Han, X. Lv, Y. Zhang, J. Wang, D. Fan and S. Dong, Anal. Chem., 2023, 95, 16725–16732 CrossRef CAS.
- H. Geng, Z. Yin, C. Zhou and C. Guo, Acta Biomater., 2020, 118, 44–53 CrossRef CAS PubMed.
- Y. Pei, T. Bian, Y. Liu, Y. Liu, Y. Xie and J. Song, Nano Lett., 2022, 22, 3003–3010 CrossRef CAS PubMed.
- M. Lv, W. Zhou, D. Fan, Y. Guo, X. Zhu, J. Ren and E. Wang, Adv. Mater., 2020, 32, 1908480 CrossRef CAS.
- K. He, H. Yang, L. Wang, J. Guan, M. Wu, H. He, S. Gunasekaran, X. Wang, Q. Wang and X. Xu, Chem. Eng. J., 2019, 368, 877–887 CrossRef CAS.
- S. Su, H. Sun, W. Cao, J. Chao, H. Peng, X. Zuo, L. Yuwen, C. Fan and L. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 6826–6833 CrossRef CAS.
- Z. Q. Bu, Q. F. Yao, Q. Y. Liu, M. X. Quan, J. Y. Lu and W. T. Huang, ACS Appl. Mater. Interfaces, 2022, 14, 8311–8321 CrossRef CAS PubMed.
- N. M. Figueiredo, I. V. Voroshylova, E. S. C. Ferreira, J. M. C. Marques and M. N. D. S. Cordeiro, Chem. Rev., 2024, 124, 3392–3415 CrossRef CAS.
- N. Xie, M. Li, Y. Wang, H. Lv, J. Shi, J. Li, Q. Li, F. Wang and C. Fan, J. Am. Chem. Soc., 2022, 144, 9479–9488 CrossRef CAS PubMed.
- D. Fan, E. Wang and S. Dong, ACS Appl. Mater. Interfaces, 2017, 9, 1322–1330 CrossRef CAS PubMed.
- W. Engelen, S. P. W. Wijnands and M. Merkx, J. Am. Chem. Soc., 2018, 140, 9758–9767 CrossRef CAS PubMed.
- A. Phillips, Nature, 2024, 625, 454–455 CrossRef CAS PubMed.
- B. Yang, D. Guneri, H. Yu, E. P. Wright, W. Chen, Z. A. E. Waller and Y. Ding, Nucleic Acids Res., 2024, 52, 2188–2197 CrossRef.
- L. Zhang, X. Ma, G. Wang, X. Liang, H. Mitomo, A. Pike, A. Houlton and K. Ijiro, Nano Today, 2021, 39, 101154 CrossRef CAS.
- X. Xiong, T. Zhu, Y. Zhu, M. Cao, J. Xiao, L. Li, F. Wang, C. Fan and H. Pei, Nat. Mach. Intell., 2022, 4, 625–635 CrossRef.
- Q. Lin, A. Wang, S. Liu, J. Li, J. Wang, K. Quan, X. Yang, J. Huang and K. Wang, Chem. Commun., 2020, 56, 5303–5306 RSC.
- J. Pan, Y. He, Z. Liu and J. Chen, Anal. Chem., 2022, 94, 714–722 CrossRef CAS PubMed.
- X. Zhang, Y. Dong, Y. Wang, Z. Zhang, X. Zhang, J. J. Zhu, Y. Tian and Q. Min, Angew. Chem. Int. Ed., 2024, 63, e202313446 CrossRef CAS PubMed.
- V. Maingi, Z. Zhang, C. Thachuk, N. Sarraf, E. R. Chapman and P. W. K. Rothemund, Nat. Commun., 2023, 14, 1532 CrossRef CAS.
- S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. D. Nijenhuis, B. Saccà, F. C. Simmel, H. Yan and P. Zhan, Nat. Rev. Methods Primers, 2021, 1, 1–24 CrossRef.
- M. DeLuca, D. Duke, T. Ye, M. Poirier, Y. Ke, C. Castro and G. Arya, Nat. Commun., 2024, 15, 3015 CrossRef CAS.
- H. D. Li, P. Q. Ma, J. Y. Wang, B. C. Yin and B. C. Ye, Small, 2023, 19, 2302301 CrossRef CAS.
- H. Lv, N. Xie, M. Li, M. Dong, C. Sun, Q. Zhang, L. Zhao, J. Li, X. Zuo, H. Chen, F. Wang and C. Fan, Nature, 2023, 622, 292–300 CrossRef CAS PubMed.
- J. J. Tabor and A. D. Ellington, Nat. Biotechnol., 2003, 21, 1013–1015 CrossRef CAS.
- T. Yuan, L. Wu, S. Li, J. Zheng, N. Li, X. Xiao, H. Zhang, T. Fei, L. Xie, Z. Zuo, D. Li, P. Huang, H. Feng, Y. Cao, N. Yan, X. Wei, L. Shi, Y. Sun, W. Wei, Y. Sun and E. Zuo, Cell Discovery, 2024, 10, 1–19 Search PubMed.
- F. Meng, H. Chai, X. Ma, Y. Tang and P. Miao, J. Mater. Chem. B, 2019, 7, 1926–1932 RSC.
- W. Chen, H. Yu, Y. Hao, W. Liu, R. Wang, Y. Huang, J. Wu, L. Feng, Y. Guan, L. Huang and K. Qian, ACS Nano, 2023, 17, 19779–19792 CrossRef CAS PubMed.
- X. Li, W. Chen, M. Wu, W. Yu, M. Wang, M. Niu, F. Meng, Y. Zhao, A. Osman, N. O. Mousa, H. Shi, K. Qian, J. Wang and L. Huang, Chem. Eng. J., 2024, 486, 150224 CrossRef CAS.
- J. Yu, T. Han, X. Lin, X. Song, H. Xie, J. He and W. Chen, J. Clin. Oncol., 2019, 37, e13051 Search PubMed.
- C. Company, M. J. Schmitt, Y. Dramaretska, M. Serresi, S. Kertalli, B. Jiang, J. A. Yin, A. Aguzzi, I. Barozzi and G. Gargiulo, Nat. Commun., 2024, 15, 897 CrossRef CAS PubMed.
- Y. Wang, S. Wang, L. Li, Y. Zou, B. Liu and X. Fang, View, 2023, 4, 20220048 CrossRef.
- C. Feng, T. Chen, D. Mao, F. Zhang, B. Tian and X. Zhu, ACS Sens., 2020, 5, 3116–3123 CrossRef CAS.
- F. Meng, W. Yu, M. Niu, X. Tian, Y. Miao, X. Li, Y. Zhou, L. Ma, X. Zhang, K. Qian, Y. Yu, J. Wang and L. Huang, J. Nanobiotechnol., 2023, 21, 104 CrossRef CAS PubMed.
- X. Q. Li, Y. L. Jia, Y. W. Zhang, H. Y. Chen and J. J. Xu, Chem. Sci., 2023, 14, 7699–7708 RSC.
- K. Wei, M. He, J. Zhang, C. Zhao, C. Nie, T. Zhang, Y. Liu, T. Chen, J. Jiang and X. Chu, Angew. Chem. Int. Ed., 2023, 62, e202307025 CrossRef CAS PubMed.
- Y. Zhang, W. Chen, Y. Fang, X. Zhang, Y. Liu and H. Ju, J. Am. Chem. Soc., 2021, 143, 15233–15242 CrossRef CAS.
- Q. Yang, F. Yang, W. Dai, X. Meng, W. Wei, Y. Cheng, J. Dong, H. Lu and H. Dong, Adv. Healthcare Mater., 2021, 10, 2101130 CrossRef CAS.
- J. Luo, B. Wang, X. Tang, P. Huang, S. Yang, S. Zhao, S. Xie, Q. Li, K. Chang and M. Chen, View, 2024, 5, 20230086 CrossRef CAS.
- J. Liu, C. Zhang, J. Song, Q. Zhang, R. Zhang, M. Zhang, D. Han and W. Tan, Adv. Sci., 2023, 10, 2206343 CrossRef CAS.
- C. Zhang, Y. Zhao, X. Xu, R. Xu, H. Li, X. Teng, Y. Du, Y. Miao, H. Lin and D. Han, Nat. Nanotechnol., 2020, 15, 709–715 CrossRef CAS PubMed.
- Q. Ma, M. Zhang, C. Zhang, X. Teng, L. Yang, Y. Tian, J. Wang, D. Han and W. Tan, Sci. Adv., 2022, 8, eade0453 CrossRef CAS PubMed.
- C. Zhang, T. Zheng, Q. Ma, L. Yang, M. Zhang, J. Wang, X. Teng, Y. Miao, H. Lin, Y. Yang and D. Han, Angew. Chem. Int. Ed., 2022, 61, e202117658 CrossRef CAS.
- F. Yin, H. Zhao, S. Lu, J. Shen, M. Li, X. Mao, F. Li, J. Shi, J. Li, B. Dong, W. Xue, X. Zuo, X. Yang and C. Fan, Nat. Nanotechnol., 2023, 18, 677–686 CrossRef CAS.
- Z. Wang, J. Yang, G. Qin, C. Zhao, J. Ren and X. Qu, Angew. Chem. Int. Ed., 2022, 61, e202204291 CrossRef CAS PubMed.
- D. Mao, Z. Dong, X. Liu, W. Li, H. Li, C. Gu, G. Chen, X. Zhu and Y. Yang, Angew. Chem. Int. Ed., 2024, 63, e202311309 CrossRef CAS PubMed.
- R. Yue, M. Chen and N. Ma, ACS Appl. Mater. Interfaces, 2020, 12, 32493–32502 CrossRef CAS PubMed.
- C. Ouyang, S. Zhang, C. Xue, X. Yu, H. Xu, Z. Wang, Y. Lu and Z. S. Wu, J. Am. Chem. Soc., 2020, 142, 1265–1277 CrossRef CAS PubMed.
- C. Li, M. Wang, P.-F. Li, J. Sheng and Q. Fu, Small, 2024, 20, e2306257 CrossRef PubMed.
- R. Fu, J. Hou, Z. Wang and Y. Xianyu, ACS Nano, 2024, 18, 14754–14763 CrossRef CAS PubMed.
- J. Chen, J. Hu and R. Kapral, Adv. Sci., 2024, 11, 2305695 CrossRef CAS.
- L. Yang, Q. Tang, M. Zhang, Y. Tian, X. Chen, R. Xu, Q. Ma, P. Guo, C. Zhang and D. Han, Nat. Commun., 2024, 15, 4583 CrossRef CAS PubMed.
- L. Yang, Y. Zhao, X. Xu, K. Xu, M. Zhang, K. Huang, H. Kang, H.-C. Lin, Y. Yang and D. Han, Angew. Chem., Int. Ed., 2020, 59, 17697–17704 CrossRef CAS PubMed.
- A. Pucchio, E. A. Eisenhauer and F. Y. Moraes, Nat. Biotechnol., 2021, 39, 388–389 CrossRef CAS.
- D. A. Winkler, Small, 2020, 16, e2001883 CrossRef PubMed.
- B. Babic, S. Gerke, T. Evgeniou and I. G. Cohen, Nat. Mach. Intell., 2021, 3, 283–287 CrossRef.
| This journal is © The Royal Society of Chemistry 2024 |