Open Access Article
Open Access ArticleCyaonoside A-loaded composite hydrogel microspheres to treat osteoarthritis by relieving chondrocyte inflammation†
Xingyan
An‡
a,
Fengjin
Zhou‡
b,
Guangfeng
Li‡
a,
Yan
Wei
a,
Biaotong
Huang
ac,
Mengmeng
Li
a,
Qin
Zhang
 a,
Ke
Xu
*ac,
Robert Chunhua
Zhao
*defg and
Jiacan
Su
a,
Ke
Xu
*ac,
Robert Chunhua
Zhao
*defg and
Jiacan
Su
 *ah
*ah
aInstitute of Translational Medicine, Shanghai University, Shanghai, 200444, China. E-mail: kexu@shu.edu.cn; drsujiacan@163.com; 13910079395@vip.163.com
bDepartment of Orthopedics, Honghui Hospital, Xi’an Jiao Tong University, Xi’an, 710000, China
cWenzhou Institute of Shanghai University, Wenzhou 325000, China
dInstitute of Basic Medical Sciences Chinese Academy of Medical Sciences, School of Basic Medicine Peking Union Medical College, Beijing, 100190, China. E-mail: zhaochunhua@ibms.pumc.edu.cn
eCenter for Excellence in Tissue Engineering, Chinese Academy of Medical Sciences, Beijing, 100730, China
fBeijing Key Laboratory of New Drug Development and Clinical Trial of Stem Cell Therapy, Beijing, 100730, China
gState Key Laboratory of Common Mechanism Research for Major Diseases, Beijing, 100005, China
hDepartment of Orthopaedics, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, 200092, China
First published on 9th April 2024
Abstract
Cyaonoside A (CyA), derived from the natural Chinese medicine, Cyathula officinalis Kuan, which was for a long time used to treat knee injuries and relieve joint pain in traditional Chinese medicine, showed an unclear mechanism for protecting cartilage. In addition, CyA was poorly hydrosoluble and incapable of being injected directly into the joint cavity, which limited its clinical application. This study reveals that CyA resisted IL-1β-mediated chondrogenic inflammation and apoptosis. Next, transcriptome sequencing is used to explore the potential mechanisms underlying CyA regulation of MSC chondrogenic differentiation. Based on these findings, CyA-loaded composite hydrogel microspheres (HLC) were developed and they possessed satisfactory loading efficiency, a suitable degradation rate and good biocompatibility. HLC increased chondrogenic anabolic gene (Acan, COL2A, and SOX9) expression, while downregulating the expression of the catabolic marker MMP13 in vitro. In the osteoarthritis mouse model, HLC demonstrated promising therapeutic capabilities by protecting the integrity of articular cartilage. In conclusion, this study provides insights into the regulatory mechanisms of CyA for chondrocytes and proposes a composite hydrogel microsphere-based advanced therapeutic strategy for osteoarthritis.
1. Introduction
Osteoarthritis (OA) is prevalent in middle-aged and older adults, with 10% of the world's population aged 60 or older facing a serious clinical problem of OA, and the incidence of the disease is increasing annually.1,2 A statistical analysis of OA-related data from 195 countries over the past 30 years showed that the number of OA patients had increased by 9.3% compared to 1990, affecting an estimated 303 million people worldwide.3 In addition, with the prevalence of obesity in the population and the increased chance of accidental joint injuries due to exercise, the age of onset of this joint disease has gradually tended to be younger, which warrants a high degree of vigilance.4–6 As a heterogeneous and complex disease with unclear mechanisms, the objective of the current clinical treatment of OA is to decelerate or inhibit joint damage as much as possible to avoid excessive wear and tear leading to joint dysfunction or amputation, which seriously affects the quality of life.7As a result of noxious mechanical stimulation or stress wear, cells in the joint cavity will secrete large amounts of inflammatory factors and matrix-degrading enzymes into the articular cavity triggering a series of downstream responses, such as interleukin-1β (IL-1β), matrix metalloproteinase 13 (MMP13), and so on.8,9 These inflammatory mediators alter the normal phenotype of chondrocytes and inhibit chondrocyte synthesis of cartilage matrix components (proteoglycans and type II collagen).10,11 Comparison of joint samples from osteoarthritis patients with normal joint samples by microarray analysis revealed the presence of 754 significantly differentially expressed genes.12 Among them, cartilage matrix degrading enzymes and related inflammatory factors were upregulated in samples from patients with OA, further confirming that chondrocyte inflammation was an accomplice in the deterioration of OA.12 However, nonsteroidal anti-inflammatory drugs (NSAIDs), which are frequently used in clinical practice, are known to have significant side effects, which may trigger gastrointestinal bleeding, cardiovascular damage, and some nephrotoxicity in patients with underlying cardiovascular disease after oral administration.13,14 Modifying the application of such drugs from oral to topical can greatly reduce side effects, but their efficacy may also be reduced when applied topically due to the structural characteristics of the knee joint.15–17 Therefore, searching for innovative drugs and designing drug delivery strategies suitable for the articular cavity microenvironment may provide new therapeutic countermeasures for OA.
Previous reports have shown that natural herbs contain a wide range of pharmacologically active components, so the development of anti-inflammatory drugs based on natural medicinal plants is a promising option.18 The Compendium of Materia Medica states that Cyathula officinalis Kuan belongs to the Amaranthaceae family which can tonify liver and kidneys, strengthen muscles and bones, and alleviate the soreness and weakness of the waist and knees.19,20 Therefore, Cyathula officinalis Kuan has long-term been used in the treatment of knee diseases and has been used in developing a variety of Chinese medicinal preparations.21 In the previous study, Cyathula officinalis Kuan extract could reverse IL-1β-induced inflammation and down-regulate the expression of apoptosis-related proteins Bax.22 In terms of mechanisms, Cyathula officinalis Kuan inhibited the activity of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) by regulating the nuclear factor kappa B (NF-κB) signaling and mitogen-activated protein kinase (MAPK) pathway to relieve inflammation progression in vitro and in vivo.19,22 As one of the main components of Cyathula officinalis Kuan, Cyaonoside A (CyA) has potential cartilage-protective effects, but its therapeutic mechanism has not yet been fully clarified.19
For diseases localized to the joints, such as OA, it is difficult to achieve drug concentration at the site of the lesion with oral administration, and the structural limitations of the drug lead to rapid clearance when administered locally.23,24 Therefore, designing an appropriate drug delivery strategy based on the drug's individual properties is a critical part of achieving efficient OA therapy.25,26 Conventional single-component hydrogels lack specific functionality, often fail to maximize their functional benefits, and are only able to load hydrophilic drug molecules.27–29 In contrast, the poor chemical and physical stability of liposomes leads to the possibility of drug leakage from liposomes during storage or transportation, which affects the efficiency of drug delivery in practical applications.30 Inspired by this, liposomes and hydrogels are combined to form composite hydrogels that maximize the superiority of both biomaterials.31,32 Composite hydrogels facilitate the encapsulation of hydrophobic and hydrophilic drugs while enhancing the mechanical properties and stability of liposomal formulations for localized, sustained drug release, thereby improving ease of administration and efficiency of delivery at the site of the lesion.33–35 Since hyaluronic acid (HA) is one of the components of synovial fluid, HA-based hydrogel microspheres not only provide better dispersion in the joint cavity, but also provide certain lubricating and cushioning properties.32,36 Meanwhile, hydrogel microspheres, which have most of the advantages of traditional hydrogels, can be applied to deliver a variety of drugs or biomolecules, offering high promise for clinical applications.37,38
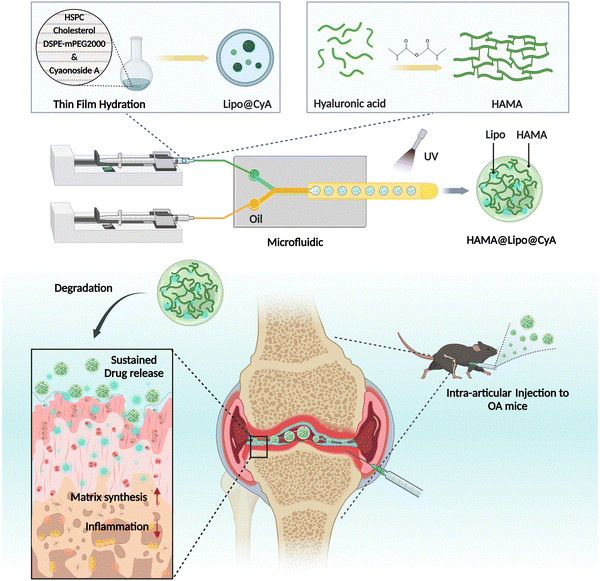
Here, we examined CyA regulation of chondrocytes in protecting cartilage and devised composite hydrogel microspheres for sustained release of CyA to alleviate osteoarthritis inflammation and remodel the cartilage matrix (Fig. 1). First, CyA was found to be effective in alleviating IL-1β-induced chondrocyte inflammation and apoptosis as well as inducing bone marrow mesenchymal stem cell (BMSC) chondrogenesis. Subsequently, CyA was encapsulated in phospholipid bilayer liposomes (Lipo@CyA) which were synthesized via thin-film hydration. Composite hydrogel microspheres (HAMA@Lipo@CyA, HLC) were prepared using a microfluidic approach in order to mix Lipo@CyA with methacrylated hyaluronic acid (HAMA) by UV cross-linking. This strategy not only overcame the self-imposed limitations of the hydrophobic drug CyA, but also allowed HLC to delay the progression of OA in DMM model rats with sustained release of the drug throughout treatment. HLC alleviated inflammation in the articular microenvironment and restored the cartilage collagen matrix, suggesting that it could be a potential alternative for OA management.
2. Materials and methods
2.1 Materials and cells
Cyaonoside A (CyA) was purchased from Chengdu HerbSubstance Co., Ltd (Chengdu, China). Hyaluronic acid methacryloyl (HAMA) was used to make hydrogel microspheres purchased from Engineering for Life (EFL, Suzhou, China). Hydrogenated soybean phosphatidylcholine (HSPC) and cholesterol were purchased from Macklin (Shanghai, China). DSPE-MPEG2000 was purchased from A.V.T. Pharmaceutical Co. Ltd (Shanghai, China). IL-1β was acquired from Sino Biological, Inc. (Beijing, China). Human primary chondrocytes and bone marrow mesenchymal stem cells (BMSCs) used in the experiment were purchased from Cyagen Biosciences Inc (Suzhou, China). The basal medium, fetal bovine serum (FBS) and penicillin/streptomycin (PS) required for cell culture were purchased from Gibco (USA). All cells were cultured in a thermostat incubator at 37 °C, saturated humidity, and 5% CO2. All the other reagents were purchased from Sigma-Aldrich (Germany) unless otherwise noted.2.2 Animals
Two-month-old male SD rats were modeled for osteoarthritis by creating destabilization of the medial meniscus (DMM) through surgery. The surgery was performed on the right knee joint of rats by injecting 10 μL of different therapeutic agents into the joint cavity using a microinjector. After 8 weeks of treatment, the rats were euthanized and tissues from the right total knee joint were removed for subsequent experiments. The experimental rats were provided by Changzhou Cavens Laboratory Animal Co. All animal experiments were performed according to the guidelines evaluated and approved by the ethics committee of Shanghai University.2.3 Synthesis of Lipo@CyA and HAMA@Lipo@CyA
According to previous reports, Liposomes were synthesized using the film hydration method.39 HSPC, cholesterol and DSPE-MPEG-2000 were dissolved in chloroform at a molar ratio of 56.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.4
38.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.3, and then spun to obtain a dried lipid film. The mixture was completely evaporated from the chloroform under vacuum, then ultrapure water was added and stirred in a 60 °C water bath for 30 minutes. The mixture was then sonicated for 30 minutes to obtain a liposome suspension. The resulting liposome suspension was passed through a polycarbonate membrane extruder (Avanti, USA) to obtain liposomes of uniform particle size. Drug-loaded liposomes were prepared in the molar ratio of HSPC
5.3, and then spun to obtain a dried lipid film. The mixture was completely evaporated from the chloroform under vacuum, then ultrapure water was added and stirred in a 60 °C water bath for 30 minutes. The mixture was then sonicated for 30 minutes to obtain a liposome suspension. The resulting liposome suspension was passed through a polycarbonate membrane extruder (Avanti, USA) to obtain liposomes of uniform particle size. Drug-loaded liposomes were prepared in the molar ratio of HSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DSPE-MPEG2000
DSPE-MPEG2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CyA = 56.3
CyA = 56.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.4
38.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.3
5.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The prepared liposomes were stored at 4 °C for subsequent experiments.
2. The prepared liposomes were stored at 4 °C for subsequent experiments.
Hydrogel microspheres were synthesized by a monodisperse microsphere fabricator developed by Suzhou Yongqinquan Intelligent Equipment Co. The HAMA, Lipo@CyA and photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) mixed solution and paraffin oil were loaded into two syringes as the internal and external phases, respectively, and passed through the central microfluidic chip and crosslinked gelation under 405 nm UV irradiation to obtain monodisperse hydrogel microspheres. In this case, the concentration of HAMA was 5%, the concentration of LAP was 0.25%, the Lipo@CyA weight ratio was 2.5%, and the paraffin oil contained 5% Span80.
2.4 Characterization of Lipo@CyA and HAMA@Lipo@CyA
To examine the physical properties of liposomes, we used a dynamic light scattering particle size dispersion instrument (Malvern, UK) to examine the particle size distribution, polydispersity index, and zeta potential of liposomes. The morphology of liposomes was observed using transmission electron microscopy (TEM, JEOL, Japan). The samples were fixed with glutaraldehyde before observation, and then the sample solution was diluted to a suitable concentration and added dropwise to the copper grid. The morphology of the hydrogel microspheres was observed using a scanning electron microscope (SEM, Hitachi, Japan). The prepared hydrogel microspheres were lyophilized before electron microscopic observation and then the samples were conductively coated using a gold sprayer etc. In addition, energy dispersive X-ray spectroscopy (EDS) mapping was performed, focusing on the C, O, P, and N elements in the microspheres. The particle size distribution of microspheres was analyzed by counting the diameters of 161 microspheres in the bright field photographs by Image J software.2.5 CyA loading and release
To further confirm the encapsulation of Lipo@CyA in composite hydrogel microspheres, the lipid-soluble dye DID (Beyotime, China) was added to label the liposomes during the synthesis of Lipo@CyA. Subsequently, the morphology of the hydrogel microspheres was observed and 3D reconstructed by laser confocal scanning microscopy (LSCM, Olympus, Japan). Samples of Lipo@CyA and HAMA@Lipo@CyA were selected for comparison, encapsulated in 10 kDa dialysis bags and immersed in PBS solution. They were placed on a constant temperature shaker at 37 °C for 28 days. Then the supernatant was aspirated to determine the amount of drug release by high performance liquid chromatography (HPLC, Shimadzu, Japan), and the total amount of drug release was used to calculate the encapsulation rate.2.6 Degradation tests
During in vitro degradation experiments, we immersed HAMA@Lipo@CyA hydrogel microspheres in 10 U mL−1 hyaluronidase solution and placed them on a constant temperature shaker at 80 rpm. The immersion liquid of the microspheres was changed every two days. The samples were taken out on days 1, 3, 7, 14, 21, and 28 to observe the morphology under a microscope. All samples were lyophilized and weighed to calculate the relative degradation rate.2.7 Cytotoxicity assay
To test the cytotoxicity of hydrogels, samples from different experimental groups were co-cultured with human chondrocytes. First, human chondrocytes were inoculated in 96-well plates at 1 × 104 cells per well. Then, the next day, complete medium was changed and hydrogel microspheres from different experimental groups were added to co-culture with the cells. The absorbance value at 450 nm of the supernatant was measured after 24 h, 48 h and 72 h using Cell Counting Kit-8 (Beyotime, China) to assess the cytotoxic effect of the hydrogel.2.8 Hemolysis assay
500 μL blood was collected from rat eyeballs, then 5 mL of PBS solution was added and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 5 min, the supernatant was discarded. Then, PBS solution was added to 10 mL and mixed well, then centrifuged at 10
000 g for 5 min, the supernatant was discarded. Then, PBS solution was added to 10 mL and mixed well, then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min until the supernatant was clarified. Resuspension of erythrocytes was obtained by adding PBS again to 10 mL. 200 μL of erythrocyte suspension were taken and incubated with microspheres on a constant temperature shaker at 100 rpm for 4 h and then observed for hemolysis. The hemolysis rate was calculated by measuring the absorbance value at 570 nm in the supernatant using a microplate reader (BioTek, USA).
000 × g for 5 min until the supernatant was clarified. Resuspension of erythrocytes was obtained by adding PBS again to 10 mL. 200 μL of erythrocyte suspension were taken and incubated with microspheres on a constant temperature shaker at 100 rpm for 4 h and then observed for hemolysis. The hemolysis rate was calculated by measuring the absorbance value at 570 nm in the supernatant using a microplate reader (BioTek, USA).
2.9 Alcian blue staining
Alcian blue is a cationic dye that reacts with proteoglycan glycans on cartilage tissue to give a dark blue color to analyze the effects of chondrocyte matrix degradation in samples. Cells were first inoculated in well plates, washed with PBS and fixed with 4% formaldehyde for 20 min. The stained tissue was incubated for 30 min using Alcian blue Staining Solution (Servicebio, China), and then washed three times with pure water for 5 min each time. Images were observed under Biotek Cytation5 (BioTek, USA).2.10 Quantitative real-time polymerase chain reaction (qRT-PCR)
Cell total RNA was extracted using RNAiso Plus (Takara, Japan) according to the manufacturer's instructions. Chloroform and isopropanol were used to purify mRNA. mRNA obtained was washed with 75% ethanol and then reverse transcribed to obtain cDNA using the SYBR PrimerScript RT-PCR kit (Takara, Japan). The reaction was carried out in a volume of 20 μL with QTOWER (Analytik Jena, Germany). qRT-PCR was performed under the following reaction conditions: 30 s at 95 °C, 1 cycle, 5 s at 95 °C, 30 s at 55 °C, and 40 cycles. The expression of each target gene was calculated using 2−ΔΔCT with GAPDH as the internal reference gene. Table S1 (ESI†) presented the primer sequences used.2.11 Western blot
After adding 100 μL RIPA protein lysate (Beyotime, China) containing protease inhibitors to the chondrocytes, total cellular proteins were scraped using a cell scraper. The supernatant was collected by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min after lysis on ice for 30 min. All samples were electrophoresed on polyacrylamide gels at 120 V for 70 min to separate proteins of different molecular weights. Subsequently all bands were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA) and blocked with skim milk powder. The diluted Acan, COL2A, SOX9, MMP13, and GAPDH antibodies (Abcam, UK) were incubated at 4 °C overnight. Subsequently, HRP-coupled secondary antibodies were incubated with the strips for 1 h and then the bands were visualized and photographed using a chemiluminescence instrument (Bio-Rad, USA). All bands were normalized and quantified using ImageJ software.
000 rpm for 10 min after lysis on ice for 30 min. All samples were electrophoresed on polyacrylamide gels at 120 V for 70 min to separate proteins of different molecular weights. Subsequently all bands were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA) and blocked with skim milk powder. The diluted Acan, COL2A, SOX9, MMP13, and GAPDH antibodies (Abcam, UK) were incubated at 4 °C overnight. Subsequently, HRP-coupled secondary antibodies were incubated with the strips for 1 h and then the bands were visualized and photographed using a chemiluminescence instrument (Bio-Rad, USA). All bands were normalized and quantified using ImageJ software.
2.12 Immunocytofluorescence
Immunocytofluorescence staining was used to detect the expression levels of COL2A and MMP13 in human chondrocytes. In brief, chondrocytes were pre-seeded on cell crawls co-cultured with different groups of hydrogel microspheres. All cells were fixed by 4% paraformaldehyde solution at 4 °C for 12 h and permeabilized with PBS containing 0.5% Triton X-100 for 10 min. The non-target protein was blocked using goat serum. Appropriately diluted COL2A-antibody (Abcam, UK) or MMP13-antibody (Abcam, UK) was added dropwise onto the crawler slides and incubated at 4 °C for 12 h. The next day the secondary antibody was added and incubated for 1 h. Finally, the slices were blocked using a DAPI-containing anti-fluorescence quencher and photographed using fluorescence microscopy.2.13 Apoptosis assay
Apoptosis of chondrocytes was measured using the Annexin V, FITC Apoptosis Detection Kit (Dojino, Japan) using flow cytometry according to the manufacturer's instructions. First, the medium was removed from the chondrocytes in different groups and washed three times with PBS. The cells were digested and centrifuged at 1000 rpm for 3 min to discard the supernatant. The final concentration of 1 × 106 cells per mL was made by adding pre-prepared 1 × Annexin V Binding Solution. 5 μL of Annexin V, FITC conjugate, and 5 μL of PI Solution were added to the cell suspension. Subsequently incubated for 15 min away from light. After incubation for 15 min away from light, the detection was carried out on flow cytometry (Beckman Coulter, USA) with an excitation wavelength of 488 nm and an emission wavelength of 530 nm.2.14 Micro-CT analysis
Knee samples from each group of rats were scanned and reconstructed using a microcomputed tomography system (Micro-CT, Bruker, Germany) after fixation with paraformaldehyde. The entire process used X-rays of 80 kV at 60 μA for scanning. Regions of interest were selected for importing into CTAn software for further analysis of the bone volume fraction (BV/TV), trabecular thickness (Tb.Th) and trabecular separation (Tb.Sp). Three-dimensional images of the entire medial tibial plateau subchondral bone sagittal plane were reconstructed with CTvol software.2.15 Histopathological and immunohistochemical studies
Rat whole knee samples were soaked in PBS solution containing 10% EDTA for 6 weeks to make them sufficiently decalcified. The decalcification solution was changed every three days during this period. Paraffin-embedded joint samples were sectioned in the sagittal plane. Hematoxylin and eosin (H&E) staining and Safranin O/Fast green (S&O) staining were performed using standardized procedures to observe cartilage integrity. Immunohistochemistry was used to detect the expression of COL-2 and MMP13 in articular cartilage.2.16 RNA-seq sequencing
Total RNA was isolated using the Trizol Reagent (Invitrogen, USA), after which the concentration, quality and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific, USA). Three micrograms of RNA were used as input material for the RNA sample preparations. Sequencing libraries were generated according to the following steps. Firstly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in an Illumina proprietary fragmentation buffer. First strand cDNA was synthesized using random oligonucleotides and Super Script II. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangings were converted into blunt ends via exonuclease/polymerase activities and the enzymes were removed. After adenylation of the 3′ ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. To select cDNA fragments of the preferred 400–500 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, USA). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using Illumina PCR Primer Cocktail in a 15-cycle PCR reaction. Products were purified and quantified using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent, USA). The sequencing library was then sequenced and analyzed on a NovaSeq 6000 platform (Illumina, USA).2.17 Statistical analysis
All statistical analyses were performed using Excel 2016 and GraphPad Prism 9. Each experimental value was expressed as the mean ± standard deviation. An unpaired t-test was performed for comparisons between two groups and one-way analysis of variance (ANOVA) was applied for pairwise comparisons among multiple groups. The significance was accepted at *p < 0.05, **p < 0.01 and ***p < 0.001. p > 0.05 was represented as no significant difference (ns).3. Results and discussion
3.1 CyA alleviated human chondrocyte inflammation and promoted chondrogenesis
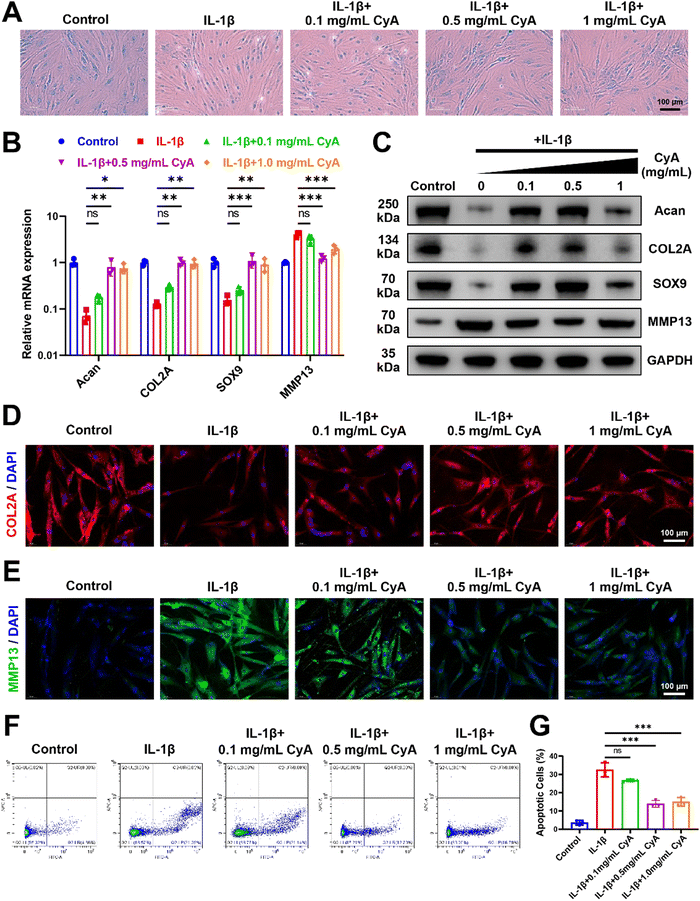
To investigate the regulatory effects of CyA on OA chondrocytes, IL-1β was used to mimic chondrocyte alterations in the inflammatory microenvironment. Addition of different concentrations of CyA restored the extracellular matrix content of human chondrocytes, as evidenced by an increase in Alcian blue stained area (Fig. 2A). As shown in Fig. 2B, CyA addition restored the expression of chondrocyte anabolic genes Aggrecan (Acan), collagen type II alpha (COL2A), and SRY-Box Transcription Factor 9 (SOX9) in an inflammatory environment. Meanwhile, relative to the IL-1β group, the expression of MMP13 was significantly reduced in the 0.5 mg mL−1 CyA group and the 1 mg mL−1 CyA group. Similar conclusions were confirmed at the protein level (Fig. 2C). CyA at 0.5 mg mL−1 had the most significant anti-inflammatory capacity, rather than the highest dose 1 mg mL−1 group (Fig. S1, ESI†). As a major component of the articular cartilage matrix, alterations in the levels of COL2A in chondrocytes were essential in responding to the severity of OA.40 In immunofluorescence images, chondrocytes in the IL-1β group barely expressed COL2A but substantially increased intracellular MMP13 levels (Fig. 2D and E). The addition of CyA restored intracellular COL2A and MMP13 levels to baseline levels. In addition to cellular inflammation, chondrocyte apoptosis triggered by the OA microenvironment was also a causative factor for articular cartilage damage.41 Previous report revealed that CyA had an inhibitory effect on rodent chondrocyte apoptosis.22 Therefore, we analyzed the apoptotic alterations in human-derived chondrocytes. Flow cytometry results showed that 0.5 mg mL−1 CyA and 1 mg mL−1 CyA had similar inhibitory abilities for IL-1β-induced apoptosis (Fig. 2F). In the 0.5 mg mL−1 CyA group, the apoptotic cell percentage decreased from 32.45% to 13.88% (Fig. 2G). More interestingly, CyA upregulated the chondrogenic differentiation capacity of human BMSCs (hBMSCs) (Fig. S2, ESI†). After 14 days of incubation with different concentrations of CyA, chondrogenic marker expression in hBMSCs was most significant in the 0.5 mg mL−1 CyA group, as evidenced by the increased levels of mRNA transcription and protein synthesis (Fig. S3 and S4, ESI†). These results indicated that 0.5 mg mL−1 CyA not only alleviated chondrocyte inflammation and apoptosis, but also promoted chondrogenic differentiation of hBMSCs, implying that CyA was a promising natural monomer compound for OA management.3.2 Potential mechanisms of CyA to promote chondrogenesis
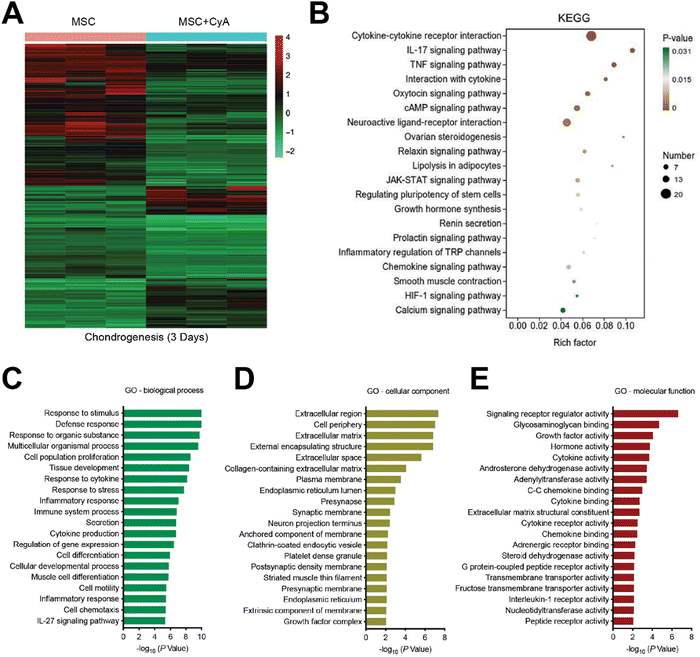
To further understand how CyA regulates chondrogenic differentiation, we used transcriptome sequencing to discuss its potential mechanisms. First, relative to normal MSC, we found 8123 differential genes after 3 days of chondrogenesis (Fig. S5, ESI†). Consistent with previous reports, the classical pathways of chondrogenesis, such as transforming growth factor-β (TGF-β) signaling pathway, Wnt signaling pathway, and Hippo signaling pathway, produced significant differences in the differentiation of MSCs to chondrocytes.42–44 After the addition of CyA, the number of differentiated genes reached 9214 for MSC chondrogenesis (Fig. S6, ESI†). After the addition of CyA, the heterogeneity of MSC chondrogenesis-related pathways in KEGG enrichment analyses all underwent diverse reductions (Fig. S7, ESI†). This implied that the regulatory effect of CyA on MSC chondrogenic differentiation was probably not through affecting these classical signaling pathways (TGF-β, Wnt, Hippo, etc.). Therefore, it was desirable to further characterize how CyA altered the process of chondrogenesis. As shown in Fig. 3A, we found 2958 statistically significant differential genes during chondrogenesis (3 days) with or without CyA supplementation. KEGG signaling pathway enrichment analysis revealed that CyA affected the process of MSC chondrogenesis predominantly through the regulation of cytokine–cytokine receptor interactions (Fig. 3B). CyA-mediated MSC chondrogenesis exhibited more pronounced alterations in cell differentiation and tissue developmental characteristics (Fig. 3C). During OA progression, extracellular matrix loss and decreased glycosaminoglycan content of articular cartilage were the primary manifestations of joint damage.45,46 Furthermore, glycosaminoglycans directly influenced and governed chondrocyte maturation and tissue development.47 Interestingly, the incorporation of CyA was more favorable for extracellular matrix construction and glycosaminoglycan binding during chondrogenesis, both of which are necessary for chondrocyte survival (Fig. 3D and E). Similar to the findings in the KEGG enrichment analyses, GO analyses for molecular function reconfirmed the regulatory capacity of CyA on cytokine–cytokine receptor activity (Fig. 3E). Taken together, CyA modulated the extracellular matrix and cytokine–cytokine receptor activity to influence MSC differentiation, suggesting its potential capacity for chondrogenic induction.3.3 Preparation and characterization of composite hydrogel microspheres
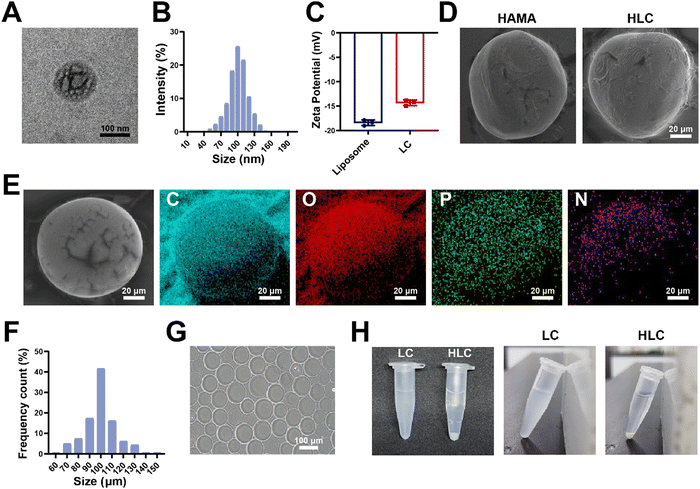
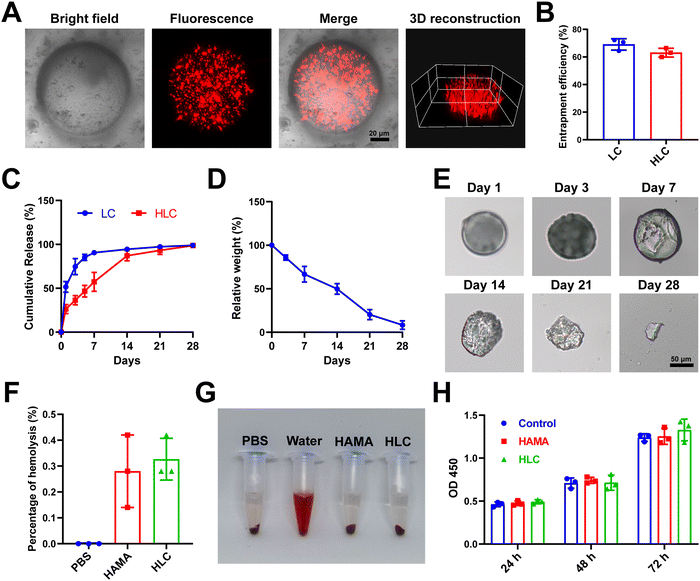
In the articular cavity, free drugs suffering from metabolism, pH and other factors, might have a short half-life and need to be administered frequently.48 This could cause inconvenience to the patient and affect compliance of therapy.49 Consequently, choosing an appropriate drug delivery manner to increase the drug retention time in the articular cavity provided an effective way of treating OA.50 For liposome preparation, HSPC, DSPE-MPEG2000, and cholesterol were used to produce Lipo@CyA (LC) by thin-film hydration combined with extrusion according to previously reported molar ratios.51 In Fig. 4A and B, the LC presented a uniformly dispersed spherical structure and particle dimension was about 100 nm. Compared to blank liposomes, LC carried less negative charge, which reduced its adsorption to serum proteins for removal (Fig. 4C).52,53 The spherical composite hydrogel microspheres loaded with LC were synthesized by microfluidics under UV light irradiation (Fig. 4D). This demonstrated that LC incorporation did not affect the morphology of the microspheres. As shown in Fig. 4E, elemental analysis revealed that phosphorus was uniformly distributed in the HLC microspheres, which indicated that phospholipid-containing LC was involved in the synthesis of the composite microspheres. Particle analysis revealed that the average particle size of HLC was 100.39 μm and uniformly sized (Fig. 4F and G). The liposomes were homogeneously dispersed in ultrapure water with milky white color, and the HLC microspheres made by mixing with HAMA could be naturally settled in water and aggregated at the bottom of the centrifuge tube (Fig. 4H). In summary, HLC microspheres were successfully prepared with a uniform size and consistent morphology for CyA loading to achieve long-term drug release.3.4 Drug loading and biological safety evaluation of composite hydrogel microspheres
The distribution and release pattern of drug-carrying liposomes in microspheres was an essential indicator for the evaluation of composite microspheres.54 In order to intuitively observe the presence of LC in HLC, we mixed DiD fluorescent dye during liposome synthesis according to the previous report, followed by the preparation of HLC composite microspheres using fluorescently labeled liposomes.55 The fluorescently labeled HLC was observed by confocal microscopy, as shown in Fig. 5A, where LC showed red dot-like fluorescence dispersed throughout the HLC. The 3D reconstructed image corroborated the spatial distribution of LC inside the entire microsphere, demonstrating good loading capacity. Compared to LC, there was no significant difference in the encapsulation rate of CyA in HLC (Fig. 5B). In contrast, HLC exhibited a completely different CyA release pattern from LC. After 72 h incubation in complete cell culture medium, LC released 74.71% of CyA, whereas HLC released only 36.39% of its contents (Fig. 5C). This suggested that HLC could continuously release CyA over 28 days, overcoming the pitfalls of repeated dosing. HA as a component of synovial fluid, HA-based biomaterials were able to be degraded to small molecules in the articular cavity and metabolized in vivo.56,57 To simulate the degradation process of hydrogels in vivo, we tested the degradation rate of HLC using a 10 U mL−1 hyaluronidase solution. The whole degradation process was performed at 37 °C in a constant temperature shaker. All samples were removed at predetermined time points lyophilized and weighed. The HLC microspheres degraded gradually over 28 days, from intact spheres to gradually being broken and disintegrated (Fig. 5D and E). In addition, since the microspheres will come into direct contact with the articular cartilage after being injected into the joint cavity, their biocompatibility should be taken into account.58,59 Hemolytic assays showed that neither blank HAMA microspheres nor HLC composite microspheres significantly triggered rat erythrocyte rupture (Fig. 5F and G). The hemolysis rate of HLC was much less than the hemolysis rate required for biomaterials used in vivo (2%), demonstrating satisfactory blood compatibility.60 Meanwhile, HLC did not affect the proliferation of human chondrocytes in Fig. 5H. In summary, HLC achieved sustained cargo release while loading CyA efficiently with ideal biocompatibility, allowing further exploration of its function at the cellular level.3.5 HLC mitigated chondrocyte inflammation and repressed apoptosis in vitro
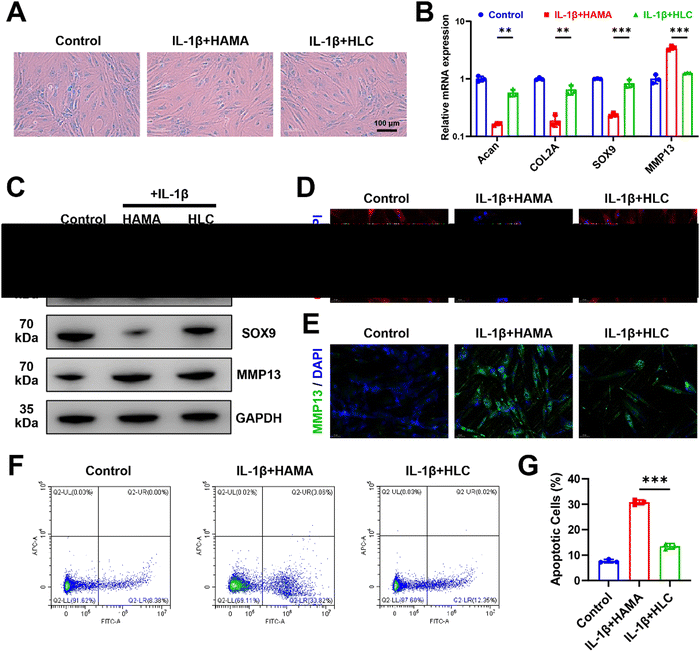
As shown in Fig. 6A, blank HAMA microspheres did not ameliorate IL-1β-induced chondrocyte damage, as evidenced by decreased Alcian blue coloration. Relative to blank microspheres, HLC restored cartilage extracellular matrix glycosaminoglycan expression by releasing CyA. At the same time, HLC increased the transcription and translation of the chondrogenic anabolic genes Acan, COL2A, and SOX9, which predicted the alleviation of chondrocyte inflammation (Fig. 6B and C). The relative expression of COL2A protein increased from 0.34 to 0.85 (Fig. S8, ESI†). Previously, several reports indicated that chondrocyte lesions in OA were characterized by a decrease in COL2A and an increase in MMP13, which were able to respond to the disease process of OA.61,62 To further validate the biological activity of HLC, cellular immunofluorescence was used to detect alterations of COL2A and MMP13 in human chondrocytes. Benefiting from the alleviation of cellular inflammation by HLC, the red fluorescent-labeled COL2A expression was significantly upregulated in chondrocytes (Fig. 6D). As an inflammatory marker, the simultaneous downregulation of intracellular MMP13 content further confirmed the effectiveness of HLC (Fig. 6E). In Fig. 6F and G, HLC lowered apoptotic chondrocytes from 30.82% to 12.35%. In addition, HLC influenced the chondrogenic differentiation of hBMSCs. As shown in Fig. S9 (ESI†), hBMSCs co-cultured with HLC exhibited more Alcian blue coloration.Consistent with the results of sustained CyA application, encapsulating CyA in composite hydrogel microspheres equally promoted the expression of chondrogenic genes (Acan, COL2A1, and SOX9) at both the transcriptional and translational levels (Fig. S10 and S11, ESI†). These data suggested that CyA-loaded HLC microspheres were able to reverse IL-1β-induced chondrocyte inflammation and upregulated hBMSC chondrogenic differentiation potential in vitro, possessing therapeutic potential for OA management.
3.6 HLC improved subchondral bone remodeling and attenuated cartilage injury in vivo
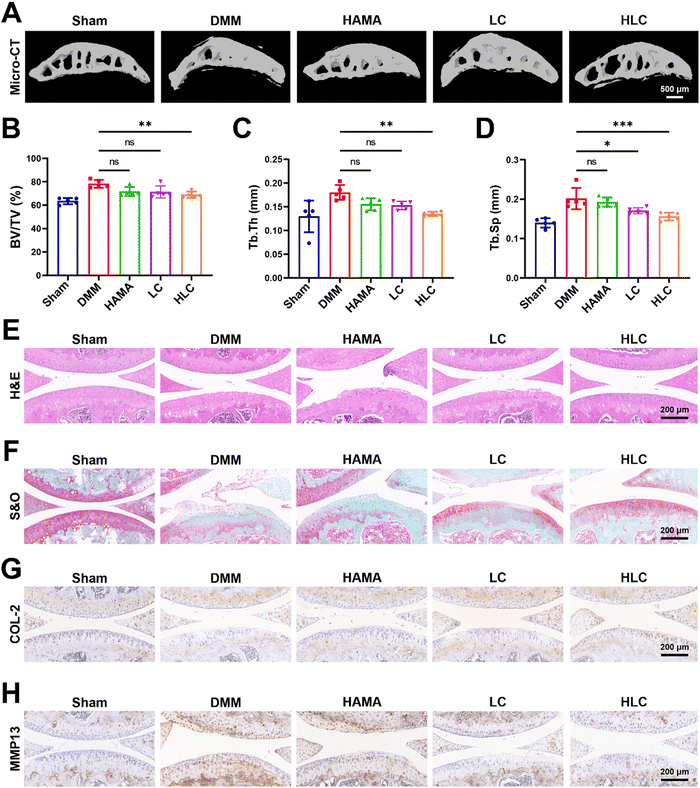
In the pathogenesis of OA, abnormal osteophytes and bone remodeling of the subchondral bone were essential in determining the severity of the joint injury.63–65 Severe ossification of the subchondral bone in OA rats resulted in the loss of normal mechanical structures (Fig. 7A). Blank HAMA or LC was not effective in alleviating bone remodeling in OA subchondral bone due to insufficient bioactivity or rapid drug release. By attenuating chondrocyte inflammation through sustained release of CyA, HLC significantly downregulated bone volume fraction (BV/TV), trabecular thickness (Tb.Th) and trabecular separation (Tb.Sp) (Fig. 7B–D). In Fig. 7E, the articular cartilage on the tibial plateau of rats with HLC injected was smoother and more intact relative to the DMM group. Similarly, the results of Safranin O/Fast green staining (S&O) demonstrated that the articular cartilage in the DMM, HAMA, and LC groups was subjected to varying degrees of wear and tear, whereas the HLC preserved the integrity of the cartilage layer in the OA rats (Fig. 7F). This was due to the slow degradation of the hydrogel microspheres and release of the drug throughout the treatment process. Additionally, the expression of cartilage-related proteins in joint tissues was of concern.66,67 Therefore, we examined the expression of COL-2 and MMP13 in articular cartilage by immunohistochemical experiments. As shown in Fig. 7G, COL-2 was highly expressed in normal articular cartilage. However, the OA inflammatory microenvironment led to impaired COL-2 synthesis in chondrocytes, as evidenced by a decrease in COL-2 positive cells in the visual field. Intra-articular injection of HLC not only restored the expression level of COL-2 in chondrocytes, but also decreased the MMP13 expression (Fig. 7H). In summary, HLC demonstrated favorable OA therapeutic ability in vivo by restoring the normal physiology of subchondral bone and protecting the integrity of hyaline cartilage.4. Conclusions
In this study, we found that CyA, a natural compound derived from traditional Chinese medicine, could modulate IL-1β-induced chondrocyte damage. 0.5 mg mL−1 CyA not only alleviated human chondrocyte inflammation and apoptosis, but also exhibited the potential to promote hBMSC chondrogenic differentiation. Transcriptome sequencing demonstrated that CyA contributed to chondrogenesis by regulating MSC extracellular matrix components and glycosaminoglycan binding activity. To overcome the excessive metabolism of CyA and achieve sustained release, HLC composite hydrogel microspheres were designed with reference to commercially available liposomes prescribed to load CyA. HLC provided suitable degradability, efficient drug encapsulation and good biocompatibility. By releasing CyA, HLC reversed IL-1β-triggered apoptosis in vitro, as evidenced by upregulated expression of chondrocyte markers such as Acan and COL2A. In in vivo experiments, HLC restored the normal structure of subchondral bone and diminished articular cartilage injury in OA rats, demonstrating desirable bioactivity. In conclusion, HLC provided new insights and is expected to be a promising platform for drug delivery in OA management.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Major Research plan of the National Natural Science Foundation of China (92249303), the Key Project of the National Natural Science Foundation of China (82230071), the CAMS Initiative for Innovative Medicine (2022-I2M-1-012) and the National Key R&D Program of China (2020YFA0113000).References
- D. Prieto-Alhambra, A. Judge, M. K. Javaid, C. Cooper, A. Diez-Perez and N. K. Arden, Ann. Rheum. Dis., 2014, 73, 1659–1664 CrossRef PubMed.
- G. B. D. O. and Collaborators, Lancet Rheumatol., 2023, 5, e508–e522 CrossRef PubMed.
- S. Safiri, A. A. Kolahi, E. Smith, C. Hill, D. Bettampadi, M. A. Mansournia, D. Hoy, A. Ashrafi-Asgarabad, M. Sepidarkish, A. Almasi-Hashiani, G. Collins, J. Kaufman, M. Qorbani, M. Moradi-Lakeh, A. D. Woolf, F. Guillemin, L. March and M. Cross, Ann. Rheum. Dis., 2020, 79, 819–828 CrossRef PubMed.
- F. Berenbaum, I. J. Wallace, D. E. Lieberman and D. T. Felson, Nat. Rev. Rheumatol., 2018, 14, 674–681 CrossRef PubMed.
- S. Y. Guan, J. X. Zheng, N. B. Sam, S. Xu, Z. Shuai and F. Pan, Autoimmun. Rev., 2023, 22, 103361 CrossRef PubMed.
- N. Chen, D. Y. T. Fong and J. Y. H. Wong, JAMA Network Open, 2023, 6, e2250674 CrossRef PubMed.
- J. N. Katz, K. R. Arant and R. F. Loeser, JAMA, 2021, 325, 568–578 CrossRef CAS PubMed.
- Q. Yao, X. Wu, C. Tao, W. Gong, M. Chen, M. Qu, Y. Zhong, T. He, S. Chen and G. Xiao, Signal Transduction Targeted Ther., 2023, 8, 56 CrossRef PubMed.
- T. Hodgkinson, D. C. Kelly, C. M. Curtin and F. J. O'Brien, Nat. Rev. Rheumatol., 2022, 18, 67–84 CrossRef PubMed.
- F. Motta, E. Barone, A. Sica and C. Selmi, Clin. Rev. Allergy Immunol., 2023, 64, 222–238 CrossRef CAS PubMed.
- L. Pang, H. Jin, Z. Lu, F. Xie, H. Shen, X. Li, X. Zhang, X. Jiang, L. Wu, M. Zhang, T. Zhang, Y. Zhai, Y. Zhang, H. Guan, J. Su, M. Li and J. Gao, Adv Healthcare Mater, 2023, 12, e2300315 CrossRef PubMed.
- S. Snelling, R. Rout, R. Davidson, I. Clark, A. Carr, P. A. Hulley and A. J. Price, Osteoarthritis Cartilage, 2014, 22, 334–343 CrossRef CAS PubMed.
- B. R. da Costa, T. V. Pereira, P. Saadat, M. Rudnicki, S. M. Iskander, N. S. Bodmer, P. Bobos, L. Gao, H. D. Kiyomoto, T. Montezuma, M. O. Almeida, P. S. Cheng, C. A. Hincapie, R. Hari, A. J. Sutton, P. Tugwell, G. A. Hawker and P. Juni, BMJ, 2021, 375, n2321 CrossRef PubMed.
- A. Lanas, J. Tornero and J. L. Zamorano, Ann. Rheum. Dis., 2010, 69, 1453–1458 CrossRef PubMed.
- C. Zeng, J. Wei, M. S. M. Persson, A. Sarmanova, M. Doherty, D. Xie, Y. Wang, X. Li, J. Li, H. Long, G. Lei and W. Zhang, Br. J. Sports Med., 2018, 52, 642–650 CrossRef PubMed.
- R. D. Altman and H. R. Barthel, Drugs, 2011, 71, 1259–1279 CrossRef CAS PubMed.
- X. Xue, H. Liu, S. C. Wang, Y. Hu, B. T. Huang, M. M. Li, J. Gao, X. H. Wang and J. C. Su, Composites, Part B, 2022, 237, 13 CrossRef.
- S. Tasneem, B. Liu, B. Li, M. I. Choudhary and W. Wang, Pharmacol. Res., 2019, 139, 126–140 CrossRef CAS PubMed.
- L. F. Yang, L. Teng, Y. H. Huang, M. Tang, W. Liao and Q. Fu, Phytochemistry, 2022, 196, 113101 CrossRef CAS PubMed.
- X. An, J. Wang, K. Xu, R. C. Zhao and J. Su, Aging Dis., 2023 DOI:10.14336/AD.2023.0817.
- Y. Huang, S. Wang, L. Liu, W. Peng, J. Wang, Y. Song, Q. Yuan, X. Yuan and C. Wu, Chin. Med., 2019, 14, 17 CrossRef CAS PubMed.
- L. Teng, Y. Shen, Y. Qu, L. Yang, Y. Yang, X. Jian, S. Fan, L. Zhang and Q. Fu, Chin. J. Nat. Med., 2023, 21, 99–112 CAS.
- F. Colella, J. P. Garcia, M. Sorbona, A. Lolli, B. Antunes, D. D'Atri, F. P. Y. Barre, J. Oieni, M. L. Vainieri, L. Zerrillo, S. Capar, S. Hackel, Y. Cai and L. B. Creemers, J. Controlled Release, 2020, 328, 985–999 CrossRef CAS PubMed.
- X. Zhang, J. Chen, Q. Jiang, X. Ding, Y. Li, C. Chen, W. Yang and S. Chen, J. Mater. Chem. B, 2020, 8, 8884–8893 RSC.
- J. Li, H. Zhang, Y. Han, Y. Hu, Z. Geng and J. Su, Theranostics, 2023, 13, 931–954 CrossRef CAS PubMed.
- N. A. Haq-Siddiqi, D. Britton and J. Kim Montclare, Adv. Drug Delivery Rev., 2023, 192, 114647 CrossRef CAS PubMed.
- Z. Li, G. Li, J. Xu, C. Li, S. Han, C. Zhang, P. Wu, Y. Lin, C. Wang, J. Zhang and X. Li, Adv. Mater., 2022, 34, e2109178 CrossRef PubMed.
- X. Ren, J. Wang, Y. Wu, Y. Zhang, J. Zhang, L. Bai, J. Liu, G. Li, P. Song, Z. Shi and J. Su, J. Mater. Sci. Technol., 2024, 188, 84–97 CrossRef.
- Q. Zhang, W. Chen, G. Li, Z. Ma, M. Zhu, Q. Gao, K. Xu, X. Liu, W. Lu, W. Zhang, Y. Wu, Z. Shi and J. Su, Small, 2024, e2306389, DOI:10.1002/smll.202306389.
- D. E. Large, R. G. Abdelmessih, E. A. Fink and D. T. Auguste, Adv. Drug Delivery Rev., 2021, 176, 113851 CrossRef CAS PubMed.
- Y. He, M. Sun, J. Wang, X. Yang, C. Lin, L. Ge, C. Ying, K. Xu, A. Liu and L. Wu, Acta Biomater., 2022, 151, 512–527 CrossRef CAS PubMed.
- Y. Lei, Y. Wang, J. Shen, Z. Cai, C. Zhao, H. Chen, X. Luo, N. Hu, W. Cui and W. Huang, Sci. Adv., 2022, 8, eabl6449 CrossRef CAS PubMed.
- G. Li, S. Liu, Y. Chen, J. Zhao, H. Xu, J. Weng, F. Yu, A. Xiong, A. Udduttula, D. Wang, P. Liu, Y. Chen and H. Zeng, Nat. Commun., 2023, 14, 3159 CrossRef CAS PubMed.
- X. Xue, H. Zhang, H. Liu, S. C. Wang, J. D. Li, Q. R. Zhou, X. Chen, X. X. Ren, Y. Y. Jing, Y. H. Deng, Z. Geng, X. H. Wang and J. C. Su, Adv. Funct. Mater., 2022, 32, 14 Search PubMed.
- Z. Zhou, P. Song, Y. Wu, M. Wang, C. Shen, Z. Ma, X. Ren, X. Wang, X. Chen, Y. Hu, Z. Li, Q. Zhang, M. Li, Z. Geng and J. Su, Mater. Horiz., 2024, 11(6), 1465–1483 RSC.
- M. S. Kalairaj, R. Pradhan, W. Saleem, M. M. Smith and A. K. Gaharwar, Adv. Healthcare Mater., 2024, e2303794, DOI:10.1002/adhm.202303794.
- G. Zuo, P. Zhuang, X. Yang, Q. Jia, Z. Cai, J. Qi, L. Deng, Z. Zhou, W. Cui and J. Xiao, Adv. Sci., 2023, e2305023, DOI:10.1002/advs.202305023.
- X. Li, X. Li, J. Yang, J. Lin, Y. Zhu, X. Xu and W. Cui, Small, 2023, 19, e2207211 CrossRef PubMed.
- Y. Barenholz, J. Controlled Release, 2012, 160, 117–134 CrossRef CAS PubMed.
- R. Xu, J. Wu, L. Zheng and M. Zhao, Ageing Res. Rev., 2023, 91, 102080 CrossRef CAS PubMed.
- H. Yang, Y. Wen, M. Zhang, Q. Liu, H. Zhang, J. Zhang, L. Lu, T. Ye, X. Bai, G. Xiao and M. Wang, Autophagy, 2020, 16, 271–288 CrossRef CAS PubMed.
- Y. Cho, S. Jeong, H. Kim, D. Kang, J. Lee, S. B. Kang and J. H. Kim, Exp. Mol. Med., 2021, 53, 1689–1696 CrossRef CAS PubMed.
- M. Wu, S. Wu, W. Chen and Y. P. Li, Cell Res., 2024, 34, 101–123 CrossRef CAS PubMed.
- Y. Gu, Y. Hu, H. Zhang, S. Wang, K. Xu and J. Su, Cell Proliferation, 2023, 56, e13517 CrossRef CAS PubMed.
- Y. Chen, K. Mehmood, Y. F. Chang, Z. Tang, Y. Li and H. Zhang, Life Sci., 2023, 335, 122243 CrossRef CAS PubMed.
- M. Rahmati, G. Nalesso, A. Mobasheri and M. Mozafari, Ageing Res. Rev., 2017, 40, 20–30 CrossRef CAS PubMed.
- M. Taieb, D. Ghannoum, L. Barre and M. Ouzzine, Cell Death Dis., 2023, 14, 355 CrossRef CAS PubMed.
- M. Rahimi, G. Charmi, K. Matyjaszewski, X. Banquy and J. Pietrasik, Acta Biomater., 2021, 123, 31–50 CrossRef CAS PubMed.
- I. A. Jones, R. Togashi, M. L. Wilson, N. Heckmann and C. T. Vangsness, Jr., Nat. Rev. Rheumatol., 2019, 15, 77–90 CrossRef PubMed.
- T. Zhao, X. Li, H. Li, H. Deng, J. Li, Z. Yang, S. He, S. Jiang, X. Sui, Q. Guo and S. Liu, Acta Pharm. Sin. B, 2023, 13, 4127–4148 CrossRef CAS PubMed.
- Y. Tao, Y. Chen, S. Wang, W. Chen, D. Zhou, D. Chen, C. Zhang, Z. Wu, J. Yan, H. Zhang, Y. Wei and J. Su, Composites, Part B, 2022, 247, 110288 CrossRef CAS.
- K. Yang, B. Mesquita, P. Horvatovich and A. Salvati, Acta Biomater., 2020, 106, 314–327 CrossRef CAS PubMed.
- D. Chatzikleanthous, S. T. Schmidt, G. Buffi, I. Paciello, R. Cunliffe, F. Carboni, M. R. Romano, D. T. O'Hagan, U. D'Oro, S. Woods, C. W. Roberts, Y. Perrie and R. Adamo, J. Controlled Release, 2020, 323, 125–137 CrossRef CAS PubMed.
- P. Song, Z. Cui and L. Hu, J. Controlled Release, 2022, 352, 946–960 CrossRef CAS PubMed.
- K. Chen, F. Wang, R. Ding, Z. Cai, T. Zou, A. Zhang, D. Guo, B. Ye, W. Cui and M. Xiang, Small, 2022, 18, e2106591 CrossRef PubMed.
- H. Yuan, L. L. E. Mears, Y. Wang, R. Su, W. Qi, Z. He and M. Valtiner, Adv. Colloid Interface Sci., 2023, 311, 102814 CrossRef CAS PubMed.
- C. Li, Z. Cao, W. Li, R. Liu, Y. Chen, Y. Song, G. Liu, Z. Song, Z. Liu, C. Lu and Y. Liu, Int. J. Biol. Macromol., 2020, 165, 1264–1275 CrossRef CAS PubMed.
- T. Wang, Y. Li, J. Liu, Y. Fang, W. Guo, Y. Liu, X. Li, G. Li, X. Wang, Z. Zheng, X. Wang and D. L. Kaplan, Biomaterials, 2022, 286, 121611 CrossRef CAS PubMed.
- Y. Yao, G. Wei, L. Deng and W. Cui, Adv. Sci., 2023, 10, e2207438 CrossRef PubMed.
- W. van Oeveren, Scientifica, 2013, 2013, 392584 CrossRef CAS PubMed.
- K. Nagata, H. Hojo, S. H. Chang, H. Okada, F. Yano, R. Chijimatsu, Y. Omata, D. Mori, Y. Makii, M. Kawata, T. Kaneko, Y. Iwanaga, H. Nakamoto, Y. Maenohara, N. Tachibana, H. Ishikura, J. Higuchi, Y. Taniguchi, S. Ohba, U. I. Chung, S. Tanaka and T. Saito, Nat. Commun., 2022, 13, 6187 CrossRef CAS PubMed.
- Q. Guo, X. Chen, J. Chen, G. Zheng, C. Xie, H. Wu, Z. Miao, Y. Lin, X. Wang, W. Gao, X. Zheng, Z. Pan, Y. Zhou, Y. Wu and X. Zhang, Cell Death Dis., 2021, 12, 13 CrossRef CAS PubMed.
- H. Zhang, L. Wang, J. Cui, S. Wang, Y. Han, H. Shao, C. Wang, Y. Hu, X. Li, Q. Zhou, J. Guo, X. Zhuang, S. Sheng, T. Zhang, D. Zhou, J. Chen, F. Wang, Q. Gao, Y. Jing, X. Chen and J. Su, Sci. Adv., 2023, 9, eabo7868 CrossRef CAS PubMed.
- J. Li, W. Zhang, X. Liu, G. Li, Y. Gu, K. Zhang, F. Shen, X. Wu, Y. Jiang, Q. Zhang, F. Zhou, K. Xu and J. Su, Cell Proliferation, 2023, 56, e13518 CrossRef CAS PubMed.
- Y. Hu, X. Chen, S. Wang, Y. Jing and J. Su, Bone Res., 2021, 9, 20 CrossRef CAS PubMed.
- Y. Yang, W. Zheng, W. Tan, X. Wu, Z. Dai, Z. Li, Z. Yan, Y. Ji, Y. Wang, W. Su, S. Zhong, Y. Li, Y. Sun, S. Li and W. Huang, Acta Biomater., 2023, 157, 321–336 CrossRef CAS PubMed.
- C. Cai, X. Zhang, Y. Li, X. Liu, S. Wang, M. Lu, X. Yan, L. Deng, S. Liu, F. Wang and C. Fan, Adv. Mater., 2022, 34, e2106564 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00294f |
| ‡ These authors contributed equally to this work |
| This journal is © The Royal Society of Chemistry 2024 |