Open Access Article
Open Access ArticlePolyNC: a natural and chemical language model for the prediction of unified polymer properties†
Haoke
Qiu
 ab,
Lunyang
Liu
ab,
Lunyang
Liu
 *a,
Xuepeng
Qiu
*a,
Xuepeng
Qiu
 bc,
Xuemin
Dai
c,
Xiangling
Ji
bc,
Xuemin
Dai
c,
Xiangling
Ji
 ab and
Zhao-Yan
Sun
ab and
Zhao-Yan
Sun
 *ab
*ab
aState Key Laboratory of Polymer Physics and Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun 130022, China. E-mail: lyliu@ciac.ac.cn; zysun@ciac.ac.cn
bSchool of Applied Chemistry and Engineering, University of Science and Technology of China, Hefei 230026, China
cCAS Key Laboratory of High-Performance Synthetic Rubber and its Composite Materials, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun 130022, China
First published on 6th December 2023
Abstract
Language models exhibit a profound aptitude for addressing multimodal and multidomain challenges, a competency that eludes the majority of off-the-shelf machine learning models. Consequently, language models hold great potential for comprehending the intricate interplay between material compositions and diverse properties, thereby accelerating material design, particularly in the realm of polymers. While past limitations in polymer data hindered the use of data-intensive language models, the growing availability of standardized polymer data and effective data augmentation techniques now opens doors to previously uncharted territories. Here, we present a revolutionary model to enable rapid and precise prediction of Polymer properties via the power of Natural language and Chemical language (PolyNC). To showcase the efficacy of PolyNC, we have meticulously curated a labeled prompt–structure–property corpus encompassing 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 polymer data points on a series of essential polymer properties. Through the use of natural language prompts, PolyNC gains a comprehensive understanding of polymer properties, while employing chemical language (SMILES) to describe polymer structures. In a unified text-to-text manner, PolyNC consistently demonstrates exceptional performance on both regression tasks (such as property prediction) and the classification task (polymer classification). Simultaneous and interactive multitask learning enables PolyNC to holistically grasp the structure–property relationships of polymers. Through a combination of experiments and characterizations, the generalization ability of PolyNC has been demonstrated, with attention analysis further indicating that PolyNC effectively learns structural information about polymers from multimodal inputs. This work provides compelling evidence of the potential for deploying end-to-end language models in polymer research, representing a significant advancement in the AI community's dedicated pursuit of advancing polymer science.
970 polymer data points on a series of essential polymer properties. Through the use of natural language prompts, PolyNC gains a comprehensive understanding of polymer properties, while employing chemical language (SMILES) to describe polymer structures. In a unified text-to-text manner, PolyNC consistently demonstrates exceptional performance on both regression tasks (such as property prediction) and the classification task (polymer classification). Simultaneous and interactive multitask learning enables PolyNC to holistically grasp the structure–property relationships of polymers. Through a combination of experiments and characterizations, the generalization ability of PolyNC has been demonstrated, with attention analysis further indicating that PolyNC effectively learns structural information about polymers from multimodal inputs. This work provides compelling evidence of the potential for deploying end-to-end language models in polymer research, representing a significant advancement in the AI community's dedicated pursuit of advancing polymer science.
1 Introduction
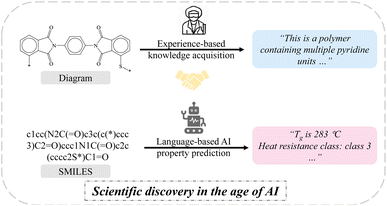
Polymers possess a large theoretically feasible chemical space. Over the past few decades, experimentalists and computational scientists have conducted extensive and valuable explorations to navigate the chemical space of polymers, accumulating invaluable data.1–7 By harnessing the wealth of available data, it's promising that precise and efficient polymer discovery guidelines can be deduced, ultimately reducing the time from the laboratory to market. Encouragingly, significant progress has been made in utilizing machine learning (ML) models to accurately and efficiently infer polymer properties.8–16 These ML models can be categorized into three main approaches: handcrafted descriptor-based, graph-based, and sequence-based methods. In the context of handcrafted descriptor-based ML, the essential steps involve the extraction of numerical representations capable of describing molecular structures, commonly leveraging well-established tools such as RDKit,17 Mordred18 and molecular fingerprints.19 These handcrafted descriptor-based models perform exceptionally well in specific polymer tasks, especially with small datasets. To strive for end-to-end learning, there has been a growing interest in graph-based approaches.20–23 Capturing end-to-end representations of polymer structures offers great flexibility for ML models to directly learn from raw data.24 Among these approaches, the graph convolutional neural network (GCN) is commonly employed. GCN treats atoms as nodes and bonds as edges, utilizing message-passing mechanisms25 to continuously aggregate neighborhood features and capture local molecular motifs and molecular topologies.26,27 Graph-based models consider more molecular topological structures, making them particularly promising for predicting alternating, random, and block copolymers.24Besides, sequence-based language models also offer a promising solution for polymer property prediction. Polymer structures can be effectively represented as language-strings such as SMILES,28 SELFIES,29 and big-SMILES,30 and language models have fewer restrictions on input format compared to conventional ML models, allowing for the utilization of custom and non-conventional polymeric patterns as inputs, especially when considering stoichiometry, molecular weight and additives. This will alleviate the burden on researchers in obtaining molecular representations that satisfy specific requirements like input shapes and makes language models highly promising for AI-driven polymer discovery. In the last five years, there has been an emergence of language-like models in the field of polymers. For instance, long short-term memory (LSTM)31,32 and recurrent neural networks (RNNs)33 have been employed to learn the sequence representation of polymer structures. More recently, transformer models specialized for sequence-based tasks have achieved significant success in addressing polymer scientific challenges incorporating self-attention mechanisms34 in a pretrained and finetune manner. Notable examples include TransPolymer15 and polyBERT.16 TransPolymer employed the RoBERTa35 model to generate machine-based fingerprints through an unsupervised approach, utilizing 5 million unlabeled polymer SMILES. Similarly, polyBERT employed a DeBERTa-based36 transformer to convert SMILES strings into numerical representations suitable for downstream multi-task regression models. These remarkable findings demonstrate the significant efficacy of the transformer family in the fields of polymers.
Indeed, polymer property prediction tasks are similar to text-based language model tasks such as machine translation and text generation. Text-based language models generate corresponding outputs based on given text inputs. Similarly, in polymer property prediction tasks, properties are predicted based on given prompts and chemical structures. In the past, attempts to solely rely on language models for polymer property prediction tasks were hindered by the scarcity and unattainability of high-quality labeled polymer datasets,37 while the availability of high-quality open-source polymer datasets is steadily increasing.38–41 More encouragingly, extensive work has shown that data augmentation-based approaches are effective in addressing the scarcity of polymer data,15,42,43 and harnessing the intelligence of general language models proves beneficial for comprehending scientific language via language models.44–47 To the best of our knowledge, a completely end-to-end language-based approach for directly predicting the properties of polymers from natural and chemical languages, rather than being used as intermediates to connect molecular structures to downstream models, is currently lacking. This concept draws parallels to how chemists can infer fundamental properties of common molecular structures through visual observation, without the need for additional analytical characterization (Fig. 1). By integrating natural language, chemical language (e.g., SMILES) and chemical knowledge (properties), language-to-property AI agents hold promise to perceive and establish a multi-domain and multi-task understanding directly from the polymer structure to its diverse properties, which presents an opportunity to drive advancements in existing robo-chemists and autonomous laboratories.48–50 In addition to regression-based property prediction tasks, we also aim for a unified model that can simultaneously handle multiple types of tasks, such as both classification and regression tasks. The ability to handle multiple types of tasks simultaneously is a capability that milestone models mentioned earlier have yet to explore owing to the challenges stemming from data distribution shifts51 and the inherent specificity of ML models themselves.
Herein, we propose the PolyNC, a fully end-to-end and multi-task language model for polymer discovery. Our model enables the execution of complex polymer discovery workflows with a single model, a previously unreported ability, surpassing even the capabilities of prevailing LLMs like ChatGPT, Claude, Llama and PaLM, due to their lack of domain knowledge. Given the limited information that can be directly extracted from SMILES by chemists, our model introduces a new paradigm for polymer discovery, design and development based on SMILES, offering remarkable convenience. In comparison to descriptor-based and graph-based models, PolyNC exhibits impressive performance across the four tasks central to our polymer research: three property prediction tasks (regression) and one polymer classification task (classification). Handling multi-task and multi-type problems is a capability hitherto unattainable by previous ML models. Notably, PolyNCs' ability to generalize to unknown structures is particularly impressive, as confirmed through experimental validation. Attention analysis reveals that this generalization capacity stems from the model's comprehension of both natural language and chemical language. This work extends the powerful natural language understanding capabilities of AI to the field of polymer research, marking an impressive step towards the development of expert models and human-level AI for understanding polymer knowledge.
2 Results and discussion
2.1D efinition of polymer-specific tasks
The properties of polymers are multi-faceted and often sparse in specific property datasets. Therefore, we focused on four significant polymer properties with publicly available datasets21,52–54 for training PolyNC. The included tasks consist of three general problems in a polymer domain (regression tasks) and one critical task specific to a particular polymer (classification task). They are as follows:(a) Glass transition temperature (Tg). Tg is a critical property that characterizes the transition from a rigid, glassy state to a more flexible, rubbery state in polymers, which is essential in understanding the processing, stability, and mechanical behavior of polymers, making it a key parameter in material design and applications.
(b) Band gap of polymer crystals (BC). The BC of polymer crystals refers to the energy difference between the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) in the crystalline state of a polymer, which is crucial for developing polymer-based electronic and photonic devices, as it influences their performance in areas like organic photovoltaics and light-emitting diodes.
(c) Atomization energy (AE). AE represents the energy required to completely separate the constituent atoms of a polymer molecule. It reflects the strength of the chemical bonds within the polymer and provides insights into its stability and reactivity. Atomization energy is relevant in various aspects of polymer chemistry, including synthesis, degradation, and understanding the relationship between structure and properties.
(d) Heat resistance class (HRC). To assess the capability of PolyNC across various task types, we also established this classification task as an example. HRC refers to the ability of polymers to withstand high temperatures without significant degradation or loss of its essential properties, particularly important in the case of high-end polymers like polyimides (PIs). Therefore, we focus on the heat resistance of PIs in this study while also investigating the performance of PolyNCs in handling tasks related to polymers. Based on the Tg of PIs and industry standards,55 we can classify them into three categories: class 1, class 2, and class 3. Class 1 indicates PIs with a Tg above 400 °C, class 2 represents PIs with a Tg ranging from 300 to 400 °C, and class 3 refers to PIs with a Tg below 300 °C.
Due to the data-hungry nature of language models, data augmentation is implemented to improve model performance under an equal mixing strategy.56 A summary of the four datasets for downstream tasks is shown in Table 1.
| Property | Source | Unit | # Entries (training/test) | # Aug. times | # Aug. entries |
|---|---|---|---|---|---|
| T g | DFT & exp. | °C | 685(615/70) | 10 | 6850 |
| BC | DFT | eV | 236(212/24) | 20 | 4720 |
| AE | DFT | eV | 390(351/39) | 15 | 5850 |
| HRC | Exp. | — | 370(333/37) | 15 | 5550 |
| Total | DFT & exp. | — | 1681 | — | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 970 |
2.2 Experimental settings
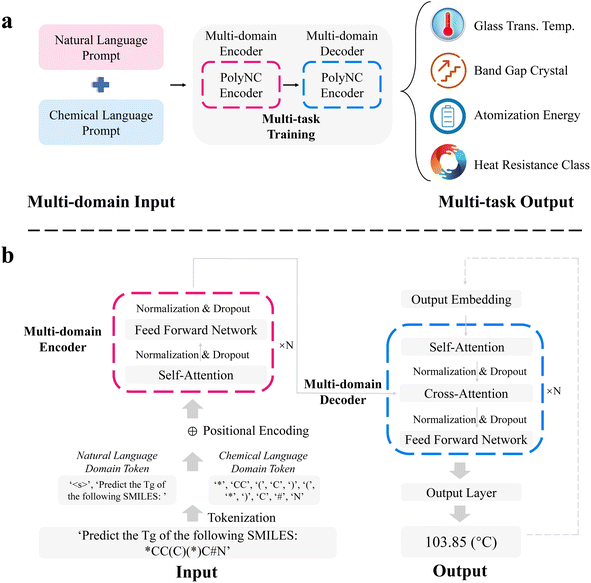
Polymeric Prompt Engineering (PPE, detailed in Methods section) has been employed to acquire the training corpus for PolyNC, enabling the model to learn natural language prompts and corresponding polymer structures. We applied data augmentation to the training corpus with the equal mixing strategy56 to ensure a relatively balanced representation of each task. The training corpus was divided into 90% training set (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 673) and 10% test set (2297). To the best of our knowledge, this is one of the largest labeled datasets available for polymer ML tasks. These input prompts are tokenized at the character level, dividing them into natural language tokens and chemical tokens. This tokenization strategy has been proven to provide better performance and stronger expressive capabilities. By observing the distribution of token sizes in the training and validation sets (depicted in the ESI, S1†), we determined to set the input token size to 150 and the output token size to 8 to accommodate all instances. To initialize our model, we chose both the t5-base and Text + Chem T5.56,57 The former is a pre-trained model based on natural language text, while the latter is a pre-trained model specifically designed for chemical text tasks such as molecular descriptions. We found that the inclusion of scientific domain knowledge weighting significantly enhances the performance of the model on polymer domain tasks (as detailed in the Ablation studies section), which implies the transferability of PolyNC to other polymeric tasks.
673) and 10% test set (2297). To the best of our knowledge, this is one of the largest labeled datasets available for polymer ML tasks. These input prompts are tokenized at the character level, dividing them into natural language tokens and chemical tokens. This tokenization strategy has been proven to provide better performance and stronger expressive capabilities. By observing the distribution of token sizes in the training and validation sets (depicted in the ESI, S1†), we determined to set the input token size to 150 and the output token size to 8 to accommodate all instances. To initialize our model, we chose both the t5-base and Text + Chem T5.56,57 The former is a pre-trained model based on natural language text, while the latter is a pre-trained model specifically designed for chemical text tasks such as molecular descriptions. We found that the inclusion of scientific domain knowledge weighting significantly enhances the performance of the model on polymer domain tasks (as detailed in the Ablation studies section), which implies the transferability of PolyNC to other polymeric tasks.
PolyNC used a whole encoder and decoder architecture each with 12 layers and 12 attention heads (Fig. 2). The encoder is responsible for extracting semantic information from the multi-domain input, while the decoder analyzes this semantic information and generates outputs based on the given conditions. Within them, self-attention was used to maintain the relationships among tokens. In the decoder, in addition to self-attention for capturing relationships within a single sequence, cross-attention is also utilized to capture relationships between the input and output sequences. This helps in learning the mapping between natural language and chemical language and their respective properties. For each output of attention block, a fully connected network is used to perform non-linear projection. We set 768 as the hidden dimension for PolyNC and 3072 for the intermediate feed forward layer with the ReLU activation function and a dropout rate of 10%. The output head for all tasks is the same as the output layer of t5-base from the huggingface transformers package.58 What sets our work apart from previous efforts (TransPolymer and polyBERT) is our ability to directly handle multitasking in a single unified model without the need for separate regression or classification heads, thus enabling PolyNC to achieve seamless end-to-end property prediction. A cosine learning rate decay strategy with a 20% warmup ratio is used to dynamically adjust the learning rate to speed up convergence and avoid skipping optimal solutions on two RTX 3090 GPUs.
2.3 Model performance on multi-task
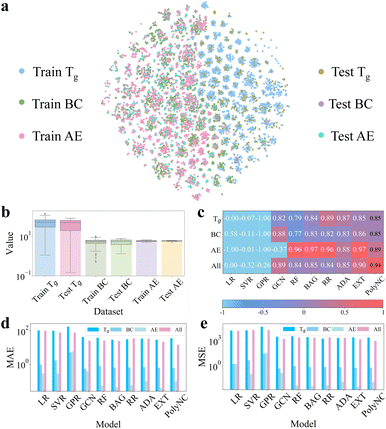 | ||
| Fig. 3 (a) Distributions of chemical space for each dataset based on t-SNE.59 It can be observed that the majority of molecules corresponding to each property are distinct, which aids the models in learning a more comprehensive mapping between molecular structures and properties from limited data. Additionally, for each individual task, the distribution of the training and testing sets is uniform, which helps assess the model's generalization ability. (b) Value distribution of each dataset. The y-axis is plotted on a logarithmic scale. This subfigure highlights the significant differences in the value ranges among the different properties. (c) R2 metric (↑). (d) MAE metric (↓). (e) RMSE metric (↓). PolyNC demonstrated impressive performance in each prediction task, particularly excelling in the multi-property prediction task, showcasing its powerful capability in handling multi-task scenarios. | ||
Under the same chemical space, we trained and validated PolyNC and these baseline models. In this study, the descriptors of baseline models implemented using the scikit-learn package were computed using RDKit, where the descriptors for each molecule were computed using the Descriptors.descList module, yielding a total of 209 descriptors, and descriptors containing missing values were excluded, resulting in a final set of 197 valid descriptors (the details are publicly available at https://github.com/HKQiu/Unified_ML4Polymers/blob/main/data/exp_val/data_with_descriptors.csv). The descriptors of the baseline model implemented using Deepchem were computed using its default settings, with 75 features for each atom (the details are publicly available at https://github.com/deepchem/deepchem/blob/master/deepchem/feat/graph_features.py#L282). The coefficient of determination (R2), mean absolute error (MAE) and mean squared error (MSE) were used as evaluation metrics for these models, as detailed in S3.1.†
Single-property prediction tasks were used to evaluate the performance of the models across different properties. And due to the interdependencies among the properties of polymers, accurately predicting multiple properties simultaneously is of interest to polymer scientists. Therefore, we employed a multi-property prediction task (denoted as ‘All’) to test the models' ability to predict multiple properties simultaneously. This task poses a challenge due to the significantly varied value ranges of different properties, as shown in Fig. 3b, thus it can serve as an indication of the model's potential in handling multi-task scenarios.
The performance comparison of the different models is shown in Fig. 3c–e and S3.1.† PolyNC demonstrated impressive performance for each single-property prediction task, demonstrating that the entirely language-based model PolyNC can achieve prediction accuracy comparable to other ML approaches. For the multi-property prediction task scenario, the predictive performances of almost all ML models exhibited a decrease compared to single-task settings. This is because traditional ML models struggle to fit datasets with different distributions.
Of note, both GCN and PolyNC showed improvements in performance for this task. In the case of GCN, GCN takes into account the topology and connectivity of molecules, allowing it to extract more useful information compared to handcrafted descriptors. This enables GCN to exhibit superior performance in multi-task settings. This finding also underscores the importance of extracting as much raw information as possible from molecules. Similarly, benefiting from its learning of both natural language and chemical language, PolyNC exhibits a deeper understanding of the structure–property mapping of polymers and facilitates PolyNC in learning multiple properties of diverse molecules simultaneously, resulting in the best performance, which highlights the significant potential of language models in constructing polymer property–structure landscapes.
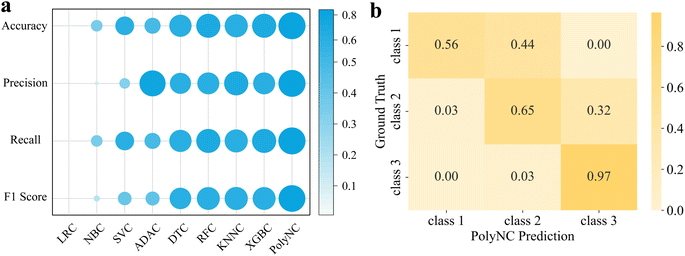
We compared the performance of PolyNC with eight baseline models for the classification task, including Logistic Regression (LRC), Naive Bayes (NBC), Support Vector Machine (SVC), AdaBoost (ADAC), Decision Tree (DTC), Random Forest (RFC), K-Nearest Neighbors (KNNC) and XGBoost (XGBC). While XGBoost was implemented using the xgboost package,62 the remaining ML models were implemented using the scikit-learn package.60 We used four evaluation metrics for classification problems, namely Accuracy, Precision, Recall, and F1 Score. The HRC task is characterized by an imbalanced dataset, which we addressed by ensuring that each class had a consistent proportion in both the training and testing sets and we assigned a weighti to class i when assessing the model performance, as detailed in S3.2.†
The performance results of each model are depicted in Fig. 4 and S3.† Impressively, PolyNC achieves the best performance in the HRC task, with all metrics exceeding 0.81, benefiting from the intelligence of language models in handling classification tasks, such as sentiment classification.63 As a point of comparison, the baseline models achieved an accuracy of around 0.7. Compared to LRC and SVC, PolyNC did not make any highly inaccurate prediction such as predicting class 3 for samples actually belonging to class 1 or vice versa though class 1 had fewer samples and class 3 had more. This demonstrates that PolyNC is not significantly affected by imbalanced sample sizes, thus avoiding the generation of biased outputs. Besides, it is worth noting that during the training phase, PolyNC learns simultaneously for all tasks (regression and classification), allowing it to capture correlations between properties. Although the mapping relationship between Tg and HRC is not explicitly informed to PolyNC, it can also spontaneously and implicitly learn these details from latent data. As evidenced in Fig. S2d,† PolyNC tends to underestimate Tg predictions, which in turn affects its classification performance, as reflected in misclassifying 44% of class 1 as class 2 and 32% of class 2 as class 3. Despite the negative impact, this also serves as evidence that PolyNC learns the correlations between different tasks. Though under these perplexing interferences, PolyNC also outperformed other baseline models in this classification task, making it a unified model capable of handling both regression and classification tasks simultaneously. As far as we know, an ML model that can simultaneously handle classification and regression problems in the field of polymers has not been seen before.
2.4 Attention analysis
The self-attention mechanism is used to efficiently consider distances between sequence elements, or the level of attention that one element pays to other elements, which can affect the representation of the entire input, thereby leading to different outputs.34,64 To unveil the intrinsic knowledge learned by PolyNC from both natural language and chemical language, we analyzed the attention scores within the encoder using the Tg task as an example. This analysis helps reveal which tokens are assigned higher correlations by PolyNC. Firstly, we constructed a Polymer Tree for all structures in the Tg task, visualizing the dataset based on the similarity of chemical structures using TMAP.65 Since the SMILES of a ring structure is more complex than the SMILES of a linear structure, to evaluate PolyNC's ability to recognize SMILES, we designed two heterocyclic structures that have never been seen before (a new branch of the Polymer Tree, denoted as PI-1 and PI-2). These two unseen molecules can also serve as a measure of the model's generalization ability to some extent. Secondly, we conducted rigorous synthesis and characterization (as detailed in S4†) for these two new structures, and found that PolyNC demonstrated optimal performance in predicting the Tg of these unknown structures, with the least deviation (5 °C and 20 °C) between predicted and ground truth values for these two samples compared to other baseline models (Fig. 5b and c).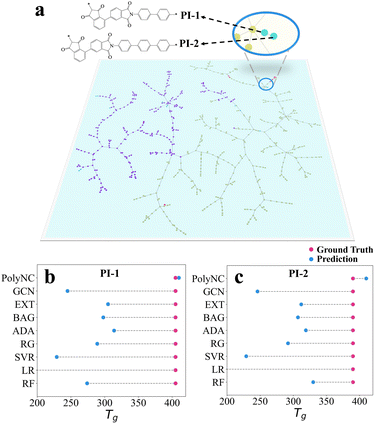 | ||
| Fig. 5 (a)The Polymer Tree of each structure within the training and test datasets of Tg. Different molecules with distinct chemical structures are located in different branches of the Polymer Tree. For instance, the left portion of the graph primarily consists of structures with fewer heterocycles, predominantly linear polymers, while the right portion comprises structures with a higher number of heterocycles (see our repository https://github.com/HKQiu/Unified_ML4Polymers/tree/main/TMAP or https://try-tmap.gdb.tools/tmap/discreet-ammonite-of-mathematics for more details of Polymer Tree). (b) and (c) Generalization ability of PolyNC in the estimation of Tg as an example. PolyNC demonstrates exceptional performance in predicting the Tg of unknown structures with a 5 °C and 20 °C deviation for each sample. The limited generalizability is a universal issue for off-the-shelf ML models,66 where PolyNC might have learned more appropriate polymer representations. | ||
Then, we analyzed the attention scores of PolyNC with respect to the input sequences corresponding to these two examples. The encoder of PolyNC consists of 12 attention heads, each focusing on different contexts to extract distinct knowledge (all the 12 attention heads of the encoder are as depicted in S5†). The attention scores for the fifth and ninth attention heads of PI-1 and PI-2 are shown in Fig. 6. It can be observed that the fifth attention head primarily attends to adjacent tokens for each token to obtain local environments, while attention head 9 mainly focuses on the tokens themselves. From Fig. 6, we can summarize PolyNC's ability to recognize complex SMILES in three aspects. (1) PolyNC exhibits higher attention scores in the feature groups (imide groups) of PI-1 and PI-2 (seen in the pink and light yellow parts in the figure). (2) PolyNC also assigns higher attention scores to the natural language part and polymerization sites corresponding to human cues and polymerization information (seen in the light purple part in the figure). (3) The structural difference between PI-1 and PI-2 is the presence of an additional benzene ring in PI-2 (seen in the green part in Fig. 6b). Adding a benzene ring changes the order of elements in the SMILES and makes it more challenging for human interpretation. However, PolyNC recognizes the benzene ring structure in complex SMILES, as evident in the corresponding attention matrices in the left panel of Fig. 6b (the black dashed box regions, which exhibit similar attention matrices along the diagonal direction). Specifically, since the purple benzene ring connects to a different chemical structure (imide structures), the lower edge of the purple attention matrix also changes accordingly. The aforementioned findings suggest that PolyNC possesses intelligent chemical perception, enabling it to pave the way for the recognition of complex SMILES expressions for further molecular property prediction and inverse design.
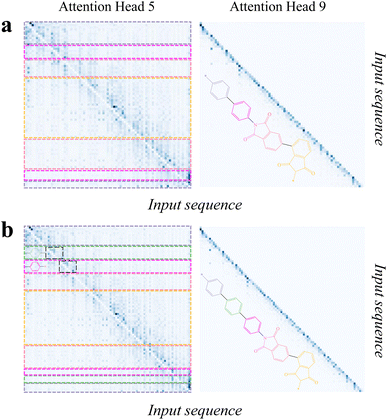 | ||
Fig. 6 The attention scores for the fifth (left) and ninth attention (right) heads of PI-1 (a) and PI-2 (b). It can be observed that the fifth attention head primarily attends to adjacent tokens for each token, while attention head 9 mainly focuses on the tokens themselves. For linear polymers, the order of elements in their SMILES representation aligns with the order of atoms in the molecular structure, making it easy to observe the molecular structure from the SMILES. However, for molecules with many rings, things become more complicated. Based on the rules of SMILES, the rings will be broken and flattened at specific atoms, causing adjacent atoms to appear in non-adjacent positions in the SMILES, which poses challenges for parsing SMILES. The input sequences of PI-1 and PI-2 were ‘Predict the Tg of the following SMILES: [*]C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C2 C(C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(N3C(C(C C(N3C(C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(C4 CC(C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(C(C5[*]) CC(C(C5[*])![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4C5 C4C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6) C6)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6C3 C6C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C O)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C C2)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1’ and ‘predict the Tg of the following SMILES: [*]C1 C1’ and ‘predict the Tg of the following SMILES: [*]C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C2 C(C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C3 C(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(N4C(C(C C(N4C(C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(C5 CC(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(C(C6[*]) CC(C(C6[*])![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5C6 C5C6![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C7) C7)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C7C4 C7C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C O)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)C C3)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C C2)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1’, respectively. PolyNC succeeded in extracting and differentiating subfunctional group information and structural difference directly from SMILES. The attention scores for all the 12 attention heads of the encoder can be found in S5.† C1’, respectively. PolyNC succeeded in extracting and differentiating subfunctional group information and structural difference directly from SMILES. The attention scores for all the 12 attention heads of the encoder can be found in S5.† | ||
2.5 Ablation studies
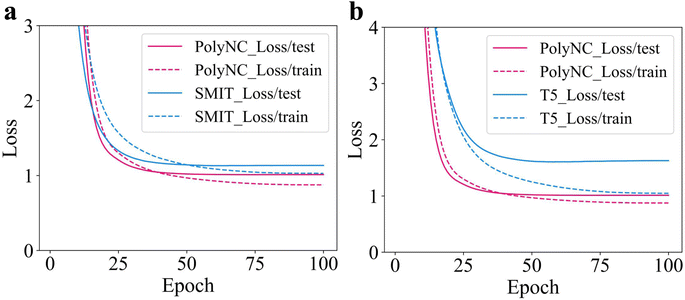
In PolyNC, we employed a character-wise tokenization approach. However, for molecular structures, there is also a group-wise tokenization approach based on functional groups. We compared the performance of these two tokenization methods. For the group-wise tokenization, we utilized the seyonec/PubChem10M_SMILES_BPE_450k tokenizer from huggingface: https://huggingface.co/seyonec/PubChem10M_SMILES_BPE_450k/tree/main (denoted as SMIT). SMIT is trained on a large number of SMILES representations of molecules and aims to tokenize SMILES based on functional groups. SMIT, through the use of the byte pair encoding (BPE) algorithm, has learned multiple functional group tokens and established a vocabulary specifically tailored to molecules. For instance, it is possible to cluster local chains composed of multiple carbon atoms in SMIT. As an illustration, a segment consisting of four carbon atoms can be tokenized as ‘CCCC’. The results can be seen in Fig. 7a. It can be observed that the character-wise tokenization method outperforms the group-wise approach. This is because the one-dimensional SMILES representation itself loses a lot of chemical information, and adjacent characters are likely not part of the same functional group. Therefore, the group-wise tokenization approach may lead to mis-tokenization. Additionally, there is a wide variety of chemical functional groups, and each functional group may have different SMILES encoding methods, making it difficult to cover all functional groups comprehensively. The complexity of molecules and the complexity of SMILES representation have led to successful language models in the field of molecular research predominantly employing character-wise tokenization.15,16,67,68To assess the transferability of domain knowledge across different domains, we tested two initial weight configurations: one based on natural language weights (t5-base) and the other based on chemical text tasks, such as molecular descriptions (Text + Chem T5).44,56 Based on these two weight configurations, PolyNC was trained from scratch under the same parameters, and their learning curves are depicted in Fig. 7b. It can be observed that PolyNC, based on the Text + Chem T5 configuration, outperforms the performance achieved by the t5-base configuration (denoted as T5). This indicates the preservation of domain knowledge during transfer, ultimately enabling the model to develop a stronger understanding of downstream tasks within the same training duration. Of particular inspiration, this finding also reveals the potential success of fine-tuning PolyNC as a foundational model in other polymeric tasks.
3 Conclusion
We propose PolyNC, a model capable of comprehending both natural language and chemical language, for polymer discovery. Diverging from conventional ML models based on descriptors or graphs, PolyNC takes an end-to-end approach to directly extract semantic information from human prompts and chemical language for property prediction. This more abstract and unbiased strategy enables PolyNC to acquire a more genuine understanding of chemical knowledge, resulting in exceptional performance in multiple polymer tasks. Notably, PolyNC is a multi-task, multi-domain model that integrates the capabilities of both classification and regression models, a distinctive feature absent in previous ML models. Moreover, due to its domain specificity, PolyNC exhibits a remarkable comprehension of chemical language that surpasses the current popular commercial LLMs (S6.1†). PolyNC contributes to inspiring modeling approaches in polymer informatics, shifting from a ”feature-extraction-first” approach to a direct language-based paradigm. This shift significantly reduces data preprocessing time, expands the scope of available data, and enhances the intelligence of the model. As the availability of high-quality polymer corpora continues to grow, PolyNC's proficiency in understanding polymer content is poised to strengthen progressively, even grooking the depths of intricate polymer structures and properties, which contributes to advancing materials research and the automation of laboratory processes. Last but not least, due to the inherent universality of language model principles, this paradigm can easily be extended and revolutionize other scientific research domains.4 Discussion
Indeed, general large language models like ChatGPT are limited by their understanding of domain-specific knowledge, which restricts their widespread application in scientific research. Our work demonstrated the success of polymer prediction tasks solely based on natural language prompts and chemical language prompts utilizing large language models. We used SMILES to describe the structures of polymers, which describes common homopolymers, regular copolymers, and stereochemistry, among others (please refer to S8† for more details). However, in reality, the structure of polymers is much more complex, involving various monomer compositions, different bonding patterns, and chain architectures, which go beyond the scope of SMILES, but they can be described using natural language with further enrichment of training data. We will continue to mine data containing these details and further empower PolyNC.Generative language models produce content probabilistically. To ensure result reproducibility, PolyNC was configured to select the token with the highest probability as the generated content during inference, while also controlling influential factors like token size. Moreover, language models should answer scientific facts objectively, even if different prompts are given. We verified the sensitivity of PolyNC to different SMILES of the same molecule, and the results (S7†) demonstrate that PolyNC gives satisfactory results for different SMILES of the same molecule to a great extent. Of note, akin to language models like ChatGPT, PolyNC does not evaluate the plausibility of the input and is capable of generating inference outcomes for any given input. Therefore, it is imperative that individuals employ PolyNC under the supervision of proficient chemists to mitigate potential risks.
5 Method
5.1 Model
Our model is implemented based on the text-to-text transfer transformer (T5),56 an encoder–decoder model belonging to the transformer34 family. T5 is specifically designed to convert various language problems into a text-to-text format, enabling it to handle multi-domain and multi-task problems simultaneously. By employing appropriate prompt engineering, T5 can be adapted to handle tasks across diverse domains. To effectively capture both natural language and chemical language, we employ a joint encoder that encompasses both domains.44 This enables our model to excel in multi-domain and multi-task scenarios. Additionally, we utilize the original T5 decoder for generating the output.5.2 Polymeric prompt engineering (PPE)
To enable our model to simultaneously predict multiple polymer properties, we employed customized polymeric prompt engineering (PPE). Different tasks may require distinct prompts, allowing us to differentiate among them. Each task-specific natural language prompt is formulated as an English sentence that does not exist in the original SMILES or the T5 tokenizer vocabulary. Thus, these prompts are added as special tokens to the vocabulary considering the relatively limited amount of available data. For instance, the prompt for predicting the glass transition temperature (Tg) is represented as “Predict the Tg of the following SMILES:” followed by the corresponding SMILES. This construction forms a multi-domain prompt that combines both natural language and chemical language which supports complex cross-modal understanding, reasoning and sophisticated multimodal content generation. In total, we considered four different polymer tasks, including Tg, band gap crystal (BC), atomization energy (AE) and heat resistance class (HRC). These tasks are sourced from publicly available datasets.21,52–545.3 Data augmentation
Recognizing that language models are data-greedy69 and existing databases are insufficient for training high-performing models, we employed data augmentation techniques; one approach involved enumerating different SMILES representations for the same molecule to enhance the training dataset,70 as different SMILES representations of the same structure are treated as different inputs by language models. We initially divided the dataset for each task into training and test sets using a ratio of 0.9/0.1. During data augmentation, we followed an equal mixing strategy56 to select different augmentation factors for each task, ensuring a balanced representation of each property, resulting in a proportional distribution of entries for each property. Through this balanced data augmentation process, we obtained a labeled dataset of polymer properties consisting of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 entries, where the ‘*’ sign in SMILES represents the polymerization points.
970 entries, where the ‘*’ sign in SMILES represents the polymerization points.
5.4 SMILES tokenization
For tokenizing SMILES, we adopted a character-level tokenization approach. This choice was motivated by the diverse and extensive range of functional groups and their combinations present in polymers, making it challenging to exhaustively enumerate them and then tokenization. Character-level tokenization allows us to minimize the size of the vocabulary as much as possible. Moreover, the superior performance of character-level tokenization has been demonstrated in multiple models15,16,45,68 and Ablation studies section of this work.Data availability
All data and code are available in this repository (https://github.com/HKQiu/Unified_ML4Polymers). PolyNC is available for trial at https://huggingface.co/hkqiu/PolyNC.Author contributions
Haoke Qiu: investigation, methodology, data curation, visualization, software and writing – original draft. Lunyang Liu: investigation, methodology and writing – review & editing. Xuepeng Qiu, Xuemin Dai and Xiangling Ji: resources, validation. Zhao-Yan Sun: conceptualization, funding acquisition, project administration, resources, supervision and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for the support from the National Key R&D Program of China (No. 2022YFB3707303), and the National Natural Science Foundation of China (No. 21833008 and 52293471). We are also grateful to the Network and Computing Center in the Changchun Institute of Applied Chemistry for the hardware support.References
- F. M. Haque and S. M. Grayson, Nat. Chem., 2020, 12, 433–444 CrossRef CAS PubMed.
- Y. Zheng, S. Zhang, J. B.-H. Tok and Z. Bao, J. Am. Chem. Soc., 2022, 144, 4699–4715 CrossRef CAS PubMed.
- M. J. Sobkowicz, Science, 2021, 374, 540 CrossRef CAS PubMed.
- Q. A. Besford, H. Yong, H. Merlitz, A. J. Christofferson, J.-U. Sommer, P. Uhlmann and A. Fery, Angew. Chem., Int. Ed., 2021, 60, 16600–16606 CrossRef CAS PubMed.
- S.-M. Wen, S.-M. Chen, W. Gao, Z. Zheng, J.-Z. Bao, C. Cui, S. Liu, H.-L. Gao and S.-H. Yu, Adv. Mater., 2023, 35, 2211175 CrossRef CAS PubMed.
- G. Wang, L.-W. Feng, W. Huang, S. Mukherjee, Y. Chen, D. Shen, B. Wang, J. Strzalka, D. Zheng, F. S. Melkonyan, J. Yan, J. F. Stoddart, S. Fabiano, D. M. DeLongchamp, M. Zhu, A. Facchetti and T. J. Marks, Proc. Natl. Acad. Sci. U.S.A., 2020, 117, 17551–17557 CrossRef CAS PubMed.
- D. J. Audus and J. J. de Pablo, ACS Macro Lett., 2017, 6, 1078–1082 CrossRef CAS PubMed.
- L. Tao, J. He, N. E. Munyaneza, V. Varshney, W. Chen, G. Liu and Y. Li, Chem. Eng. J., 2023, 465, 142949 CrossRef CAS.
- S. Wu, Y. Kondo, M.-a. Kakimoto, B. Yang, H. Yamada, I. Kuwajima, G. Lambard, K. Hongo, Y. Xu, J. Shiomi, C. Schick, J. Morikawa and R. Yoshida, npj Comput. Mater., 2019, 5, 66 CrossRef.
- R. Ma, H. Zhang and T. Luo, ACS Appl. Mater. Interfaces, 2022, 14, 15587–15598 CrossRef CAS PubMed.
- M. A. Webb, N. E. Jackson, P. S. Gil and J. J. de Pablo, Sci. Adv., 2020, 6, eabc6216 CrossRef PubMed.
- Y. Zhao, R. J. Mulder, S. Houshyar and T. C. Le, Polym. Chem., 2023, 14, 3325–3346 RSC.
- H. Qiu, W. Zhao, H. Pei, J. Li and Z.-Y. Sun, Polymer, 2022, 256, 125216 CrossRef CAS.
- E. R. Antoniuk, P. Li, B. Kailkhura and A. M. Hiszpanski, J. Chem. Inf. Model., 2022, 62(22), 5435–5445 CrossRef CAS PubMed.
- C. Xu, Y. Wang and A. Barati Farimani, npj Comput. Mater., 2023, 9, 64 CrossRef.
- C. Kuenneth and R. Ramprasad, Nat. Commun., 2023, 14, 4099 CrossRef CAS PubMed.
- G. Landrum et al. , Rdkit: A software suite for cheminformatics, computational chemistry, and predictive modeling, Greg Landrum, 2013, vol. 8, p. 31 Search PubMed.
- H. Moriwaki, Y.-S. Tian, N. Kawashita and T. Takagi, J. Cheminf., 2018, 10, 4 Search PubMed.
- H. L. Morgan, J. Chem. Doc., 1965, 5, 107–113 CrossRef CAS.
- D. Duvenaud, D. Maclaurin, J. Aguilera-Iparraguirre, R. Gómez-Bombarelli, T. Hirzel, A. Aspuru-Guzik and R. Adams, Adv. Neural Inf. Process. Sys., 2015, 2015, 2224–2232 Search PubMed.
- H. Qiu, X. Qiu, X. Dai and Z.-Y. Sun, J. Mater. Chem. C, 2023, 11, 2930–2940 RSC.
- P. Veličković, G. Cucurull, A. Casanova, A. Romero, P. Liò and Y. Bengio, Int. Conf. Learn. Represent., 2017 Search PubMed.
- K. Yang, K. Swanson, W. Jin, C. Coley, P. Eiden, H. Gao, A. Guzman-Perez, T. Hopper, B. Kelley, M. Mathea, A. Palmer, V. Settels, T. Jaakkola, K. Jensen and R. Barzilay, J. Chem. Inf. Model., 2019, 59, 3370–3388 CrossRef CAS PubMed.
- M. Aldeghi and C. W. Coley, Chem. Sci., 2022, 13, 10486–10498 RSC.
- L. Zhang, M. Chen, A. Arnab, X. Xue and P. H. S. Torr, IEEE Trans. Pattern Anal. Mach. Intell., 2022, 1–17 Search PubMed.
- R. A. Patel, C. H. Borca and M. A. Webb, Mol. Syst. Des. Eng., 2022, 7, 661–676 RSC.
- S. Mohapatra, J. An and R. Gómez-Bombarelli, Mach. Learn.: Sci. Technol., 2022, 3, 015028 Search PubMed.
- D. Weininger, J. Chem. Inf. Model., 1988, 28, 31–36 CrossRef CAS.
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Mach. Learn.: Sci. Technol., 2020, 1, 045024 Search PubMed.
- T.-S. Lin, C. W. Coley, H. Mochigase, H. K. Beech, W. Wang, Z. Wang, E. Woods, S. L. Craig, J. A. Johnson, J. A. Kalow, K. F. Jensen and B. D. Olsen, ACS Cent. Sci., 2019, 5, 1523–1531 CrossRef CAS PubMed.
- M. A. Webb, N. E. Jackson, P. S. Gil and J. J. De Pablo, Sci. Adv., 2020, 6, eabc6216 CrossRef PubMed.
- L. Simine, T. C. Allen and P. J. Rossky, Proc. Natl. Acad. Sci. U.S.A., 2020, 117, 13945–13948 CrossRef CAS PubMed.
- D. Bhattacharya, D. C. Kleeblatt, A. Statt and W. F. Reinhart, Soft Matter, 2022, 18, 5037–5051 RSC.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. Kaiser and I. Polosukhin, Adv. Neural Inf. Process. Syst., Red Hook, NY, USA, 2017, pp. 6000–6010 Search PubMed.
- Y. Liu, M. Ott, N. Goyal, J. Du, M. Joshi, D. Chen, O. Levy, M. Lewis, L. Zettlemoyer and V. Stoyanov, CoRR, 2019, 471–484 CAS.
- P. He, X. Liu, J. Gao, W. Chen, Deberta: decoding-enhanced bert with disentangled attention, OpenReview.net (2021), Austria, 2020, https://openreview.net/forum?id=XPZIaotutsD Search PubMed.
- A. J. Gormley and M. A. Webb, Nat. Rev. Mater., 2021, 6, 642–644 CrossRef CAS PubMed.
- T. D. Huan, A. Mannodi-Kanakkithodi, C. Kim, V. Sharma, G. Pilania and R. Ramprasad, Sci. Data, 2016, 3, 160012 CrossRef CAS PubMed.
- J. Yang, L. Tao, J. He, J. R. McCutcheon and Y. Li, Sci. Adv., 2022, 8, eabn9545 CrossRef CAS PubMed.
- R. Ma and T. Luo, J. Chem. Inf. Model., 2020, 60, 4684–4690 CrossRef CAS PubMed.
- N. Andraju, G. W. Curtzwiler, Y. Ji, E. Kozliak and P. Ranganathan, ACS Appl. Mater. Interfaces, 2022, 14, 42771–42790 CrossRef CAS PubMed.
- S. Lo, M. Seifrid, T. Gaudin and A. Aspuru-Guzik, J. Chem. Inf. Model., 2023, 63, 4266–4276 CrossRef CAS PubMed.
- J. G. Ethier, R. K. Casukhela, J. J. Latimer, M. D. Jacobsen, B. Rasin, M. K. Gupta, L. A. Baldwin and R. A. Vaia, Macromolecules, 2022, 55, 2691–2702 CrossRef CAS.
- D. Christofidellis, G. Giannone, J. Born, O. Winther, T. Laino and M. Manica, Int. Conf. Mach. Learn., 2023 Search PubMed.
- J. Lu and Y. Zhang, J. Chem. Inf. Model., 2022, 62, 1376–1387 CrossRef CAS PubMed.
- R. Luo, L. Sun, Y. Xia, T. Qin, S. Zhang, H. Poon and T.-Y. Liu, Briefings Bioinf., 2022, 23, bbac409 CrossRef PubMed.
- W. Ahmad, E. Simon, S. Chithrananda, G. Grand and B. Ramsundar, ChemBERTa-2: Towards Chemical Foundation Models, 2022 Search PubMed.
- M. Seifrid, R. Pollice, A. Aguilar-Granda, Z. Morgan Chan, K. Hotta, C. T. Ser, J. Vestfrid, T. C. Wu and A. Aspuru-Guzik, Acc. Chem. Res., 2022, 55, 2454–2466 CrossRef CAS PubMed.
- Q. Zhu, F. Zhang, Y. Huang, H. Xiao, L. Zhao, X. Zhang, T. Song, X. Tang, X. Li, G. He, B. Chong, J. Zhou, Y. Zhang, B. Zhang, J. Cao, M. Luo, S. Wang, G. Ye, W. Zhang, X. Chen, S. Cong, D. Zhou, H. Li, J. Li, G. Zou, W. Shang, J. Jiang and Y. Luo, Natl. Sci. Rev., 2022, 9, nwac190 CrossRef CAS PubMed.
- G. Turon, J. Hlozek, J. G. Woodland, A. Kumar, K. Chibale and M. Duran-Frigola, Nat. Commun., 2023, 14, 5736 CrossRef CAS PubMed.
- Q. Dou, D. Coelho de Castro, K. Kamnitsas and B. Glocker, Adv. Neural Inf. Process. Sys., 2019, 32, 6450–6461 Search PubMed.
- M. A. F. Afzal, A. R. Browning, A. Goldberg, M. D. Halls, J. L. Gavartin, T. Morisato, T. Hughes, D. J. Giesen and J. E. Goose, ACS Appl. Polym. Mater., 2020, 3, 620–630 CrossRef.
- C. Kuenneth, A. C. Rajan, H. Tran, L. Chen, C. Kim and R. Ramprasad, Patterns, 2021, 2, 100238 CrossRef CAS PubMed.
- D. Kamal, H. Tran, C. Kim, Y. Wang, L. Chen, Y. Cao, V. R. Joseph and R. Ramprasad, J. Chem. Phys., 2021, 154, 174906 CrossRef CAS PubMed.
- P. Ma, C. Dai, H. Wang, Z. Li, H. Liu, W. Li and C. Yang, Compos. Commun., 2019, 16, 84–93 CrossRef.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, J. Mach. Learn Res., 2020, 21, 1–67 Search PubMed.
- D. Christofidellis, G. Giannone, J. Born, O. Winther, T. Laino and M. Manica, Unifying Molecular and Textual Representations via Multi-task Language Modelling, 2023 Search PubMed.
- T. Wolf, L. Debut, V. Sanh, J. Chaumond, C. Delangue, A. Moi, P. Cistac, T. Rault, R. Louf, M. Funtowicz, J. Davison, S. Shleifer, P. von Platen, C. Ma, Y. Jernite, J. Plu, C. Xu, T. Le Scao, S. Gugger, M. Drame, Q. Lhoest and A. Rush, Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, 2020, pp. 38–45 Search PubMed.
- L. Van der Maaten and G. Hinton, J. Mach. Learn Res., 2008, 9, 2579–2605 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, J. Mach. Learn Res., 2011, 12, 2825–2830 Search PubMed.
- B. Ramsundar, P. Eastman, P. Walters, V. Pande, K. Leswing and Z. Wu, Deep Learning for the Life Sciences, O'Reilly Media, 2019 Search PubMed.
- T. Chen and C. Guestrin, Proceedings of the 22nd ACM SIGKDD Int. Conf. on Knowledge Discovery and Data Mining, 2016, pp. 785–794 Search PubMed.
- Y. Wang, M. Huang, X. Zhu and L. Zhao, Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, 2016, pp. 606–615 Search PubMed.
- P. Shaw, J. Uszkoreit and A. Vaswani, Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2018, vol. 2, Short Papers, pp. 464–468 Search PubMed.
- D. Probst and J.-L. Reymond, J. Cheminf., 2020, 12, 12 Search PubMed.
- T. Stuyver and C. W. Coley, J. Chem. Phys., 2022, 156, 084104 CrossRef CAS PubMed.
- P. Schwaller, T. Laino, T. Gaudin, P. Bolgar, C. A. Hunter, C. Bekas and A. A. Lee, ACS Cent. Sci., 2019, 5, 1572–1583 CrossRef CAS PubMed.
- Z. Cao, R. Magar, Y. Wang and A. Barati Farimani, J. Am. Chem. Soc., 2023, 145, 2958–2967 CrossRef CAS PubMed.
- K. Anoop, G. P. Manjary, P. Deepak, V. L. Lajish, Responsible Data Science, Springer, Singapore, 2022, vol. 940, pp. 13–45 Search PubMed.
- E. J. Bjerrum, arXiv, preprint, arXiv:1703.07076, 2017, DOI:10.48550/arXiv.1703.07076.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05079c |
| This journal is © The Royal Society of Chemistry 2024 |