Differentiation of snake venom using Raman spectroscopic analysis†
Vera
Mozhaeva
 *ab,
Vladislav
Starkov
b,
Denis
Kudryavtsev
*ab,
Vladislav
Starkov
b,
Denis
Kudryavtsev
 b,
Kirill
Prokhorov
a,
Sergey
Garnov
a and
Yuri
Utkin
b,
Kirill
Prokhorov
a,
Sergey
Garnov
a and
Yuri
Utkin
 b
b
aProkhorov General Physics Institute of the Russian Academy of Sciences, Moscow, 119991, Russian Federation. E-mail: VeraMozhaev@yandex.ru
bShemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, 117997, Russian Federation
First published on 31st May 2023
Abstract
Snake venoms are complex mixtures of different substances, proteins being their predominant components. To study the composition of venoms, methods based on chromatographic separation and mass spectrometric analysis are currently used, requiring the application of a number of sophisticated instruments. To assess the composition of snake venoms, we propose an alternative method based on Raman spectroscopy, which is an express method to study the structural features of different substances, including proteins. The method does not require preliminary preparation of the samples, which are used in small quantities; this makes Raman spectroscopy extremely attractive for venom research. In this work, we have carried out Raman spectroscopic studies on a number of dry venoms from various venomous snakes. Based on the obtained Raman spectra, with the help of mathematical methods of dimensionality reduction and clustering, differentiation of venoms reflecting their composition and the assignment of the venom producing snake to the corresponding family or even genus were performed. The proposed method can be used to analyze both the composition of and variations in venoms of different snake species, including rare and endangered ones.
Introduction
Among the several thousand known snake species, not all are venomous; those that are venomous mainly belong to the families Viperidae and Elapidae.1 The venom of these snakes is of the greatest medical importance, as their bites are dangerous to humans and often lead to disability and/or death. On the other hand, some components of venom have potential as pharmaceuticals.2,3 The venoms are mixtures of various compounds, such as proteins (peptides), lipids, nucleotides, some inorganic salts, etc. Compounds of a protein nature prevail in the composition of snake venom, and their content is about 90%.4 Among venom proteins, usually called toxins, the following main families are distinguished: secreted phospholipases A2 (PLA2), three-finger toxins (3FTx), snake venom metalloproteinases (SVMP), snake venom serine proteases (SVSP), L-amino acid oxidases (LAAO), cysteine-rich secretory protein (CRiSP), etc. Homologues of Kunitz-type serine protease inhibitors (KuIn), which predominate in the venom of some mambas, should also be mentioned.The venoms of different snake species have different ratios of these proteins. Thus, as a rule, 3FTx and PLA2 prevail in the venom of Elapidae, while SVMP, PLA2, and SVSP prevail in the venom of Viperidae.4 This explains different effects of venoms on the victim organism: in general, Elapidae bites cause neurotoxic syndrome (with systemic toxicity) with minimal local tissue damage, while Viperidae bites are accompanied by deep local tissue damage and hemotoxic syndrome.5 The composition of venom depends on the species of snake, as well as on other factors, such as the habitat (particularly in relation to diet6) and age.7 It is known that even among snakes of the same species, but living in different geographical locations, the composition of venom can differ significantly.4,8 Thus, the venom of Bungarus fasciatus (BuFa) from Thailand contains about 65% PLA2 and 20% KuIn, that from Indonesia contains 30% PLA2, 15% 3FTx and 50% KuIn, and venom of BuFa from China is dominated by PLA2 (more than 90%).
Determination of both interspecific and intraspecific variability of venoms, as well as the dependence of their composition on the geographic origin of the snake, is an important task. From a therapeutic point of view, knowledge of such variability will allow the treatment of bite victims more effectively, and in particular, it may help in the selection (especially when only monospecific antivenoms are available9) and production of a suitable antivenom (bites by conspecific populations may differ in symptomatology and require different treatments6). The knowledge of snake venom composition is also important for studying the evolution of venomous snakes and phylogenetic characterization of snakes at the level of species and subspecies.8 Differences in the properties and composition of venoms can be used to identify subspecies of snakes, as well as distinguish between morphologically similar species. It is also interesting to discover differences in the composition of the venoms of snakes of the same species living in different locations or over a vast territory. There is also an inverse problem of determining the type of snake or its habitat (place of capture) by analyzing its venom, for example, because of illegal transport and trade of venoms.
Raman spectroscopy is a rapid and versatile method for studying different materials, which allows exploration of their characteristics, e.g., of proteins, with micrograms of samples available. The Raman spectrum of a protein contains unique information about its structure.10 In addition to data on the primary structure, the Raman spectrum provides information on the secondary structure and some other features of protein folding. The low consumption of material makes the method extremely attractive for the study of those venoms which are difficult to obtain in significant quantities. The reduction in the dimension of spectral data and their presentation in a visual form using principal component analysis (PCA)11 and clustering methods allow quick classification of samples of protein nature.12
In this work, we used Raman spectroscopy to classify the venoms of several species of snakes from the families Elapidae and Viperidae, based on the spectra recorded from micrograms of dry venoms.
Experimental
Materials and methods
| Abbreviation | Species | Origin | Family |
|---|---|---|---|
| AgCo | Agkistrodon contortrix | Not known | Viperidae (V) |
| GlSa | Gloydius saxatilis | Primorsky Territory, Russia | V |
| BuFa | Bungarus fasciatus | Vietnam | Elapidae (E) |
| BuMu | Bungarus multicinctus | Vietnam | E |
| CrAd | Crotalus adamanteus | Southeast USA | V |
| DeAn | Dendroaspis angusticeps | Kenya | E |
| EcCa | Echis carinatus | Turkmenistan | V |
| EcMu | Echis multisquamatus | Not known | V |
| MaLeO | Macrovipera lebetina obtusa | Armenia | V |
| NaNa | Naja naja | Sri Lanka | E |
| NaKa | Naja kaouthia | Not known | E |
| ViNi | Vipera nikolskii | Krasivaya Mecha, Russia | V |
| ViRa | Montivipera raddei | Armenia | V |
| WaAe | Walterinnesia aegyptia | Iran | E |
| NaOx | Naja oxiana | Turkmenistan | E |
| ViAm | Vipera ammodytes | Bulgaria | V |
| ViUr | Vipera ursinii | Krasnodar Territory, Russia | V |
The Bungarus fasciatus and B. multicinctus krait venoms were obtained from the authorized, licensed farm for snake breeding and venom production located in Vinh Shon (Vinh Tuong, Vinh Phuc province, Vietnam). The certificate of the farm has been registered in accordance with the Government's Decree No. 82/2006/ND-CP of 10 August 2006 by the Department of Agriculture and Rural Development of the Socialist Republic of Vietnam. These venoms were collected and pooled from several tens of snake specimens. Venoms of Crotalus adamanteus, were kind gift from Professor T. Morita (Meiji Pharmaceutical University, Tokyo, Japan), venom of Dendroaspis angusticeps was a kind gift from Doctor E. Karlsson (Uppsala University, Uppsala, Sweden) and venoms of Macrovipera lebetina obtusa were a kind gift from Professor N. Aivazyan (Orbeli Institute of Physiology, Yerevan, Armenia). The numbers of snakes from which these venoms were obtained are not known. Cobra (Naja kaouthia, N. naja, N. oxiana and Walterinnesia aegyptia), viper (Vipera ammodytes, V. nikolskii, V. ursinii, Echis carinatusand E. multisquamatus) and pit viper (Agkistrodon contortrix and Gloydius saxatilis) venoms were obtained by milking the snakes kept in captivity. The venoms of cobras Naja kaouthia, N. naja and N. oxiana were collected and pooled from not less than 5 snake specimens. The venom of cobra Walterinnesia aegyptia was collected after milking several times from 2 specimens. The venoms of vipers were collected and pooled from not less than 10 snake specimens. The venoms of pit vipers were collected after milking several times from 3 snake specimens. Venoms obtained were dried over anhydrous CaCl2 and stored at −20 °C until use. All procedures with snakes were approved by the commission for the maintenance and use of laboratory animals of the Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences. (protocol no. 324/2021 of June 23, 2021.)
K-means clustering of PCA scores was performed in Python 3.9 using the “Scikit-learn” library. The number of clusters was determined by the elbow method14 (elbow criteria, sometimes the name “knee” is used) with the help of package “kneed”.
Results
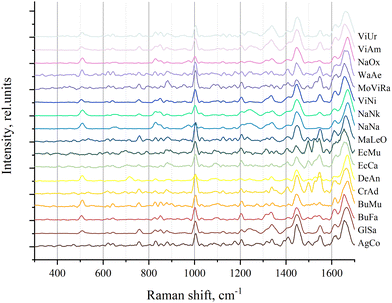
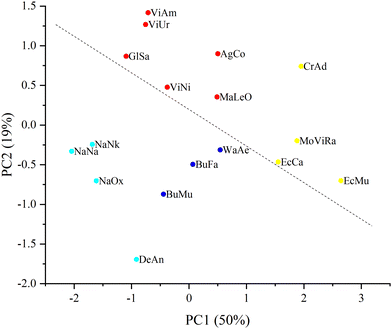
The Raman spectra were recorded from dry venom samples. The processed spectra of the samples are shown in Fig. 1.These processed spectra were subjected to PCA. The resulting PCA score plot is shown in Fig. 2 where, for clarity, 2 principal components (PCs) out of 3 used are presented. Loadings for PC1 and PC2 are shown in Fig. S2 (ESI†).
Furthermore, in order to identify groups of venom samples, we clustered our samples using the k-means method. Since k-means clustering requires a priori specification of the number of clusters, we used the elbow method, which, according to the dynamics of the sum of the squared error (SSE) (Fig. S1, ESI†), allowed us to determine the number of clusters (that is, their optimal number, with an additional increase of which SSE reduction is already negligible). As a result, the samples were divided into four clusters, which are shown in different colors in Fig. 2.
From the plot in Fig. 2 it can be seen that the samples can be divided into 2 large groups corresponding to Elapidae and Viperidae venoms; the dashed line crossing the figure illustrates this division. It is possible to achieve a more stringent separation by applying the discriminant function analysis (DFA) method (namely, principal component DFA (PC-DFA)) and it was applied in this study.
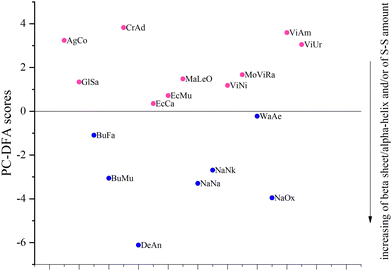
About 40% of the samples named train were used to train the PC-DFA algorithm (Table 2), that is, we indicated in advance which family (Elapidae (E) or Viperidae (V)) they belonged to. To ensure the diversity of the training set, 3 and 4 training samples were selected respectively from the venoms of Elapidae and Viperidae families. After that, based on the already trained model, the other samples named test were automatically assigned to one of the two families. The obtained results are shown in Fig. 3 (note that in this plot, only the vertical distance between the samples is significant; the distance along the horizontal axis is not important: the samples are separated by horizontal distances to avoid overlapping).
| Sample | Family | Set |
|---|---|---|
| AgCo | Test | |
| GlSa | Test | |
| BuFa | E | Train |
| BuMu | Test | |
| CrAd | V | Train |
| DeAn | Test | |
| EcCa | V | Train |
| EcMu | Test | |
| MaLeO | Test | |
| NaNa | E | Train |
| NaNk | Test | |
| ViNi | V | Train |
| MoViRa | Test | |
| WaAe | E | Train |
| NaOx | Test | |
| ViAm | V | Train |
| ViUr | Test |
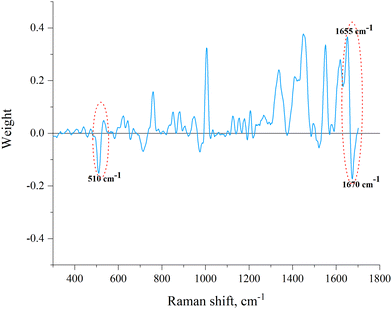
It can be seen that all test samples were correctly assigned to one of the two groups (which correspond to families): Elapidae venoms have a negative value on the PC-DFA score scale, and Viperidae venoms have a positive value. We were interested to see which spectral regions are responsible for such a separation. This information can be obtained from an analysis of the PC-DFA loadings, which show the effect of different Raman spectrum frequencies on the DFA scores (Fig. 4). It can be seen that the frequencies responsible for assigning the venom to the Elapidae family, that is, having a negative weight, correspond to peaks around 510 and 1670 cm−1.
We also performed a direct analysis of those regions of the Raman spectra that were identified as responsible for the division of samples into groups E and V through PC-DFA (Fig. 5). Conclusions about the secondary structure can be drawn from the data on the position of the peak maximum in the amide I band (Fig. 5(a) and Table 3). For the S–S region (Fig. 5(b)) we even managed to determine the boundary intensity of the peak maximum (∼0.18 rel. units, horizontal straight line in Fig. 5(b) and Table 3), above which are the maxima of the spectra of E venoms, while below it are the maxima of the spectra of V venoms.
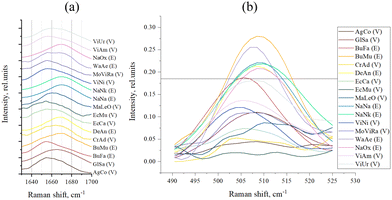 | ||
| Fig. 5 (a) Amide I band of sample spectra; and (b) S–S band of sample spectra (the horizontal line, intensity = 0.18 rel. units, separates venoms E and V). | ||
| Sample | Snake family | Amide I maximum, cm−1 | The S–S band intensity, rel. units |
|---|---|---|---|
| AgCo | V | 1654.5 | 0.04 |
| GlSa | V | 1664.5 | 0.11 |
| BuFa | E | 1654.5 | 0.19 |
| BuMu | E | 1669.5 | 0.28 |
| CrAd | V | 1663.5 | 0.05 |
| DeAn | E | 1669.5 | 0.21 |
| EcCa | V | 1654.5 | 0.07 |
| EcMu | V | 1654.5 | 0.02 |
| MaLeO | V | 1660.5 | 0.09 |
| NaNa | E | 1669.5 | 0.22 |
| NaNk | E | 1669.5 | 0.22 |
| ViNi | V | 1656 | 0.12 |
| ViRa | V | 1656 | 0.11 |
| WaAe | E | 1656 | 0.26 |
| NaOx | E | 1669.5 | 0.21 |
| ViAm | V | 1656 | 0.14 |
| ViUr | V | 1660.5 | 0.18 |
To evaluate the contribution of each secondary structural component to the overall structure of samples, we additionally performed curve-fitting analysis of the amide I band for each sample. Initially, the number of features (peaks) for each amide I band was chosen to be 3 according to the following possible structures: α-helix, nonregular and β-sheet structure. In some cases, the method detected a smaller number of hidden peaks and the fitting was not possible with 3 functions. Then their number was reduced to 2 or 1. The graphical results of the analysis are shown in Fig. S3 (ESI†). For each sample the frequencies of the maxima of all functions used to fit each amide I band, as well as the areas under these curves in percentages that correspond to a particular structure are given in Table 4. The types of secondary structures that are characteristic of the corresponding frequencies and number of fitting functions are also indicated.
| Sample | Number of fitting functions | Structure type | Frequency (cm−1) | Area (%) |
|---|---|---|---|---|
| AgCo | 3 | α-Helix | 1652 | 63 |
| Nonregular | 1663 | 21 | ||
| β-Sheet | 1673 | 16 | ||
| GlSa | 3 | α-Helix | 1656 | 56 |
| Nonregular | 1666 | 4 | ||
| β-Sheet | 1675 | 40 | ||
| BuFa | 3 | α-Helix | 1654 | 79 |
| Nonregular | 1664 | 2 | ||
| β-Sheet | 1674 | 19 | ||
| BuMu | 3 | α-Helix | 1652 | 26 |
| Nonregular | 1664 | 19 | ||
| β-Sheet | 1674 | 55 | ||
| CrAd | 3 | α-Helix | 1654 | 66 |
| Nonregular | 1663 | 5 | ||
| β-Sheet | 1672 | 29 | ||
| DeAn | 3 | α-Helix | 1654 | 30 |
| Nonregular | 1664 | 22 | ||
| β-Sheet | 1674 | 48 | ||
| EcCa | 3 | α-Helix | 1654 | 86 |
| Nonregular | 1663 | 2 | ||
| β-Sheet | 1671 | 12 | ||
| EcMu | 3 | α-Helix | 1652 | 92 |
| Nonregular | 1663 | 2 | ||
| β-Sheet | 1670 | 6 | ||
| MaLeO | 3 | α-Helix | 1656 | 59 |
| Nonregular | 1664 | 15 | ||
| β-Sheet | 1674 | 26 | ||
| NaNa | 1 | β-Sheet | 1669 | 100 |
| NaNk | 1 | β-Sheet | 1669 | 100 |
| ViNi | 2 | α-Helix | 1655 | 76 |
| β-Sheet | 1673 | 24 | ||
| MoViRa | 2 | α-Helix | 1654 | 90 |
| β-Sheet | 1671 | 10 | ||
| WaAe | 3 | α-Helix | 1655 | 72 |
| Nonregular | 1662 | 4 | ||
| β-Sheet | 1673 | 24 | ||
| NaOx | 1 | β-Sheet | 1668 | 100 |
| ViAm | 2 | α-Helix | 1656 | 81 |
| β-Sheet | 1676 | 19 | ||
| ViUr | 2 | α-Helix | 1657 | 73 |
| β-Sheet | 1675 | 27 |
Discussion
In general, Raman spectroscopy was proposed as a method to study the structural characteristics of different matter including biological materials.15 Still playing an important role in the structural characterization of substances, this method has significantly expanded the scope of application. Nowadays, it is used for early detection of cancer,16 bacteria detection,17 to study the pathophysiology of brain diseases,18 to determine the biochemical composition of plant,19etc. It also demonstrates the possibility of working with complex samples (multicomponent mixtures), for example, a study reported the classification of blood samples obtained from 11 animal classes and human subjects by statistical analysis of their Raman spectra.20 So, this is a very versatile method, and in this work we applied it for the classification of snake venoms. It should be mentioned that earlier Raman spectroscopy was used for the analysis of individual venom components. For example, the presence of β-structure was shown for neurotoxins of Pelamis platurus21 and for cobramine B, a small basic protein from cobra venom.22PCA serves primarily as a dimensionality reduction tool. The spectral data simplified in this way can then be analyzed by other methods, in particular, by classification methods, for the application of which, as a rule, it is necessary that the number of samples exceeds the number of variable parameters (characteristics in which the samples differ). Also, PCA makes it possible to visualize relationships (similarities and differences) between samples in this new reduced dimensional space (using the distance between samples as a measure of their difference). In addition to the obvious simplification of data, PCA allows small spectral noise to be ignored, while retaining important information.
Previously, using Raman spectroscopy and PCA we had classified protein toxins including those from snake venoms in accordance with their structural features; moreover, using this approach it was possible to distinguish the disulfide isomers of the peptide toxin.12 In the present work, we have carried out Raman spectroscopic studies on a number of dry venoms from various venomous snakes supplemented with data analysis by PCA. The resulting PCA score plot (Fig. 2) shows the differentiation of all studied venoms into several clusters. Cluster 1 (red) includes the venoms of representatives of the Vipera, Agkistrodon and Gloydius genus and Levantine viper. Cluster 2 (yellow) includes samples from the Echis genus, Montivipera genus, and the eastern diamondback rattlesnake (CrAd). All venoms in cluster 2 are united by the predominance of a component called SVMP.4 The CrAd venom sample is quite distinct from other samples of this group, which can be explained by the fact that the CrAd species is USA endemic and its venom has specific features. Cluster 3 (blue) combines the venoms of kraits (Bungarus genus) and the venom of the desert cobra (WaAe). Interestingly, in contrast to the venom of most Elapidae, the venom of some representatives of the Bungarus genus is dominated by PLA2. Probably, this feature distinguishes venom samples of the 3rd and 4th clusters. Cluster 4 (light blue) consists of true cobra venoms and DeAn venom, the main components of which are 3FTx. Within this cluster, cobra venoms are quite clearly distinguished as a separate subgroup, while the venom of mamba DeAn is fairly different from them.
This PC-DFA method23 allows us to build a boundary between known classes of training objects. And if up to this point the entire analysis of the spectra (clustering) was carried out without any a priori information about the venoms (in particular about the producing snakes), the DFA requires training on certain samples (training set), for which one has information about whether its samples belong to one or another of two groups (classes). A sample inclusion in one of the two groups is determined by whether its discriminant score is positive or negative, which is calculated during DFA for each sample. We used a modification of the DFA method – PC-DFA. In this case, only the new variables (principal components) are subjected to DFA since we are applying this analysis to the data obtained after PCA.
As mentioned in the introduction, proteins are the predominant components of venom and therefore, they make the main contribution to the spectra of the studied samples (resonance effects that significantly enhance the spectrum of some other components of the venom were not found). Therefore, the sample spectrum can be interpreted as superposition of the spectra of various proteins that make up venom. It is known that the spectral region of 490–550 cm−1 represents the disulfide bond band, and the oscillation at a frequency of around 510 cm−1 corresponds to the main (gauche–gauche–gauche) conformation of C–C–S–S–C–C linkage.24,25 Accordingly, the presence of a peak at this frequency in the Raman spectrum indicates the presence of disulfide bonds in the sample (protein), and the intensity of the peak reflects their number. The 1600–1690 cm−1 region, the so-called the amide I band, contains information about the secondary structure of proteins and vibrations at a frequency of about 1670 cm−1 are characteristic to β-sheets in their structure.10 Vibrations at a frequency of about 1655 cm−1 are characteristic to α-helical structural elements of the protein, and, as can be seen from PC-DFA loadings (Fig. 4), the presence of α-helices increases the “probability” of assigning the sample to the Viperidae family.
Since no other sufficiently intense “negative peaks” were observed except for peaks around 510 and 1670 cm−1 (Fig. 4), it can be assumed that the main criteria for assigning a venom to the E or V family are the number of disulfide bonds and the amount of β-sheets. The more of these structural elements are present in the venom, the greater is the negative contribution to the value of its PC-DFA score and the more likely it is to be assigned to the E family. It can also be assumed that the lower the venom sample is located in the plot of Fig. 3, the greater is the relative amount of disulfide bonds and/or β-sheets in the proteins that make it up (the corresponding direction is marked in Fig. 3 with an arrow).
These results may be interpreted in terms of the quantitative content of specific proteins in venoms. Since the content of 3FTx usually dominates in Elapidae venoms,4,26 the greater amount of β-structural elements in their samples can be attributed precisely to 3FTx: it is known25,27 that the structure of most of these toxins is dominated by β-sheets that form loops. At the same time, a feature of some venoms from snakes of the genus Bungarus from the Elapidae family is a relatively low content of 3FTx with a predominance of PLA2.4 In particular, the main components of BuFa venom from Vietnam are as follows:4 67% PLA2 and 7% LAAO. Since α-helixes predominate in the PLA2 structure,28 we can attribute the increase in the amount of α-helixes to these proteins. This is reflected in the plot in Fig. 3: the BuFa sample from the Elapidae family is close to the border with Viperidae. From the data in this figure, it can also be concluded that the ratio of β/α structures is also reduced in WaAe venom, compared with other samples of the Elapidae family. The opposite situation is observed for DeAn, which is indeed characterized by an extremely high content of 3FTx.4
The shift of the amide I band maximum towards higher (>1665 cm−1) frequencies (Fig. 5(a) and Table 3) is typical for all E venoms, except for BuFa and WaAe, which we discussed above. In the case of the S–S region (Fig. 5(b) and Table 3), we see that indeed, as mentioned above, E venoms have a more intense peak here (at ∼510 cm−1), that is, a greater number of disulfide bonds.
Thus, it can be argued that the number of disulfide bonds in the venom can determine which family its source-snake belongs to, which is not a priori obvious: the amount of disulfide may be a characteristic of different venoms.
It should be mentioned that the two PCA with k-means clustering and PC-DFA methods used provide grouping of venoms (Fig. 2 and 3) based on the analysis of the entire spectra and, thus, take into account many more features of the sample spectra than are detected by direct analysis of specific parts of the spectra (Fig. 5), and the plots obtained as a result of clustering combine all the features of the sample spectra in a convenient visual format.
When interpreting the fitting results (Table 4 and Fig. S3, ESI†), we assumed that α-helix structure peaks centered around 1652–1656 cm−1 and β-sheet structure peaks were around 1669–1675 cm−1. Peaks below 1666 cm−1 but higher than 1662 cm−1 were associated with the presence of disordered structures.10,24,29
From the data presented in Table 4, it can be seen that, as was concluded from the analysis of the maximum position of the entire amide I band (Table 3 and Fig. 5(a)), α-helical structural elements dominate in the samples belonging to the Viperidae family and β-sheets dominate in the samples belonging to the Elapidae family. The exceptions are also BuFa and WaAe. An interesting result is that venoms of snakes belonging to Vipera and Montivipera genus have a minimum number of nonregular structures (the corresponding peak was not found) and in the venoms of true cobras, the predominant structural component is β-sheets (a single peak corresponding to this structure was found).
We were able to find only two papers describing the crude venom study by Raman spectroscopy. Thus, Raman spectroscopy was used to characterize biochemical alterations in the mouse gastrocnemius that was injected with Bothrops jararacussu venom, as well as, this venom was detected by Raman spectroscopy in the mouse muscle tissue.30 In another paper,31 surface-enhanced Raman spectroscopy (SERS) was used to compare and analyze two venoms: Trimeresurus stejnegeri from the Elapidae family and Bungarus multicinctus from the Viperidae family. The SERS fingerprints were different and enough to distinguish the two venoms studied. In SERS, nanoparticles (usually gold or silver) are used to amplify the signal. We believe that conventional Raman spectroscopy is more preferable to this method, primarily because it has a much greater reproducibility of results (spectra), even if they are obtained in different laboratories. In turn, SERS spectra depend significantly on the properties of nanoparticles (geometry, surface chemistry, and others). In addition, since the signal from the regions of molecules located near the surface of the nanoparticle is mainly amplified, the spectra obtained characterize not so much the whole venom sample itself but the nature of the interaction of its individual proteins with nanoparticles. At the same time, conventional Raman spectra contain information about the complete amino acid sequence32 and the 3D structure of all venom proteins. It should be noted, that protein classification by Raman spectroscopy is not affected by sample drying.33
The method proposed here facilitates fast analysis by comparing a large number of dry venoms that do not require a special sample preparation and are needed in minimal quantities (∼10 μg). Thus, the venom obtained from one specimen (individual) of the snake is quite sufficient. This fact, along with the speed of analysis, is an advantage of this method.
Currently, one of the main methods for assessing the composition of crude venom (“direct” approach) is liquid chromatography with tandem mass spectrometry (LC/MS/MS), which makes it possible to determine what protein families are present in the venom.34,35 This method is not without drawbacks: many proteins required de novo MS sequence analysis with homology searches to place protein in a particular venom toxin family; venom gland transcriptome data, although of great utility in MS identification of venom proteins do not necessarily reflect the proteome of the venom.34 Techniques such as chromatography and gel electrophoresis are used to separate venom proteins and typically require following mass spectrometric analysis for protein identification.36
Our method based on Raman spectroscopy can be considered as an alternative way to analyze particular venom by establishing its similarity with other characterized venoms and thus assessing its composition. The method can be considered as the first step in the development of new methods for determining the type of venom using a swab from the bite site, for the correct selection of antivenom for the victim. Snakebite envenoming is an important cause of preventable death. The World Health Organization (WHO) set a goal to halve snakebite mortality by 2030. In 2019, 63,400 people died globally from snakebites.37 An estimated 5.4 million people are bitten each year with up to 2.7 million envenoming (https://www.who.int/). South Asia had the greatest burden. Since compact Raman spectrometers are available,38 it is possible to carry out analysis outside the laboratory, which is important for work in developing countries where mortality from snake bites is quite high.9
Conclusions
In this work, for the first time, a Raman spectroscopic study of crude venoms from various snake species of different genera and families was carried out. The study showed the applicability of this spectroscopic technique for investigation of such complex protein mixtures as venoms and the usefulness of dimensionality reduction and clustering methods for interpretation of complex spectral data. Based on the obtained spectral data, the venom samples were divided into two groups corresponding to the Viperidae and Elapidae families, which venom-producing snakes belong to. A more detailed classification into four groups, reflecting the composition of venoms, was also carried out.Thus, we have proposed an extremely economical express method, which made it possible to quickly analyze a large number of venoms, and the low consumption of materials made it possible to work with small quantities of samples, which is important when studying the venoms of unproductive (low yielding) snakes, as well as rare and endangered species of snakes. Our approach provided a way to evaluate the compositions of venoms by comparison with those already characterized.
Author contributions
Vera Mozhaeva: conceptualization, investigation, methodology, supervision, and writing – original draft; Vladislav Starkov: resources and methodology; Denis Kudryavtsev: conceptualization, validation, and writing – review and editing; Kirill Prokhorov: methodology and writing – review and editing; Sergey Garnov: supervision; Yuri Utkin: conceptualization, resources, validation, writing – original draft, and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a grant from the Ministry of Science and Higher Education of the Russian Federation (075-15-2022-315) for the organization and development of a World-class research center ≪Photonics≫.Notes and references
- A. Munawar, S. A. Ali, A. Akrem and C. Betzel, Toxins, 2018, 10, 474 CrossRef CAS PubMed.
- K. de, C. F. Bordon, C. T. Cologna, E. C. Fornari-Baldo, E. L. Pinheiro-Júnior, F. A. Cerni, F. G. Amorim, F. A. P. Anjolette, F. A. Cordeiro, G. A. Wiezel, I. A. Cardoso, I. G. Ferreira, I. S. de Oliveira, J. Boldrini-França, M. B. Pucca, M. A. Baldo and E. C. Arantes, Front. Pharmacol., 2020, 11, 1132 CrossRef PubMed.
- T. M. A. El-Aziz, A. G. Soares and J. D. Stockand, Toxins, 2019, 11, 564 CrossRef PubMed.
- A. L. Oliveira, M. F. Viegas, S. L. da Silva, A. M. Soares, M. J. Ramos and P. A. Fernandes, Nat. Rev. Chem., 2022, 6, 451–469 CrossRef PubMed.
- M. Tednes and T. L. Slesinger, Evaluation and Treatment of Snake Envenomations, 2022 Search PubMed.
- J. C. Daltry, W. Wüster and R. S. Thorpe, Nature, 1996, 379, 537–542 CrossRef CAS PubMed.
- H. A. Reid and R. D. G. Theakston, Am. J. Trop. Med. Hyg., 1978, 27, 1053–1057 CrossRef CAS PubMed.
- J. P. Chippaux, V. Williams and J. White, Toxicon, 1991, 29, 1279–1303 CrossRef CAS PubMed.
- J. M. Gutiérrez, J. J. Calvete, A. G. Habib, R. A. Harrison, D. J. Williams and D. A. Warrell, Nat. Rev. Dis. Prim., 2017, 3, 17063 CrossRef PubMed.
- A. Rygula, K. Majzner, K. M. Marzec, A. Kaczor, M. Pilarczyk and M. Baranska, J. Raman Spectrosc., 2013, 44, 1061–1076 CrossRef CAS.
- A. Tharwat, Int. J. Appl. Pattern Recognit., 2016, 3, 197–240 CrossRef.
- V. Mozhaeva, D. Kudryavtsev, K. Prokhorov, Y. Utkin, S. Gudkov, S. Garnov, I. Kasheverov and V. Tsetlin, Spectrochim. Acta, Part A, 2022, 278, 121276 CrossRef CAS PubMed.
- R. M. Jarvis, D. Broadhurst, H. Johnson, N. M. O’Boyle and R. Goodacre, Bioinformatics, 2006, 22, 2565–2566 CrossRef CAS PubMed.
- V. Satopää, J. Albrecht, D. Irwin and B. Raghavan, in Proceedings – International Conference on Distributed Computing Systems, 2011, pp. 166–171.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner, P. L. Martin-Hirsch, M. J. Walsh, M. R. McAinsh, N. Stone and F. L. Martin, Nat. Protoc., 2016, 11, 664–687 CrossRef CAS PubMed.
- R. Shaikh, A. Daniel and F. M. Lyng, Molecules, 2023, 28, 2502 CrossRef CAS PubMed.
- L. Liu, W. Ma, X. Wang and S. Li, Biosensors, 2023, 13, 350 CrossRef CAS PubMed.
- O. Terrones, J. Olazar-Intxausti, I. Anso, M. Lorizate, J. A. Nieto-Garai and F. X. Contreras, Int. J. Mol. Sci., 2023, 24, 2384 CrossRef PubMed.
- A. Zavafer and M. C. Ball, Funct. Plant Biol., 2023, 50, 1–16 CrossRef PubMed.
- G. McLaughlin, K. C. Doty and I. K. Lednev, Anal. Chem., 2014, 86, 11628–11633 CrossRef CAS PubMed.
- A. T. Tu, B. H. Jo and N.-T. Yu, Int. J. Pept. Protein Res., 1976, 8, 337–343 CrossRef CAS PubMed.
- N. T. Yu, B. H. Jo and D. C. O’Shea, Arch. Biochem. Biophys., 1973, 156, 71–76 CrossRef CAS PubMed.
- P. S. Gromski, H. Muhamadali, D. I. Ellis, Y. Xu, E. Correa, M. L. Turner and R. Goodacre, Anal. Chim. Acta, 2015, 879, 10–23 CrossRef CAS PubMed.
- E. Li-Chan, S. Nakai and M. Hirotsuka, in Protein Structure-Function Relationships in Foods, ed. R. Y. Yada, R. L. Jackman and J. L. Smith, Springer US, Boston, MA, 1994, pp. 163–197 Search PubMed.
- T. Takamatsu, I. Harada and K. Hayashi, BBA – Protein Struct., 1980, 622, 189–200 CrossRef CAS PubMed.
- Y. N. Utkin, Toxicon, 2013, 62, 50–55 CrossRef CAS PubMed.
- P. Kessler, P. Marchot, M. Silva and D. Servent, J. Neurochem., 2017, 142, 7–18 CrossRef CAS PubMed.
- P. G. Ojeda, D. Ramírez, J. Alzate-Morales, J. Caballero, Q. Kaas and W. González, Toxins, 2018, 10 Search PubMed.
- I. Chourpa, V. Ducel, J. Richard, P. Dubois and F. Boury, Biomacromolecules, 2006, 7, 2616–2623 CrossRef CAS PubMed.
- W. F. Vieira, B. Kenzo-Kagawa, M. H. M. Britto, H. J. Ceragioli, K. K. Sakane, V. Baranauskas and M. A. da Cruz-Höfling, Lasers Med. Sci., 2018, 33, 503–512 CrossRef PubMed.
- C. C. Huang, Y. S. Lai, C. H. Wang, L. K. Chau and W. Chen, J. Mater. Chem. B, 2020, 8, 10744–10753 RSC.
- V. J. C. Lin and J. L. Koenig, Biopolymers, 1976, 15, 203–218 CrossRef CAS PubMed.
- G. Devitt, W. Rice, A. Crisford, I. Nandhakumar, A. Mudher and S. Mahajan, ACS Chem. Neurosci., 2019, 4593–4611 CrossRef CAS PubMed.
- J. W. Fox and S. M. T. Serrano, Proteomics, 2008, 8, 909–920 CrossRef CAS PubMed.
- N. van de Merbel, Sample Prep. LC-MS Bioanal., 2019, 304–318 CAS.
- S. M. T. Serrano, J. D. Shannon, D. Wang, A. C. M. Camargo and J. W. Fox, Proteomics, 2005, 5, 501–510 CrossRef CAS PubMed.
- N. L. S. Roberts, E. K. Johnson, S. M. Zeng, E. B. Hamilton, A. Abdoli, F. Alahdab, V. Alipour, R. Ancuceanu, C. L. Andrei, D. Anvari, J. Arabloo, M. Ausloos, A. F. Awedew, A. D. Badiye, S. M. Bakkannavar, A. Bhalla, N. Bhardwaj, P. Bhardwaj, S. Bhaumik, A. Bijani, A. Boloor, T. Cai, F. Carvalho, D. T. Chu, R. A. S. Couto, X. Dai, A. A. Desta, H. T. Do, L. Earl, A. Eftekhari, F. Esmaeilzadeh, F. Farzadfar, E. Fernandes, I. Filip, M. Foroutan, R. C. Franklin, A. M. Gaidhane, B. G. Gebregiorgis, B. Gebremichael, A. Ghashghaee, M. Golechha, S. Hamidi, S. E. Haque, K. Hayat, C. Herteliu, O. S. Ilesanmi, M. M. Islam, J. Jagnoor, T. Kanchan, N. Kapoor, E. A. Khan, M. N. Khatib, R. Khundkar, K. Krishan, G. A. Kumar, N. Kumar, I. Landires, S. S. Lim, M. Madadin, V. Maled, N. Manafi, L. B. Marczak, R. G. Menezes, T. J. Meretoja, T. R. Miller, A. Mohammadian-Hafshejani, A. H. Mokdad, F. N. P. Monteiro, M. Moradi, V. C. Nayak, C. T. Nguyen, H. L. T. Nguyen, V. Nuñez-Samudio, S. M. Ostroff, J. R. Padubidri, H. Q. Pham, M. Pinheiro, M. Pirestani, Z. Quazi Syed, N. Rabiee, A. Radfar, V. Rahimi-Movaghar, S. J. Rao, P. Rastogi, D. L. Rawaf, S. Rawaf, R. C. Reiner, A. Sahebkar, A. M. Samy, M. Sawhney, D. C. Schwebel, S. Senthilkumaran, M. A. Shaikh, V. Y. Skryabin, A. A. Skryabina, A. Soheili, M. A. Stokes, R. Thapar, M. R. Tovani-Palone, B. X. Tran, R. S. Travillian, D. Z. Velazquez, Z. J. Zhang, M. Naghavi, R. Dandona, L. Dandona, S. L. James, D. M. Pigott, C. J. L. Murray, S. I. Hay, T. Vos and K. L. Ong, Nat. Commun., 2022, 13, 6160 CrossRef PubMed.
- D. Dhankhar, A. Nagpal and P. M. Rentzepis, Rev. Sci. Instrum., 2021, 92, 054101 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb00829k |
| This journal is © The Royal Society of Chemistry 2023 |