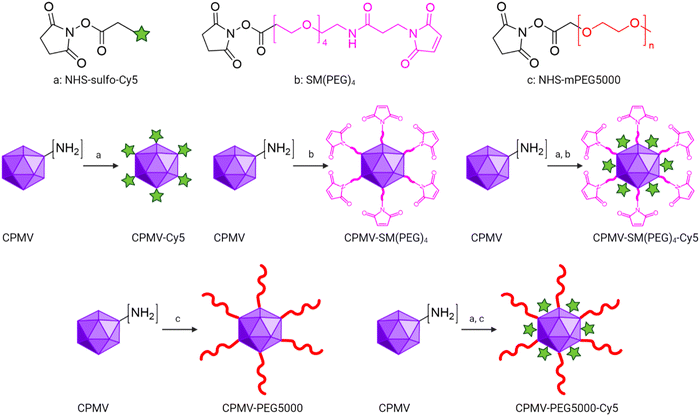
A co-formulated vaccine of irradiated cancer cells and cowpea mosaic virus improves ovarian cancer rejection†
Zhongchao
Zhao
 abc,
Oscar A.
Ortega-Rivera
abc,
Oscar A.
Ortega-Rivera
 a,
Young Hun
Chung
a,
Young Hun
Chung
 cd,
Andrea
Simms
a and
Nicole F.
Steinmetz
cd,
Andrea
Simms
a and
Nicole F.
Steinmetz
 *abcdefg
*abcdefg
aDepartment of NanoEngineering, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA. E-mail: nsteinmetz@ucsd.edu
bCenter for Nano-ImmunoEngineering, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
cMoores Cancer Center, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
dDepartment of Bioengineering, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
eDepartment of Radiology, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
fInstitute for Materials Discovery and Design, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
gCenter for Engineering in Cancer, University of California, San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA
First published on 2nd March 2023
Abstract
Ovarian cancer ranks fifth in cancer deaths amongst women, and most patients are diagnosed with late-stage and disseminated diseases. Surgical debulking and chemotherapy remove most of the tumor burden and provide a short period of remission; however, most patients experience cancer relapse and eventually succumb to the disease. Therefore, there is an urgent need for the development of vaccines to prime anti-tumor immunity and prevent its recurrence. Here we developed vaccine formulations composed of a mixture of irradiated cancer cells (ICCs, providing the antigen) and cowpea mosaic virus (CPMV) adjuvants. More specifically we compared the efficacy of co-formulated vs. mixtures of ICCs and CPMV. Specifically, we compared co-formulations where the ICCs and CPMV are bonded through natural CPMV–cell interactions or chemical coupling vs. mixtures of PEGylated CPMV and ICCs, where PEGylation of CPMV prevents ICC interactions. Flow cytometry and confocal imaging provided insights into the composition of the vaccines and their efficacy was tested using a mouse model of disseminated ovarian cancer. 67% of the mice receiving the co-formulated CPMV–ICCs survived the initial tumor challenge, and 60% of the surviving mice rejected tumors in a re-challenge experiment. In stark contrast, simple mixtures of the ICCs and (PEGylated) CPMV adjuvants were ineffective. Overall, this study highlights the importance of the co-delivery of cancer antigens and adjuvants in ovarian cancer vaccine development.
10th Anniversary StatementThe Journal of Materials Chemistry B is one of the most important journals for researchers in the field of development and application of biomaterials and nanomaterials for medical purposes to publish research works about drug delivery systems and nanomedicines. The development of multi-functional and sophisticated virus-based nanomaterials as cancer immunotherapeutics has been a hot topic in the cancer immunotherapy field and has grown with the Journal of Materials Chemistry B. I sincerely celebrate the 10th anniversary of the Journal of Materials Chemistry and would like to contribute further to the growth of the journal. |
Introduction
Ovarian cancer is one of the most lethal gynecological cancers in women.1,2 In 2022, ∼20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 women were diagnosed with ovarian cancer and ∼13
000 women were diagnosed with ovarian cancer and ∼13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 women died due to ovarian cancer in the United States.3 Many cases are diagnosed at the late-stage with disseminated, metastatic disease due to nonspecific symptoms such as bloating, weight gain, abdominal pain, bowel changes, and fatigue. The prognosis is poor and the 5 year survival rate is only ∼30%.4 The standard of care for late-stage ovarian cancer involves (i) surgical debulking to remove most of the tumor tissues from the peritoneal cavity and (ii) platinum-based chemotherapy.5 This treatment often leads to a short period of cancer remission, but most patients experience cancer relapse and eventually succumb to this disease. Therefore, developing a vaccine that would elicit potent anti-tumor immunity during the remission period to prevent ovarian cancer relapse is an important goal to improve survival rates.
000 women died due to ovarian cancer in the United States.3 Many cases are diagnosed at the late-stage with disseminated, metastatic disease due to nonspecific symptoms such as bloating, weight gain, abdominal pain, bowel changes, and fatigue. The prognosis is poor and the 5 year survival rate is only ∼30%.4 The standard of care for late-stage ovarian cancer involves (i) surgical debulking to remove most of the tumor tissues from the peritoneal cavity and (ii) platinum-based chemotherapy.5 This treatment often leads to a short period of cancer remission, but most patients experience cancer relapse and eventually succumb to this disease. Therefore, developing a vaccine that would elicit potent anti-tumor immunity during the remission period to prevent ovarian cancer relapse is an important goal to improve survival rates.
The approval of immune checkpoint blockade (ICB) drugs targeting CLTA4, PD1 and PDL1 established immunotherapy as a new pillar for cancer treatment.6–8 While clinical success has been observed for many tumor types, ICB has little efficacy in women with ovarian cancer,9,10 likely due to the highly immunosuppressive tumor microenvironment (TME).11 Therapeutic vaccines have been investigated for cancer treatment and prevention of recurrence – these include cell-based vaccines using dendritic cells (DCs) loaded with tumor lysates or tumor antigens,12,13 and subunit vaccines that deliver the tumor associated antigens (TAAs) as a protein, peptide antigen,14,15 or gene.16,17 However, many cancer vaccines have shown only moderate efficacy in clinical trials.18 A highly potent adjuvant is needed to prime potent and durable adaptive anti-tumor immunity.
Our vaccine strategy utilizes plant viruses (specifically cowpea mosaic virus, CPMV) as immunostimulants and adjuvants and we make use of irradiated tumor cells as the antigen/neoantigen source. Irradiation is a suitable method to produce the source of antigen, because irradiation can cause slow immunogenic cell death while maintaining cancer neoantigens.19 In the translational setting, ovarian tumor tissues collected during surgery could be irradiated, mixed with the CPMV antigen and then administered to boost the patient's immune system to launch anti-tumor immunity. This is a personalized approach utilizing the patient's own tumor.13,20
Immunostimulatory adjuvants, e.g., TLR agonists CpG ODNs21 and poly-IC,22 have previously been applied in combination with tumor cell lysates or irradiated cancer cells (ICCs), but clinical success however was limited. This in part may be explained that the small molecule agonists do not mimic the 3D architecture of natural pathogen-associated molecular patterns (PAMPs); also, the low molecular weight may lead to rapid clearance. To overcome these challenges we turned toward CPMV as an adjuvant: CPMV is a multi-toll-like receptor agonist that signals through TLRs 2, 4, and 7,23 thereby eliciting immunomodulation via multiple pathways. The multivalent architecture functions as a danger signal and provides signal enhancement through avidity effects; the nanoscale size allows for effective tissue retention and uptake by draining lymph nodes.24 Furthermore, T helper cell epitopes within the capsid structure further boost immunity.25 CPMV has already demonstrated utility as a potent adjuvant in infectious diseases and cancer. For example, we produced subunit vaccine candidates for COVID-19 by displaying peptide antigens on CPMV. The formulation elicited neutralizing antibodies against SARS-CoV-2.26 We also developed CPMV cancer vaccines displaying HER224,27 or NY-ESO-128 peptides to prime humoral and cellular anti-tumor immune responses for breast cancer treatment. Finally, CPMV is uniquely potent as an in situ vaccine for cancer treatment including ovarian cancer in mouse models29–31 and cancer in canine patients.32 Intratumorally injected CPMV activates and polarizes innate immune cells to antitumor phenotypes within the TME, resulting in a durable cytotoxic T-lymphocyte (CTL) response and immune memory.23,29,30,33–35 Therefore, we hypothesized that CPMV could function as a potent adjuvant for the formulation of ovarian cancer cell vaccines.
In a previous study, we demonstrated that irradiated murine ovarian cancer ID8-Defb29/Vegf-a-Luc cells (ICCs) mixed with CPMV protect mice from the ovarian tumor challenge – the CPMV outperformed other adjuvants such as monophosphoryl lipid A (MPLA).36
In the present work, we set out to investigate the vaccine formulation in more detail; we hypothesized that the exceptional efficacy of the ICC + CPMV vaccine candidate is, in part, a result of the co-delivery of the antigens (ICCs) and adjuvants (CPMV). A growing body of data indicates that synchronous delivery of antigens and adjuvants to antigen presenting cells (APCs) reduces adverse effects and focuses immune responses on the target by significantly boosting the stimulation of dendritic cells (DCs) and induction of CTL responses.37–40 To test our hypothesis that co-delivery of ICCs and CPMV is required for potency, we formulated a series of vaccine candidates using unmodified /modified CPMV that would either naturally bind to, chemically conjugate to, or be shielded from ICCs; vaccines were formulated at 4 °C or 37 °C to promote cell binding or uptake. Vaccine efficacy was studied using the murine ID8-Defb29/Vegf-a-Luc ovarian cancer model and C57BL/6 mice. Flow cytometry and confocal imaging were used to investigate whether co-delivery was achieved or prevented, and immunological assays were used to assay tumor-specific CTL responses.
Materials and methods
Production of CPMV vaccine candidates
CPMV was propagated in black eyed pea no. 5 plants and purified as previously reported.41 Following purification, an additional step using a PD-10 desalting column (Cytiva) was added to buffer exchange to PBS pH 7.4 (Corning), which was used throughout this study. CPMV was then filtered using a 33 mm-diameter membrane syringe-driven filter device with 0.22 μm pore size (MilliporeSigma) to remove any aggregates and contaminants. The concentration of CPMV was determined using ultraviolet-visible (UV-vis) spectroscopy (NanoDrop 2000, Thermo Scientific) and CPMV's extinction coefficient (ε) at 260 nm = 8.1 mg (mL × cm)−1. Lastly, potential endotoxin contamination was examined using a Pierce Chromogenic Endotoxin Quant Kit (ThermoFisher). For CPMV preps having endotoxin higher than 12.3 EU mg−1, endotoxin was removed using the established Triton X-114 method for CPMV.34 CPMV was stored at 4 °C at 10 mg mL−1.CPMV-SM(PEG)4, CPMV-PEG5000, CPMV-Cy5, CPMV-SM(PEG)4-Cy5, and CPMV-PEG5000-Cy5 were synthesized by covalently conjugating the N-hydroxysuccinimide (NHS) ester groups of Sulfo-Cy5 (777.95 g mol−1, NHS-Sulfo-Cy5, Lumiprobe), SM(PEG)4 (513.5 g mol−1, Thermo Fisher), and PEG5000 (5000 g mol−1, NHS-mPEG5000, Nanocs) to CPMV's solvent-exposed surface lysine residues. First, 50 mg mL−1 NHS-Sulfo-Cy5 and 20 mg mL−1 SM(PEG)4 were prepared by dissolving them in Ultra-Pure DMSO (VWR), followed by 50 mg mL−1 NHS-mPEG5000 in PBS (Corning). To synthesize CPMV-Cy5, CPMV-SM(PEG)4, and CPMV-PEG5000, a 3000-molar excess of NHS-Sulfo-Cy5, SM(PEG)4, and NHS-mPEG5000 was mixed with 2 mg mL−1 (final concentration) of CPMV (molecular weight = 5.6 × 106 g mol−1) in PBS for 2 h at room temperature. To synthesize CPMV-SM(PEG)4-Cy5 or CPMV-PEG5000-Cy5, a 1500 molar excess of NHS-Sulfo-Cy5 and a 3000 molar excess of SM(PEG)4 or a 1500 molar excess of NHS-Sulfo-Cy5 and a 5000-molar excess of NHS-mPEG5000 were mixed simultaneously with 2 mg mL−1 of CPMV in PBS for 2 h at room temperature. There were two major steps to purify conjugated CPMV particles: (i) the first step utilized Amicon Ultra-0.5 mL Centrifugal Filters with 100 kDa cutoff (MilliporeSigma) and 3 washes in PBS to remove excess reagents; (ii) conjugated CPMV particles were passed through a PD MidiTrap G-25 column (Cytiva) for further purification according to the manufacturer's protocol.
Characterization of CPMV vaccine candidates
After the last purification step, all CPMV vaccine concentrations were adjusted to 1 mg mL−1 for characterization using UV-vis spectroscopy, dynamic light scattering (DLS), SDS-PAGE, agarose gel electrophoresis, size exclusion chromatography (SEC), and transmission electron microscopy (TEM).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L mol−1 cm−1) were used at 647 nm.
000 L mol−1 cm−1) were used at 647 nm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×.
000×.
Cell lines and cell culture
All cells were maintained at 37 °C with 5% CO2. The murine ovarian cancer cell line ID8-Defb29/Vegf-a-Luc42 was cultured using RPMI 1640 medium with L-glutamine (Corning) supplemented with 10% (v/v) fetal-bovine serum (FBS) (VWR), 1% (v/v) penicillin/streptomycin (Pen/Strep) (Cytiva), 1 mM sodium pyruvate (ThermoFisher Scientific), and 0.05 mM β-mercaptoethanol (ThermoFisher Scientific). The murine colon cancer cell line CT26 was purchased from ATCC and cultured using RPMI 1640 medium with L-glutamine supplemented with 10% (v/v) FBS and 1% (v/v) Pen/Strep. Trypsin-EDTA (Corning) was used to harvest ID8-Defb29/Vegf-a-Luc and CT26 cells. PBS was purchased from Corning to wash cells. Cell dissociation buffer (Gibco) was used when harvesting cells for flow cytometry experiments.Cancer cell vaccine formulation
Each vaccine formulation contained 5 × 106 irradiated cancer cells (ICCs) and 100 μg of CPMV in 200 μL of PBS. Vaccine formulations were prepared as previously established.36 First, CPMV, CPMV-SM(PEG)4 and CPMV-PEG5000 were adjusted to 1 mg mL−1 in PBS. ID8-Defb29/Vegf-a-Luc cells were harvested, washed once using PBS, and then resuspended in PBS giving 50 × 106 cells per mL. To obtain ICCs, ID8-Defb29/Vegf-a-Luc cells were irradiated with 70 Gray using a Cs-137 source (10 Gy per 1.18 min for 8.26 min). ICCs were then mixed with CPMV, CPMV-SM(PEG)4, and CPMV-PEG5000 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, which were termed as ICCs–CPMV, ICCs–CPMV-SM(PEG)4, and ICCs–CPMV-PEG5000, respectively, and kept on ice for 30 min prior to intraperitoneal (i.p.) injection. Two additional vaccine formulations were prepared by mixing ID8-Defb29/Vegf-a-Luc cells with CPMV first in a 1
1 ratio, which were termed as ICCs–CPMV, ICCs–CPMV-SM(PEG)4, and ICCs–CPMV-PEG5000, respectively, and kept on ice for 30 min prior to intraperitoneal (i.p.) injection. Two additional vaccine formulations were prepared by mixing ID8-Defb29/Vegf-a-Luc cells with CPMV first in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and incubated at 4 °C or 37 °C for 30 min followed by irradiation. These two formulations were termed CPMV 4 °C-ICCs and CPMV 37 °C-ICCs.
1 ratio and incubated at 4 °C or 37 °C for 30 min followed by irradiation. These two formulations were termed CPMV 4 °C-ICCs and CPMV 37 °C-ICCs.
Mice
All mouse studies were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the University of California, San Diego (UCSD), and were approved by the Animal Ethics Committee of UCSD. Because ovarian cancer only afflicts women, all experiments were conducted using female C57BL/6 mice purchased from Jackson Laboratories.Vaccination and tumor challenge in mice
All mice were subjected to a prime and boost vaccination regimen. In detail, on day −14 and day −7, all mice (n = 5 per group) were i.p. injected with 5 different formulations of vaccines: ICCs–CPMV, ICCs–CPMV-SM(PEG)4, ICCs–CPMV-PEG5000, CPMV 4 °C-ICCs, and CPMV 37 °C-ICCs in 200 μL with 5 × 106 ICCs and 100 μg of CPMV per mouse. 200 μL of PBS served as a control group. On day 0, ID8-Defb29/Vegf-a-Luc cells were harvested, washed once using PBS, and then adjusted to 5 × 106 cells in 200 μL of PBS and were i.p. injected to each mouse for tumor challenge. Mice were monitored 3 times per week for tumor progression measured by increases in body weight and circumference. Mice were euthanized when their body weight reached 35 g or circumference reached 9 cm.Detection of a CTL response via ELISpot and the enzyme-linked immunosorbent assay (ELISA)
3 mice were immunized with PBS (control) or 5 × 106 ICCs mixed with 100 μg of CPMV (ICCs–CPMV vaccine) in 200 μL of PBS using the prime-boost regimen. On day 10, spleens were collected from all mice and single cell splenocyte suspensions were prepared using the Spleen Dissociation Kit for the mouse and a gentleMACS Dissociator (Miltenyi Biotec) according to the manufacturer's protocols.ELISpot assay
To assay for tumor-specific CTL responses, ELISpot assays were carried out using a Mouse interferon (IFN)-γ single color ELISPOT kit with pre-coated 96-well plates (Cellular Technology Limited) as done previously.26 Splenocytes were adjusted to 5 × 106 cells per mL in CTL test media. ID8-Defb29/Vegf-a-Luc cells and CT26 cells were harvested and resuspended in CTL test media at 5 × 106 cells per mL. 100 μL of the splenocyte suspension was aliquoted into each well (5 × 105 cells per well), and then 100 μL of 5 × 106 cells per L of ID8-Defb29/Vegf-a-Luc cells, 100 μL of 10 μg mL−1 CPMV, 100 μL of 5 × 106 cells per mL of CT26 cells (specificity control), 100 μL of 50 ng mL−1 phorbol 12-myristate 13-acetate (PMA) with 1 μg mL−1 ionomycin (Sigma-Aldrich) (positive control), or 100 μL of CTL test media (negative control) were added. The plates were incubated at 37 °C and 5% CO2 for 48 h. After incubation, bound cancer cells were gently removed using a stainless-steel spatula and the plates were washed with PBST (0.05% (v/v) Tween-20). Following the manufacturer's protocol and reagents, the plates were processed to develop IFN-γ spots. The colored spots were quantified using an Immunospot S6 Entry analyzer (Cellular Technology Limited). The splenocytes were evaluated per animal and tested in triplicate for each stimulant.ELISA assay
ELISA was performed using the IFN-γ Mouse Uncoated ELISA Kit (ThermoFisher Scientific). Similar to the ELISpot setup, 1 × 106 cells of each splenocyte were mixed with 1 × 106 ID8-Defb29/Vegf-a-Luc or CT26 cells, 10 μg of CPMV, 50 ng of PMA with 1 μg of ionomycin (positive control), or CTL test media (negative control) in a 24-well plate (1 mL per well) to stimulate IFN-γ production. The plates were incubated at 37 °C and 5% CO2 for 43 h, and the medium from each well was collected and subjected to IFN-γ detection using an ELISA kit as per the manufacturer's protocol.Detection of free thiols on the ID8-Defb29/Vegf-a-Luc cell surface
ID8-Defb29/Vegf-a-Luc cells were harvested and resuspended in PBS, and then mixed with 0.1 μg mL−1 Oregon Green 488 (OG488) or OG488 Maleimide in PBS (both from ThermoFisher Scientific) in a 96-well V-shape plate (Fisher Scientific) (1 × 106 cells in 100 μL per well). The plate was incubated at room temperature for 15 min. The cells were washed three times using PBS to remove excess dye, resuspended in 200 μL of FACS buffer (2% (v/v) FBS and 0.09% (w/v) NaN3 in PBS), and immediately analyzed by flow cytometry. All cells were analyzed using a BD Accuri C6 Plus Flow Cytometer (BD Biosciences). 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded for each sample, and all events were analyzed using FlowJo 10.7 software.
000 events were recorded for each sample, and all events were analyzed using FlowJo 10.7 software.
Interactions between cancer cells and CPMV vaccine candidates
ID8-Defb29/Vegf-a-Luc cells (4 × 106 cells per mL) were resuspended in FBS-free culture media and irradiated with 70 Gy to prepare ICCs. Then, 37.2 μg mL−1 (or 4 × 1012 particles per mL) CPMV-Cy5, CPMV-SM(PEG)4-Cy5, and CPMV-PEG5000-Cy5 were prepared in FBS-free media.Flow cytometry
100 μL of the ID8-Defb29/Vegf-a-Luc cells or ICCs were aliquoted into each well (400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) of a 96-well V-shape plate, and then mixed with 100 μL of each CPMV formulation (1 × 106 CPMV per cell) and incubated at 4 °C or 37 °C for 30 min. The samples were analyzed in duplicate, and experiments were repeated 3 times. After incubation, the cells were washed thrice using PBS (and centrifuged at 500 × g for 5 min at 4 °C) to remove excess CPMV. Samples were divided and one half was resuspended in 200 μL of FACS buffer and the other half was first treated with 1 mg mL−1 pronase (Sigma-Aldrich) in PBS for 15 min at room temperature. Pronase was washed away with PBS, and the cells were resuspended into 200 μL of FACS buffer and analyzed immediately. All cells were analyzed using a BD Accuri C6 Plus flow cytometer (BD Biosciences). 50
000 cells per well) of a 96-well V-shape plate, and then mixed with 100 μL of each CPMV formulation (1 × 106 CPMV per cell) and incubated at 4 °C or 37 °C for 30 min. The samples were analyzed in duplicate, and experiments were repeated 3 times. After incubation, the cells were washed thrice using PBS (and centrifuged at 500 × g for 5 min at 4 °C) to remove excess CPMV. Samples were divided and one half was resuspended in 200 μL of FACS buffer and the other half was first treated with 1 mg mL−1 pronase (Sigma-Aldrich) in PBS for 15 min at room temperature. Pronase was washed away with PBS, and the cells were resuspended into 200 μL of FACS buffer and analyzed immediately. All cells were analyzed using a BD Accuri C6 Plus flow cytometer (BD Biosciences). 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded for each sample, and all events were analyzed using FlowJo 10.7 software.
000 events were recorded for each sample, and all events were analyzed using FlowJo 10.7 software.
Confocal microscopy
The cells not analyzed by flow cytometry were used to prepare slides for confocal microscopy. Briefly, the cells were fixed with 4% (v/v) paraformaldehyde (Electron Microscopy Sciences) in PBS for 10 min at room temperature and washed 3 times with PBS followed by centrifugation at 500 × g for 5 min at 4 °C and resuspension in 100 μL of PBS. The cells were spun onto Superfrost Plus Microscope slides (Fisher Scientific) using a Statspin Cytofuge Cytocentrifuge (BECKMAN COULTER) at 27 × g for 4 min. Cell membranes were first stained using Alexa Fluor 555-conjugated wheat germ agglutinin (WGA555, ThermoFisher Scientific) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution with 5% (v/v) goat serum albumin (GSA) in PBS) for 45 min at room temperature and washed 3 times using PBS. The cell nucleus was then stained with DAPI (ThermoFisher Scientific) (1
1000 dilution with 5% (v/v) goat serum albumin (GSA) in PBS) for 45 min at room temperature and washed 3 times using PBS. The cell nucleus was then stained with DAPI (ThermoFisher Scientific) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7500 dilution in PBS) for 5 min at room temperature and washed 3 times using PBS. After the cells were air dried, 4 μL of Fluoroshield (Millipore Sigma) was applied to a circular cover glass with a diameter of 12 mm (Fisherbrand), which was then mounted onto each slide. Fluorescence images were obtained using a Nikon A1R Confocal microscope with an Apo TIRF 100×/1.49 oil objective (Nikon). Collected images were analyzed using NIS-Elements AR Analysis 5.30 software (Nikon).
7500 dilution in PBS) for 5 min at room temperature and washed 3 times using PBS. After the cells were air dried, 4 μL of Fluoroshield (Millipore Sigma) was applied to a circular cover glass with a diameter of 12 mm (Fisherbrand), which was then mounted onto each slide. Fluorescence images were obtained using a Nikon A1R Confocal microscope with an Apo TIRF 100×/1.49 oil objective (Nikon). Collected images were analyzed using NIS-Elements AR Analysis 5.30 software (Nikon).
Results and discussion
Production and characterization of CPMV particles
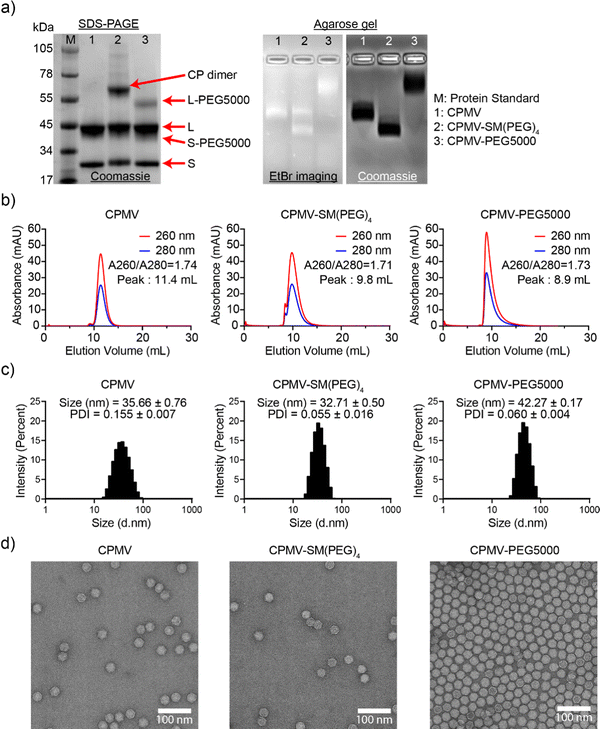
CPMV was propagated and purified from black eyed pea no. 5 plants as previously reported41 with a yield of 1 mg of CPMV per gram of infected leaves. CPMV is ∼30 nm in diameter with a pseudo-T = 3 symmetry.43 Each CPMV particle is comprised of 60 copies each of a small (S, 24 kDa) and large (L, 42 kDa) coat protein (CP); it has a bipartite RNA genome with RNA-1 and RNA-2 packaged in separate yet identical particles.43 CPMV presents with 300 solvent-exposed lysine residues, which can be modified using N-hydroxysuccinimide (NHS) ester bioconjugation.29,44 Here we modified CPMV with dyes, PEGs, and SM(PEG)4 linker molecules. The dye (sulfo-cyanine 5, sulfo-Cy5) was used for imaging, the PEG shields the CPMV from cell interactions, and the SM(PEG)4 linker served to conjugate CPMV to the ICCs. Specifically, NHS-Sulfo-Cy5, SM(PEG)4, and NHS-mPEG5000 were conjugated yielding CPMV-SM(PEG)4 and CPMV-PEG5000 for efficacy studies and CPMV-Cy5, CPMV-SM(PEG)4-Cy5, and CPMV-PEG5000-Cy5 for mechanistic investigation (Fig. 1).CPMV nanoparticles were purified through dialysis and then biochemically characterized (Fig. 2 and Fig. S1, ESI†). First, the concentration of CPMV and the degree of Cy5 labeling were determined using UV-vis spectroscopy and the CPMV extinction coefficient at 260 nm was found to be 8.1 mL mg−1 cm−1 and the Cy5-specific extinction coefficient was found to be 271![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L mol−1 cm−1 at 647 nm, respectively. CPMV-Cy5, CPMV-SM(PEG)4-Cy5, and CPMV-PEG5000-Cy5 displayed 48, 33, and 37 Cy5 per particle, respectively (Fig. S1b, ESI†). To confirm the covalent conjugation of Cy5, PEG5000, and the SM(PEG)4 linker, all samples were analyzed using denaturing SDS-PAGE and native agarose gel electrophoresis (Fig. 2(a) and Fig. S1a, ESI†). SDS-PAGE revealed the presence of S and L CP as well as modified CPs: CPMV-SM(PEG)4 showed a distinct CP dimer band (∼66 kDa); we often observe this for CPMV-SM(PEG)4 – while we cannot rule out crosslinking, it is likely that CPs are associated non-covalently, because the introduced maleimide remains available for conjugation. Conjugation of PEG5000 was apparent as higher molecular weight bands were detected for the L and S protein at < 55 kDa and < 45 kDa; the bands were more clearly detectable for L-PEG5000, because S-PEG5000 overlaid with the native L CP. Gel densitometry analysis using ImageJ showed that ∼28 PEG5000 molecules were conjugated to each CPMV. Native agarose gels also indicated the conjugation of Cy5, SM(PEG)4, and PEG5000; small molecule conjugation to CPMV results in increased mobility toward the anode due to the reduction of positive lysine side chains, while larger molecules, such as PEG, reduced mobility due to molecular weight increases. The banding pattern is consistent with SM(PEG)4 and PEG5000 conjugation (Fig. 2(a)). The RNA (EtBr imaging), Cy5, and protein bands colocalized under native gel electrophoresis and therefore indicate that conjugated CPMV remained intact. Fluorescence imaging of the agarose gel and SDS-PAGE also revealed the co-localization of Cy5 with CPs indicating covalent attachment (Fig. S1a and b, ESI†).
000 L mol−1 cm−1 at 647 nm, respectively. CPMV-Cy5, CPMV-SM(PEG)4-Cy5, and CPMV-PEG5000-Cy5 displayed 48, 33, and 37 Cy5 per particle, respectively (Fig. S1b, ESI†). To confirm the covalent conjugation of Cy5, PEG5000, and the SM(PEG)4 linker, all samples were analyzed using denaturing SDS-PAGE and native agarose gel electrophoresis (Fig. 2(a) and Fig. S1a, ESI†). SDS-PAGE revealed the presence of S and L CP as well as modified CPs: CPMV-SM(PEG)4 showed a distinct CP dimer band (∼66 kDa); we often observe this for CPMV-SM(PEG)4 – while we cannot rule out crosslinking, it is likely that CPs are associated non-covalently, because the introduced maleimide remains available for conjugation. Conjugation of PEG5000 was apparent as higher molecular weight bands were detected for the L and S protein at < 55 kDa and < 45 kDa; the bands were more clearly detectable for L-PEG5000, because S-PEG5000 overlaid with the native L CP. Gel densitometry analysis using ImageJ showed that ∼28 PEG5000 molecules were conjugated to each CPMV. Native agarose gels also indicated the conjugation of Cy5, SM(PEG)4, and PEG5000; small molecule conjugation to CPMV results in increased mobility toward the anode due to the reduction of positive lysine side chains, while larger molecules, such as PEG, reduced mobility due to molecular weight increases. The banding pattern is consistent with SM(PEG)4 and PEG5000 conjugation (Fig. 2(a)). The RNA (EtBr imaging), Cy5, and protein bands colocalized under native gel electrophoresis and therefore indicate that conjugated CPMV remained intact. Fluorescence imaging of the agarose gel and SDS-PAGE also revealed the co-localization of Cy5 with CPs indicating covalent attachment (Fig. S1a and b, ESI†).
SEC, DLS, and TEM were in agreement indicating the production of intact and monodisperse particles. The SEC elution profiles of CPMV and CPMV-Cy5 were comparable and indicated the co-elution of RNA (260 nm), protein (280 nm), and Cy5 (647 nm, for Cy5-conjugated particles). CPMV and CPMV-Cy5 eluted at ∼10 mL while CPMV-PEG5000 and CPMV-PEG5000-Cy5 eluted at ∼9 mL indicating the increased hydrodynamic radius post conjugation – this is in agreement with the native agarose gel electrophoresis (Fig. 2(b) and Fig. S1c, ESI†). All samples showed the characteristic absorbance ratio of ∼1.8 between 260 and 280 nm at the elution peak, a sign of intact CPMV with packaged RNA.45 DLS corroborated this result (Fig. 2(c) and Fig. S1c, ESI†). DLS data displayed a single peak with a homogeneous distribution for all samples; broken particles or aggregated particles were not apparent in any of the preparations. CPMV, CPMV-SM(PEG)4, CPMV-Cy5, and CPMV-SM(PEG)4-Cy5 measured 31–35 nm in diameter (Fig. 2(c) and Fig. S1d, ESI†). CPMV-PEG5000 and CPMV-PEG5000-Cy5 exhibited an increased hydrodynamic diameter of ∼42 nm indicating the display of PEG5000. Lastly, TEM showed monodisperse and intact conjugated CPMV nanoparticles with a morphology similar to that of native CPMV (Fig. 2(d) and Fig. S1e, ESI†).
ICCs + CPMV vaccine formulations: co-delivery vs. mixture
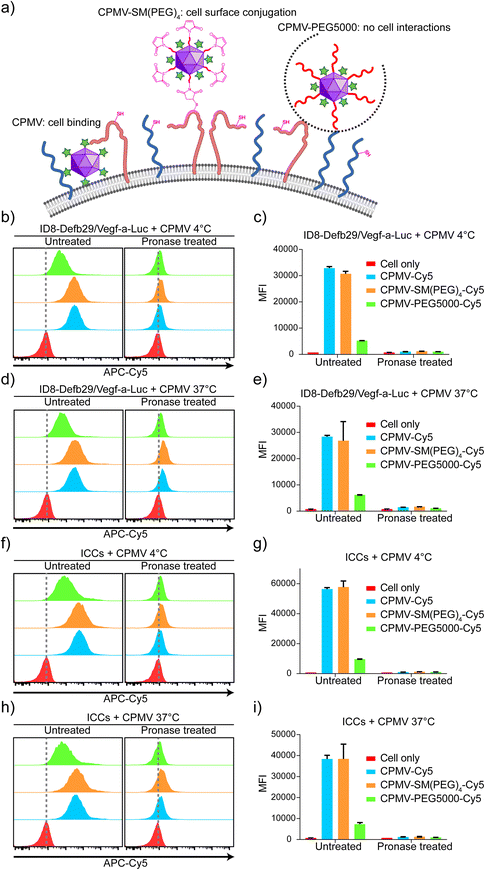
We prepared three types of ICCs + CPMV vaccine formulations: mixtures of ICCs and native CPMV, ICCs covalently conjugated with CPMV-SM(PEG)4, and ICCs and CPMV-PEG5000; for the latter PEGylation was applied to reduce any natural CPMV–ICC interactions. The CPMV-SM(PEG)4 formulation is conjugated to the ICC surface through exposed thiols from cell surface membrane proteins;46,47 this formulation affords the co-delivery of the antigen and the adjuvant. PEGylation is known to reduce interactions between cells and nanoparticles/biologics from cell interactions;48–50 therefore, we expect this formulation to keep adjuvants and antigens separate and non-interacting. We hypothesized that in the mixture of ICCs and native CPMV, CPMV is bound to and/or possibly taken up by the cells – therefore behaving more like ICCs conjugated with CPMV-SM(PEG)4, and that adjuvant–antigen (CPMV–ICCs) co-delivery through cell binding/uptake would result in increased efficacy over the simple mixture using CPMV-PEG5000 (Fig. 3(a)).First, we confirmed that ID8-Defb29/Vegf-a-Luc cells possess reduced free thiols for maleimide conjugation with CPMV-SM(PEG)4 using flow cytometry. We first incubated ID8-Defb29/Vegf-a-Luc cells with maleimide-functionalized or non-functionalized OG488 dyes. Fluorescence signals were only observed when cells were probed with maleimide-OG488 indicating that free thiols are available for conjugation; non-specific cell interactions with OG488 were not observed (Fig. S2a and b, ESI†).
Next, to examine the proposed interactions between cancer cells and CPMV-Cy5, CPMV-SP(PEG)4-Cy5, or CPMV-PEG5000-Cy5, we incubated these formulations with live ID8-Defb29/Vegf-a-Luc cells or ICCs. To differentiate CPMV binding vs. internalization, we performed studies at 4 °C or 37 °C. ICCs were also prepared by irradiating cancer cells with 70 Gy using a Cs-137 source to confirm that irradiation does not affect the interactions between CPMV and cancer cells for vaccine formulation. Post CPMV incubation, all cells were washed 3 times vigorously to remove unbonded CPMV and then were analyzed by flow cytometry and confocal microscopy. In flow cytometry (Fig. 3(b)–(i)), ID8-Defb29/Vegf-a-Luc cells and ICCs showed strong Cy5 signals when incubated with CPMV-Cy5 and CPMV-SM(PEG)4-Cy5 compared to cells only; whereas the cells incubated with CPMV-PEG5000-Cy5 showed a detectable but negligible Cy5 signal, suggesting that the PEGylation of CPMV can significantly reduce the interaction between CPMV and the cells but not completely abolish the interaction. Since both CPMV-Cy5 and CPMV-SM(PEG)4-Cy5 showed similar interaction patterns with cells, we could not rule out that the conjugation of SM(PEG)4 has a negligible effect on the CPMV and cell interaction because native CPMV already effectively interacts with the cells. The median fluorescence intensity (MFI) of Cy5 for each cell group was compared, showing that CPMV-Cy5 and CPMV-SM(PEG)4-Cy5 strongly interacted with cells (with a MFI of ∼27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–58
000–58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for CPMV-Cy5 and CPMV-SM(PEG)4-Cy5 under all conditions) whereas CPMV-PEG5000-Cy5 hardly interacted with cells (∼5000–9000 under all conditions). To investigate whether CPMV attached to the cell surface or being internalized, incubation at 4 vs. 37 °C was compared. The cells were also treated with pronase to remove all surface proteins and surface-bound CPMV.51 After pronase treatment, Cy5 signals were lost with all MFI dropping back to baseline (MFI below ∼1600; MFI of ∼800 for cells only) (Fig. 3(c), (e), (g), and (i)). Data indicate that cell uptake was not apparent when analyzing cells incubated at 37 °C (Fig. 3(e) and (i)). Together data indicate that during the 30 min incubation period, the CPMV adjuvant binds to the cell surface but is not internalized. In contrast, CPMV-PEG5000 barely interacts with the cells due to shielding by PEG (Fig. 3(b)–(i)). These data also indicate that ICCs and live ID8-Defb29/Vegf-a-Luc cells have similar nanoparticle interactions when comparing various adjuvant formulations.
000 for CPMV-Cy5 and CPMV-SM(PEG)4-Cy5 under all conditions) whereas CPMV-PEG5000-Cy5 hardly interacted with cells (∼5000–9000 under all conditions). To investigate whether CPMV attached to the cell surface or being internalized, incubation at 4 vs. 37 °C was compared. The cells were also treated with pronase to remove all surface proteins and surface-bound CPMV.51 After pronase treatment, Cy5 signals were lost with all MFI dropping back to baseline (MFI below ∼1600; MFI of ∼800 for cells only) (Fig. 3(c), (e), (g), and (i)). Data indicate that cell uptake was not apparent when analyzing cells incubated at 37 °C (Fig. 3(e) and (i)). Together data indicate that during the 30 min incubation period, the CPMV adjuvant binds to the cell surface but is not internalized. In contrast, CPMV-PEG5000 barely interacts with the cells due to shielding by PEG (Fig. 3(b)–(i)). These data also indicate that ICCs and live ID8-Defb29/Vegf-a-Luc cells have similar nanoparticle interactions when comparing various adjuvant formulations.
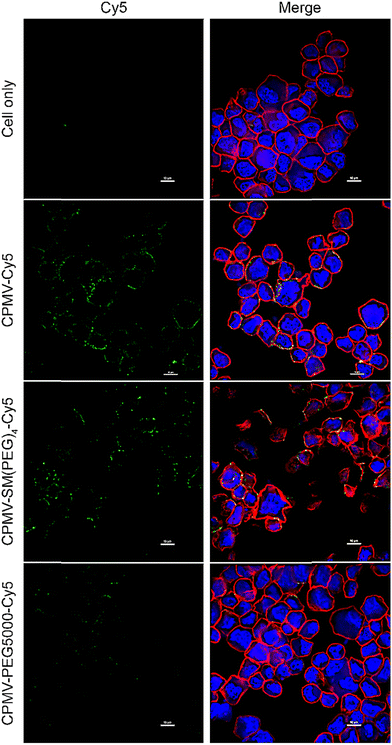
These results were further validated using confocal microscopy (Fig. 4 and Fig. S3, ESI†). Here we imaged the interaction of the vaccine formulations with live ID8-Defb29/Vegf-a-Luc cells and incubation at 4 °C. Live cells were chosen for ease of handling – we assumed this was sufficient as there were no differences comparing live vs. ICCs in flow cytometry. Additionally, we also did a pilot confocal study using ICCs and data indicated a similar behavior independent of irradiation (data not shown). The confocal images demonstrate that CPMV-Cy5 and CPMV-SM(PEG)4-Cy5 co-localize with the cell membrane (stained with WGA-AlexaFluor555), while the CPMV-PEG5000-Cy5 formulation was shielded from interacting with the cells. These results were consistent with our previous published data analyzing CPMV and CPMV-PEG formulations.33,49,50
Co-delivered vaccines outperform simple mixtures in an ovarian tumor challenge
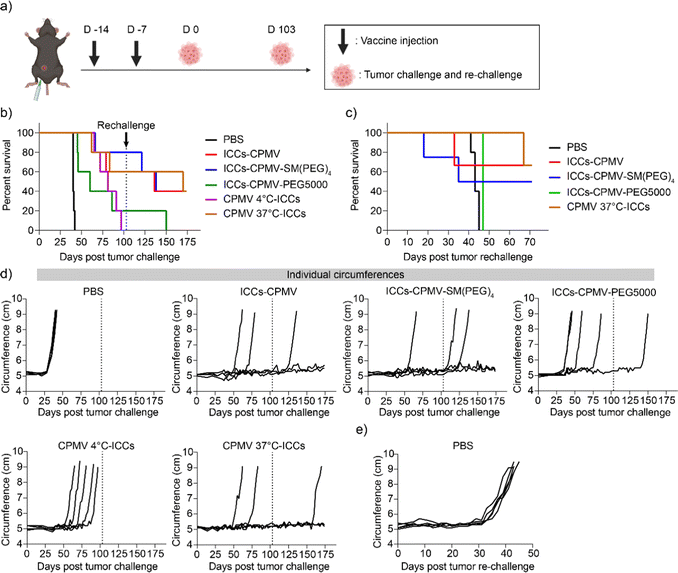
ICCs were prepared as mentioned above and then mixed with CPMV, CPMV-SM(PEG)4, and CPMV-PEG5000, and termed as ICCs–CPMV, ICCs–CPMV-SM(PEG)4, and ICCs–CPMV-PEG5000, respectively; vaccines were kept on ice for 30 min to enable CPMV and CPMV-SM(PEG)4 interactions with ICCs and hence co-formulation. Two additional vaccines were prepared by mixing live ID8-Defb29/Vegf-a-Luc cells with CPMV, and then incubating at 4 °C or 37 °C for 30 min followed by irradiation; the formulations are termed CPMV 4 °C-ICCs and CPMV 37 °C-ICCs. With the latter formulations we set out to test whether CPMV should be added pre-or post-irradiation.To compare efficacy, we vaccinated C57BL/6 mice (n = 5) using i.p. injections and a prime and boost regimen with injections spaced one week apart (Fig. 5(a)). Each vaccine formulation contained 5 × 106 ICCs and 100 μg of CPMV in 200 μL of PBS; PBS served as a control. 7 days after the second vaccine, all mice were challenged with 5 × 106 ID8-Defb29/Vegf-a-Luc cells in 200 μL of PBS through i.p. injection. Tumor progression was monitored by the increase in the body weight (Fig. S4, ESI†) and circumference (Fig. 5(c)) resulting from the tumor burden and development of ascites. The mice were euthanized when body weight reached 35 g or circumference reached 9 cm, and the survival rate was recorded (Fig. 5(b) and (c)). Mice that survived 100 days post tumor challenge were re-challenged using 5 × 106 ID8-Defb29/Vegf-a-Luc cells again while 5 additional naïve, age-matched mice served as a control (Fig. 5(c) and (e)). All untreated mice (PBS control) showed severe tumor burden with steep tumor progression 5 weeks post tumor challenge and all animals were sacrificed within 45 days (Fig. 5(b) and (c)). While the tumor onset was delayed in mice that were vaccinated using a mixture of ICCs + CPMV-PEG5000 (Fig. 5(d)), the mixture formulation failed to suppress tumor growth with 4 out of 5 mice reaching the endpoint within 100 days. The surviving mouse succumbed to the re-challenge by day 150 (Fig. 5(b)).
When comparing the CPMV 4 °C-ICC and CPMV 37 °C-ICC groups it was somewhat surprising that while flow cytometry did not indicate differential and energy-dependent binding/uptake of CPMV, differences in anti-tumor efficacy were observed: in the CPMV 4 °C-ICC group all mice (n = 5) reached endpoint within 100 days. In stark contrast, potent efficacy was observed for the CPMV 37 °C-ICCs, ICCs–CPMV, and ICCs–CPMV-SM(PEG)4 groups with 10 out of 15 mice surviving the initial tumor challenge, and 6 out of 10 mice surviving the tumor re-challenge (Fig. 5(b) and (c)). Upon completion of the study (70 days post tumor re-challenge), 2 mice from the ICCs–CPMV group, 2 mice from the ICCs–CPMV-SM(PEG)4 group, and 2 mice from the CPMV 37 °C-ICC group remained tumor-free and healthy. The median survival is as follows: 170, 136, 138, 81, 60, and 40 days for CPMV 37 °C-ICCs, ICCs–CPMV, ICCs–CPMV-SM(PEG)4, CPMV 4 °C-ICCs, ICCs–CPMV-PEG5000, and PBS groups. A summary of the median survival per vaccine group is shown in Table 1.
| Vaccine group | Formulation | Media survival (days) |
|---|---|---|
| a It is hypothesized that the CPMV–cell interactions induced at 4 °C are not sufficiently strong to maintain bonding during irradiation and subsequent sample processing. | ||
| ICCs–CPMV | Co-delivery by natural CPMV–cell interactions | 136 |
| ICCs–CPMV-SM(PEG)4 | Co-delivery by conjugation | 138 |
| ICCs–CPMV-PEG5000 | Mixture; no evidence of association between CPMV and ICCs | 81 |
| CPMV 4 °C-ICCs | Co-delivery by natural CPMV–cell interactionsa | 60 |
| CPMV 37 °C-ICCs | Co-delivery by natural CPMV–cell interactions | 170 |
Taking together these data show that cellular interactions and therefore antigen/adjuvant co-delivery are important for vaccine efficacy. The most potent formulation was CPMV 37 °C-ICCs – here CPMV was mixed with live cancer cells prior to irradiation, which may help to crosslink the particles to the cells, thereby conferring enhanced in vivo stability. The “CPMV 4 °C-ICC” vaccine group was treated like a co-formulation group – based on flow cytometry analysis (see Fig. 3); however, efficacy data indicated that this formulation was not effective to protect mice against the tumor challenge. We speculate that although incubating CPMV and ID8-Defb29/Vegf-a-Luc cells at 4 °C leads to CPMV–cell interactions, this bonding may be labile (uptake is energy-dependent and not observed at 4 °C). It is possible that the weak CPMV–cell interactions induced at 4 °C may disrupt during irradiation and subsequent processing steps. Therefore, incubating CPMV and tumor cells at 37 °C prior to irradiation may be necessary to achieve the most effective vaccine formulation. Future studies should elaborate the formulation steps and chemistry further – data from this study clearly indicate that co-formulation is needed to stimulate potent anti-tumor responses using the ICC vaccine candidate and CPMV adjuvant.
ICCs–CPMV vaccination induces adaptive CTL anti-tumor immune responses
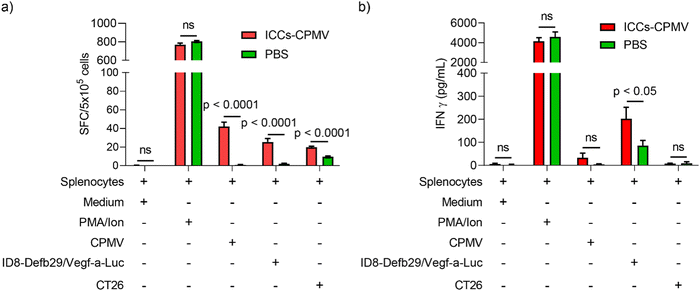
In our efficacy study, we observed that the mice that received co-delivered CPMV–ICC vaccine formulations survived tumor challenge and re-challenge, indicating the establishment of adaptive anti-tumor immunity. To confirm this hypothesis, we analyzed the CTL response against ID8-Defb29/Vefg-a-Luc cancer cells post vaccination with the co-delivered vaccine. Here we chose ICCs–CPMV vaccine for our investigation because all CPMV 37 °C-ICCs, ICCs–CPMV, and ICCs–CPMV-SM(PEG)4 groups showed similar survival post re-challenge. Splenocytes from vaccinated and non-vaccinated animals were harvested 10 days after the second dose of ICCs–CPMV and pulsed using ID8-Defb29/Vegf-a-Luc cells, CT26 cells (non-target colon cancer cells), CPMV, and positive control phorbol 12-myristate 13-acetate and ionomycin (PMV/Ion). CTL stimulation was quantified by measuring Interferon (IFN)-γ using ELISpot (Fig. 6(a)) and ELISA assays (Fig. 6(b)). INF-γ is a cytokine mainly secreted by cytotoxic T cells and T helper cells to mediate a pro-inflammatory, anti-tumor Th1 type immune response.30,52 Therefore, INF-γ secretion is used to measure CTL activity against the target antigen, e.g., ID8-Defb29/Vegf-a-Luc cells.ELISpot confirmed that the ICCs–CPMV vaccine formulation elicited tumor specific CTL responses against the target ID8-Defb29/Vegf-a-Luc cells. There was an over 12-fold increase in IFN-γ spot-forming colonies (SFCs) against ID8-Defb29/Vegf-a-Luc cells compared to unvaccinated mice (p < 0.0001) (Fig. 6(a)). We observed a similar result from the ELISA with more IFN-γ released from splenocytes after incubation with ID8-Defb29/Vegf-a-Luc cells (p < 0.05) (Fig. 6(b)). Unsurprisingly, splenocytes from the vaccinated mice showed significant SFCs when pulsed with CPMV as we previously observed after CPMV vaccination; CPMV is immunogenic and cross presentation leads to humoral and cellular immunity against the plant virus adjuvant.53
While data confirm that the ICCs–CPMV vaccine formulation elicits anti-tumor immunity with CTL responses against the target tumor cells, ID8-Defb29/Vegf-a-Luc, we also noted an ∼2-fold increase in IFN-γ SFCs following incubation with non-target CT26 cells – this was only observed using ELISpot and not ELISA (Fig. 6(a) and (b)). We suspect that the SFCs resulting from CT26 were mostly due to the background from the experiment itself, because even splenocytes from naïve mice showed obvious IFN-γ SFCs upon CT26 cell simulation.
Conclusion
In this work, we formulated 5 distinct ovarian cancer vaccines (Table 1) (ICCs–CPMV, ICCs–CPMV-SM(PEG)4, ICCs–CPMV-PEG5000, CPMV 4 °C-ICCs, and CPMV 37 °C-ICCs) using ICCs and CPMV and demonstrated that the co-delivery of the ICCs + CPMV, where ICCs and CPMV are bonded, taken up, or coupled, significantly enhances protection against the ovarian cancer challenge. In vitro incubation of live ID8-Defb29/Vegf-a-Luc cells or ICCs with CPMV and CPMV-SM(PEG)4 confirmed CPMV–cell interactions while PEGylation (CPMV-PEG5000) blocked cellular interactions. Vaccination using formulations in which the antigen (ICCs) and the adjuvant (CPMV) were co-delivered outperformed simple mixtures and protected mice from the tumor challenge; protection was durable, and mice showed long-term protection also in a re-challenge study. ELISpot and ELISA assays show that vaccination with co-delivered CPMV and ICCs induces adaptive anti-tumor immunity. Overall, our data demonstrate the potent efficacy of the vaccines in rejecting ovarian cancer and highlight the importance of co-delivery of cancer antigens and immunostimulatory adjuvants for cancer vaccine development.Conflicts of interest
Dr Steinmetz is a co-founder of, has equity in, and has a financial interest with Mosaic ImmunoEngineering Inc. Dr Steinmetz serves as Director, Board Member, and Acting Chief Scientific Officer, and paid consultant to Mosaic. The other authors declare no potential conflicts of interest.Acknowledgements
This work was supported in part by the NIH (R01-CA253615) and the Shaughnessy Family Fund for Nano-ImmunoEngineering (nanoIE) at the UCSD.References
- PDQATE Board, Ovarian Epithelial, Fallopian Tube, and Primary Peritoneal Cancer Treatment (PDQ®). In PDQ Cancer Information Summaries [Internet], National Cancer Institute (US), 2021.
- T. Arora, S. Mullangi and M. R. Lekkala, Ovarian cancer, StatPearls, 2022 Search PubMed.
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, Cancer statistics, 2022, Ca-Cancer J. Clin., 2022, 72(1), 7–33 CrossRef PubMed.
- R. L. Siegel, K. D. Miller and A. Jemal, Cancer statistics, 2020, Ca-Cancer J. Clin., 2020, 70(1), 7–30 CrossRef PubMed.
- C. Della Pepa, G. Tonini, C. Pisano, M. Di Napoli, S. C. Cecere, R. Tambaro, G. Facchini and S. Pignata, Ovarian cancer standard of care: are there real alternatives?, Chin. J. Cancer, 2015, 34(1), 17–27 CrossRef CAS PubMed.
- A. J. Korman, S. C. Garrett-Thomson and N. Lonberg, The foundations of immune checkpoint blockade and the ipilimumab approval decennial, Nat. Rev. Drug Discovery, 2021, 1–20 Search PubMed.
- N. Lonberg and A. Korman, Masterful antibodies: Checkpoint blockade, Cancer Immunol. Res., 2017, 5(4), 275–281, DOI:10.1158/2326-6066 ; CIR-17-0057.[Abstract][CrossRef][Google Scholar].
- D. R. Littman, Releasing the brakes on cancer immunotherapy, Cell, 2015, 162(6), 1186–1190 CrossRef CAS PubMed.
- J. Hamanishi, M. Mandai, T. Ikeda, M. Minami, A. Kawaguchi, T. Murayama, M. Kanai, Y. Mori, S. Matsumoto and S. Chikuma, Safety and antitumor activity of anti–PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer, J. Clin. Oncol., 2015, 33(34), 4015–4022 CrossRef CAS PubMed.
- A. Leary, D. Tan and J. Ledermann, Immune checkpoint inhibitors in ovarian cancer: where do we stand?, Ther. Adv. Med. Oncol., 2021, 13 DOI:10.1177/17588359211039899.
- T. Worzfeld, E. Pogge von Strandmann, M. Huber, T. Adhikary, U. Wagner, S. Reinartz and R. Müller, The unique molecular and cellular microenvironment of ovarian cancer, Front. Oncol., 2017, 7, 24 Search PubMed.
- P. P. Bapsy, B. Sharan, C. Kumar, R. P. Das, B. Rangarajan, M. Jain, V. S. S. Attili, S. Subramanian, S. Aggarwal and M. Srivastava, Open-label, multi-center, non-randomized, single-arm study to evaluate the safety and efficacy of dendritic cell immunotherapy in patients with refractory solid malignancies, on supportive care, Cytotherapy, 2014, 16(2), 234–244 CrossRef CAS PubMed.
- C. L.-L. Chiang, L. E. Kandalaft, J. Tanyi, A. R. Hagemann, G. T. Motz, N. Svoronos, K. Montone, G. M. Mantia-Smaldone, L. Smith and H. L. Nisenbaum, A Dendritic Cell Vaccine Pulsed with Autologous Hypochlorous Acid-Oxidized Ovarian Cancer Lysate Primes Effective Broad Antitumor Immunity: From Bench to BedsideHOCl-Oxidized Lysate-Pulsed DC Elicits Potent TH1 Responses, Clin. Cancer Res., 2013, 19(17), 4801–4815 CrossRef CAS PubMed.
- K. Odunsi, J. Matsuzaki, S. R. James, P. Mhawech-Fauceglia, T. Tsuji, A. Miller, W. Zhang, S. N. Akers, E. A. Griffiths and A. Miliotto, Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer, Cancer Immunol. Res., 2014, 2(1), 37–49 CrossRef CAS PubMed.
- O. E. Rahma, E. Ashtar, M. Czystowska, M. E. Szajnik, E. Wieckowski, S. Bernstein, V. E. Herrin, M. A. Shams, S. M. Steinberg and M. Merino, A gynecologic oncology group phase II trial of two p53 peptide vaccine approaches: subcutaneous injection and intravenous pulsed dendritic cells in high recurrence risk ovarian cancer patients, Cancer Immunol. Immunother., 2012, 61(3), 373–384 CrossRef CAS PubMed.
- J. L. Gulley, P. M. Arlen, K.-Y. Tsang, J. Yokokawa, C. Palena, D. J. Poole, C. Remondo, V. Cereda, J. L. Jones and M. P. Pazdur, A pilot study to evaluate the safety and clinical outcomes of vaccination with recombinant CEA-MUC-1-TRICOM (PANVAC) poxviral-based vaccines in patients with metastatic carcinoma. Clinical cancer research: an official journal of the American Association for, Cancer Res., 2008, 14(10), 3060 CAS.
- E. Jäger, J. Karbach, S. Gnjatic, A. Neumann, A. Bender, D. Valmori, M. Ayyoub, E. Ritter, G. Ritter and D. Jäger, Recombinant vaccinia/fowlpox NY-ESO-1 vaccines induce both humoral and cellular NY-ESO-1-specific immune responses in cancer patients, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(39), 14453–14458 CrossRef PubMed.
- S. A. Rosenberg, J. C. Yang and N. P. Restifo, Cancer immunotherapy: moving beyond current vaccines, Nat. Med., 2004, 10(9), 909–915 CrossRef CAS PubMed.
- M. Obeid, T. Panaretakis, N. Joza, R. Tufi, A. Tesniere, P. Van Endert, L. Zitvogel and G. Kroemer, Calreticulin exposure is required for the immunogenicity of γ-irradiation and UVC light-induced apoptosis, Cell Death Differ., 2007, 14(10), 1848–1850 CrossRef CAS PubMed.
- L. Vandenberk, A. D. Garg, T. Verschuere, C. Koks, J. Belmans, M. Beullens, P. Agostinis, S. De Vleeschouwer and S. W. Van Gool, Irradiation of necrotic cancer cells, employed for pulsing dendritic cells (DCs), potentiates DC vaccine-induced antitumor immunity against high-grade glioma, Oncoimmunology, 2016, 5(2), e1083669 CrossRef PubMed.
- D. E. Speiser, D. Liénard, N. Rufer, V. Rubio-Godoy, D. Rimoldi, F. Lejeune, A. M. Krieg, J.-C. Cerottini and P. Romero, Rapid and strong human CD8+ T cell responses to vaccination with peptide, IFA, and CpG oligodeoxynucleotide 7909, J. Clin. Invest., 2005, 115(3), 739–746 CrossRef CAS PubMed.
- H. Okada, L. H. Butterfield, R. L. Hamilton, A. Hoji, M. Sakaki, B. J. Ahn, G. Kohanbash, J. Drappatz, J. Engh and N. Amankulor, Induction of robust type-I CD8+ T-cell responses in WHO grade 2 low-grade glioma patients receiving peptide-based vaccines in combination with poly-ICLC, Clin. Cancer Res., 2015, 21(2), 286–294 CrossRef CAS PubMed.
- C. Mao, V. Beiss, J. Fields, N. F. Steinmetz and S. Fiering, Cowpea mosaic virus stimulates antitumor immunity through recognition by multiple MYD88-dependent toll-like receptors, Biomaterials, 2021, 275, 120914 CrossRef CAS PubMed.
- S. Shukla, J. T. Myers, S. E. Woods, X. Gong, A. E. Czapar, U. Commandeur, A. Y. Huang, A. D. Levine and N. F. Steinmetz, Plant viral nanoparticles-based HER2 vaccine: Immune response influenced by differential transport, localization and cellular interactions of particulate carriers, Biomaterials, 2017, 121, 15–27 CrossRef CAS PubMed.
- M. O. Mohsen, G. Augusto and M. F. Bachmann, The 3Ds in virus-like particle based-vaccines: Design, Delivery and Dynamics, Immunol. Rev., 2020, 296(1), 155–168 CrossRef CAS PubMed.
- O. A. Ortega-Rivera, M. D. Shin, A. Chen, V. Beiss, M. A. Moreno-Gonzalez, M. A. Lopez-Ramirez, M. Reynoso, H. Wang, B. L. Hurst and J. Wang, Trivalent subunit vaccine candidates for COVID-19 and their delivery devices, J. Am. Chem. Soc., 2021, 143(36), 14748–14765 CrossRef CAS PubMed.
- H. Cai, S. Shukla, C. Wang, H. Masarapu and N. F. Steinmetz, Heterologous prime-boost enhances the antitumor immune response elicited by plant-virus-based cancer vaccine, J. Am. Chem. Soc., 2019, 141(16), 6509–6518 CrossRef CAS PubMed.
- B. K. Patel, C. Wang, B. Lorens, A. D. Levine, N. F. Steinmetz and S. Shukla, Cowpea Mosaic Virus (CPMV)-Based Cancer Testis Antigen NY-ESO-1 Vaccine Elicits an Antigen-Specific Cytotoxic T Cell Response, ACS Appl. Bio Mater., 2020, 3(7), 4179–4187 CrossRef CAS PubMed.
- S. Shukla, C. Wang, V. Beiss, H. Cai, T. Washington 2nd, A. A. Murray, X. Gong, Z. Zhao, H. Masarapu, A. Zlotnick, S. Fiering and N. F. Steinmetz, The unique potency of Cowpea mosaic virus (CPMV) in situ cancer vaccine, Biomater. Sci., 2020, 8(19), 5489–5503 RSC.
- P. H. Lizotte, A. M. Wen, M. R. Sheen, J. Fields, P. Rojanasopondist, N. F. Steinmetz and S. Fiering, In situ vaccination with cowpea mosaic virus nanoparticles suppresses metastatic cancer, Nat. Nanotechnol., 2016, 11(3), 295–303 CrossRef CAS PubMed.
- Y. H. Chung, J. Park, H. Cai and N. F. Steinmetz, S100A9-Targeted Cowpea Mosaic Virus as a Prophylactic and Therapeutic Immunotherapy against Metastatic Breast Cancer and Melanoma. Advanced, Science, 2021, 8(21), 2101796 CAS.
- P. J. Hoopes, R. J. Wagner, K. Duval, K. Kang, D. J. Gladstone, K. L. Moodie, M. Crary-Burney, H. Ariaspulido, F. A. Veliz, N. F. Steinmetz and S. N. Fiering, Treatment of Canine Oral Melanoma with Nanotechnology-Based Immunotherapy and Radiation, Mol. Pharmaceutics, 2018, 15(9), 3717–3722 CrossRef CAS PubMed.
- S. Shukla, C. Wang, V. Beiss and N. F. Steinmetz, Antibody response against cowpea mosaic viral nanoparticles improves in situ vaccine efficacy in ovarian cancer, ACS Nano, 2020, 14(3), 2994–3003 CrossRef CAS PubMed.
- C. Wang, V. Beiss and N. F. Steinmetz, Cowpea Mosaic Virus Nanoparticles and Empty Virus-Like Particles Show Distinct but Overlapping Immunostimulatory Properties, J. Virol., 2019, 93(21), e00129–19 CAS.
- C. Wang, S. N. Fiering and N. F. Steinmetz, Cowpea Mosaic Virus Promotes Anti-Tumor Activity and Immune Memory in a Mouse Ovarian Tumor Model, Adv. Ther., 2019, 2(5), 1900003 CrossRef PubMed.
- C. T. Stump, G. Ho, C. Mao, F. A. Veliz, V. Beiss, J. Fields, N. F. Steinmetz and S. Fiering, Remission-Stage Ovarian Cancer Cell Vaccine with Cowpea Mosaic Virus Adjuvant Prevents Tumor Growth, Cancers, 2021, 13(4), 627 CrossRef CAS PubMed.
- C. E. Callmann, L. E. Cole, C. D. Kusmierz, Z. Huang, D. Horiuchi and C. A. Mirkin, Tumor cell lysate-loaded immunostimulatory spherical nucleic acids as therapeutics for triple-negative breast cancer, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(30), 17543–17550 CrossRef CAS PubMed.
- S. Hamdy, O. Molavi, Z. Ma, A. Haddadi, A. Alshamsan, Z. Gobti, S. Elhasi, J. Samuel and A. Lavasanifar, Co-delivery of cancer-associated antigen and Toll-like receptor 4 ligand in PLGA nanoparticles induces potent CD8+ T cell-mediated anti-tumor immunity, Vaccine, 2008, 26(39), 5046–5057 CrossRef CAS PubMed.
- J. A. Hubbell, S. N. Thomas and M. A. Swartz, Materials engineering for immunomodulation, Nature, 2009, 462(7272), 449–460 CrossRef CAS PubMed.
- M. Pei, H. Li, Y. Zhu, J. Lu and C. Zhang, In vitro evidence of oncofetal antigen and TLR-9 agonist co-delivery by alginate nanovaccines for liver cancer immunotherapy, Biomater. Sci., 2022, 10(11), 2865–2876 RSC.
- A. M. Wen, K. L. Lee, I. Yildiz, M. A. Bruckman, S. Shukla and N. F. Steinmetz, Viral nanoparticles for in vivo tumor imaging, J. Visualized Exp., 2012, 69, e4352 Search PubMed.
- R. Patel, A. E. Czapar, S. Fiering, N. L. Oleinick and N. F. Steinmetz, Radiation Therapy Combined with Cowpea Mosaic Virus Nanoparticle in Situ Vaccination Initiates Immune-Mediated Tumor Regression, ACS Omega, 2018, 3(4), 3702–3707 CrossRef CAS PubMed.
- T. Lin, Z. Chen, R. Usha, C. V. Stauffacher, J.-B. Dai, T. Schmidt and J. E. Johnson, The refined crystal structure of cowpea mosaic virus at 2.8 Å resolution, Virology, 1999, 265(1), 20–34 CrossRef CAS PubMed.
- A. Chatterji, W. F. Ochoa, M. Paine, B. R. Ratna, J. E. Johnson and T. Lin, New addresses on an addressable virus nanoblock; uniquely reactive Lys residues on cowpea mosaic virus, Chem. Biol., 2004, 11(6), 855–863 CrossRef CAS PubMed.
- I. Yildiz, K. L. Lee, K. Chen, S. Shukla and N. F. Steinmetz, Infusion of imaging and therapeutic molecules into the plant virus-based carrier cowpea mosaic virus: cargo-loading and delivery, J. Controlled Release, 2013, 172(2), 568–578 CrossRef CAS PubMed.
- M. T. Stephan, J. J. Moon, S. H. Um, A. Bershteyn and D. J. Irvine, Therapeutic cell engineering with surface-conjugated synthetic nanoparticles, Nat. Med., 2010, 16(9), 1035–1041 CrossRef CAS PubMed.
- M. T. Stephan and D. J. Irvine, Enhancing cell therapies from the outside in: cell surface engineering using synthetic nanomaterials, Nano Today, 2011, 6(3), 309–325 CrossRef CAS PubMed.
- K. L. Lee, S. Shukla, M. Wu, N. R. Ayat, C. E. El Sanadi, A. M. Wen, J. F. Edelbrock, J. K. Pokorski, U. Commandeur and G. R. Dubyak, Stealth filaments: Polymer chain length and conformation affect the in vivo fate of PEGylated potato virus X, Acta Biomater., 2015, 19, 166–179 CrossRef CAS PubMed.
- N. F. Steinmetz and M. Manchester, PEGylated viral nanoparticles for biomedicine: the impact of PEG chain length on VNP cell interactions in vitro and ex vivo, Biomacromolecules, 2009, 10(4), 784–792 CrossRef CAS PubMed.
- N. F. Steinmetz, C. F. Cho, A. Ablack, J. D. Lewis and M. Manchester, Cowpea mosaic virus nanoparticles target surface vimentin on cancer cells, Nanomedicine, 2011, 6(2), 351–364 CrossRef CAS PubMed.
- P. Lam and N. F. Steinmetz, Delivery of siRNA therapeutics using cowpea chlorotic mottle virus-like particles, Biomater. Sci., 2019, 7(8), 3138–3142 RSC.
- M. B. Fuertes, A. K. Kacha, J. Kline, S.-R. Woo, D. M. Kranz, K. M. Murphy and T. F. Gajewski, Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells, J. Exp. Med., 2011, 208(10), 2005–2016 CrossRef CAS PubMed.
- O. A. Ortega-Rivera, S. Shukla, M. D. Shin, A. Chen, V. Beiss, M. A. Moreno-Gonzalez, Y. Zheng, A. E. Clark, A. F. Carlin and J. K. Pokorski, Cowpea Mosaic Virus Nanoparticle Vaccine Candidates Displaying Peptide Epitopes Can Neutralize the Severe Acute Respiratory Syndrome Coronavirus, ACS Infect. Dis., 2021, 7(11), 3096–3110 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb02355e |
| This journal is © The Royal Society of Chemistry 2023 |