Selective organ targeting nanoparticles: from design to clinical translation
Jian
Li
 ab and
Hai
Wang
ab and
Hai
Wang
 *ab
*ab
aCAS Key Laboratory for Biomedical Effects of Nanomaterials & Nanosafety, CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology, Beijing, 100190, China. E-mail: wanghai@nanoctr.cn
bUniversity of Chinese Academy of Sciences, Beijing, 100049, China
First published on 27th June 2023
Abstract
Targeting nanoparticle is a very promising therapeutic approach that can precisely target specific sites to treat diseases. Research on nanoscale drug delivery systems has made great progress in the past few years, making targeting nanoparticles a promising prospect. However, selective targeting nanoparticles designed for specific organs still face several challenges, one of which is the unknown fate of nanoparticles in vivo. This review starts with the in vivo journey of nanoparticles and describes the biological barriers and some targeting strategies for nanoparticles to target specific organs. Then, through the collection of literature in recent years, the design of selective targeting nanoparticles for various organs is illustrated, which provides a reference strategy for people to study the design of selective organ targeting nanoparticles. Ultimately, the prospect and challenge of selective organ targeting nanoparticles are discussed by collecting the data of clinical trials and marketed drugs.
1. Introduction
Targeting nanoparticles, which can deliver therapeutic molecules to specific sites in the body, are a very promising drug delivery system (DDS).1 Especially after the COVID-19 pandemic, the widespread application of lipid nanoparticles (LNPs) has attracted more and more people's attention to this field, and targeting nanoparticles to increase efficiency and reduce toxicity has become a hot spot sought after by capital and the market.2 However, the fate of nanoparticles in vivo is very complex, current targeting strategies lack accuracy, and off-target effects and side effects are still serious.3 To successfully deliver nanoparticles to target sites, delivery systems must overcome limitations that render therapeutic molecules ineffective, such as biological barriers and low biodistribution at desired sites.4 In order to solve the problem of limited delivery of nanoparticles, a large number of scientists around the world are paying attention to the fate of nanoparticles in the body.There are many influencing factors that determine the fate of nanoparticles in the body, including physical, chemical and biological barriers.4 There are physical and chemical barriers on the surface of various organs, mainly composed of epithelial cells and their secretions, which prevent foreign matter from entering the body and damaging health.5 After nanoparticles enter the systemic circulation, they need to overcome biological barriers such as the blood-brain barrier (BBB) to enter the target organ for targeted delivery. During this process, nanoparticles are affected by biological processes in the body such as inflammation,6 nutritional deficiency,7 immune phagocytosis,8 and liver/kidney clearance,9 which ultimately determine their distribution and fate in the body. Given that the physical and chemical properties of nanoparticles determine how they are affected by the aforementioned biological processes, understanding these processes and their mechanisms is important for the design of organ targeting nanoparticles.9
Before designing organ targeting nanoparticles, it is necessary to understand a key pharmacological concept, namely organ targeting. Organ targeting is not about delivering all the doses to the target organs, but about delivering enough of dose to achieve the desired biological effects while limiting off-target cumulative toxicity.9 Although most of the injected doses do not reach the target organs, it should also play an important role in fully exerting physiological and therapeutic effects. Organ targeting has been studied for a long time, and most of the nanoparticles will be enriched in the liver after entering the systemic circulation. Targeting organs other than the liver has been an unsolved problem for a long time, and extrahepatic targeting will be a very important direction for the future research of targeting nanoparticles.1 In the past two years, a delivery strategy called selective organ targeting has been reported.10 LNPs can be selectively targeted to liver, spleen, lungs and other organs by adding an auxiliary component called selective organ targeting molecules to lipid nanoparticles.10 The report of this work makes the study of organ targeting nanoparticles attract more scientists, thus forming the concept of endogenous targeting.
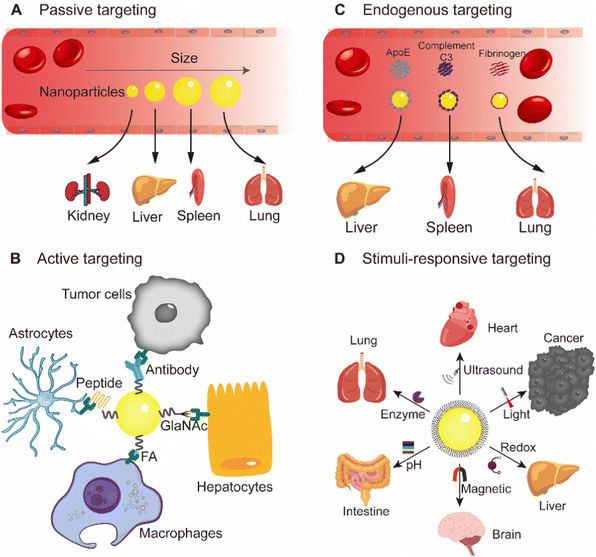
This review mainly focuses on selective organ targeting nanoparticles, briefly describes a series of barriers experienced by nanoparticles after entering the body, and expounds the mechanism of nanoparticle organ targeting, including active targeting, passive targeting, endogenous targeting and stimuli-responsive targeting (Fig. 1). Examples are given to illustrate recent advances in nanoparticles targeting specific organs such as brain, heart, liver, spleen, lungs, and kidneys. The promise and clinical translation of selective organ targeting nanoparticles are also discussed. Since many reviews summarized strategies for targeting tumors with nanomaterials, we did not address tumors as a specific target in this review.
2. Several barriers to organ-targeted nanoparticle delivery
Reaching the target is a very difficult process for nanoparticles, which need to pass various barriers to be effective. For targeted nanoparticles, the first thought is through injection, and oral administration of targeted nanoparticles has long been considered impractical.3 However, oral administration has always been the most widely used route of administration.11 Orally administrated drugs need to pass through a series of barriers to enter the systemic circulation.12 The intestinal barrier through which oral drugs pass is composed of several parts: the physical barrier formed by the tight junction between intestinal mucosal epithelial cells, the chemical barrier composed of mucus secreted by intestinal epithelial cells, digestive juices secreted by gastrointestinal tract and bacteriostatic substances produced by intestinal symbiotic bacteria, the immune barrier composed of intestinal associated lymphoid tissue and its immunoreactive substances, and the microbial barrier composed of various viruses, bacteria and their secretions (Fig. 2).13 After oral administration of micro/nanoparticles through intestinal epithelial cells, they tend to leak out through lymphatic vessels and enter the systemic circulation through the subclavian vein.3 Other non-intravascular administrations, such as intramuscular and subcutaneous injections, need to overcome several barriers before entering systemic circulation. For example, in intramuscular injection, nanoparticles need to overcome the extracellular barrier and vascular endothelial barrier before entering systemic circulation to avoid being phagocytized by non-targeted cells.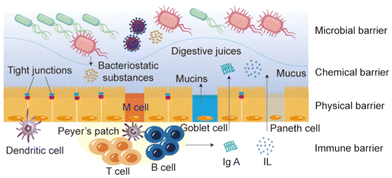 | ||
| Fig. 2 The intestinal barrier that oral drugs need to pass before entering the systemic circulation. M cell, microfold cell; IL, Interleukin. | ||
After the nanoparticles enter the systemic circulation, they will experience the same biological barrier as the intravascular nanoparticles (Fig. 3). When nanoparticles enter blood vessels, various enzymes in the blood vessels may degrade the active components of the nanoparticles, such as nucleic acid.14 In addition, they are usually cleared by the reticuloendothelial system (RES); nanoparticles smaller than 6 nm are filtered through the kidneys, nanoparticles smaller than 25 nm are taken up by Kupffer cells in the liver, and nanoparticles larger than 150 nm are more likely to be filtered and phagocytosed by macrophages in the spleen.15 During this process, the accumulation of nanoparticles in the target and off-target organs, phagocytosis by the mononuclear macrophage system (MPS), renal filtration clearance, and hepatobiliary system excretion occur simultaneously and compete dynamically.16 In addition to various enzymes in blood vessels, when nanoparticles flow in blood vessels, blood,17 immune cells18 and vascular endothelial cells19 also interact with nanoparticles and affect the fate of nanoparticles. A study showed that blood flow conditions had little effect on the binding of nanoparticles to B cells and that charged and large nanoparticles were more likely to bind to B cells in the blood.20 Another study showed that thiol-reactive nanoparticles can bind to various cells and some plasma proteins in human blood and trigger the activation of the corresponding cells.17 In short, nanoparticles will experience a variety of events in blood vessels, interact with plasma proteins, enzymes, blood cells, antibodies, immune cells, etc., and affect the circulation of nanoparticles in the body.
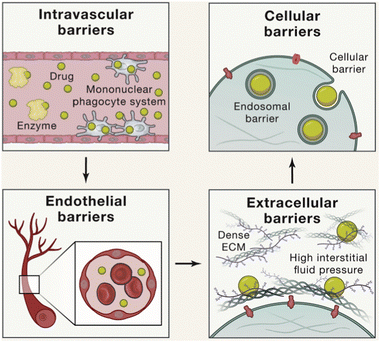 | ||
| Fig. 3 Intravascular nanoparticles encounter serial biological barriers, including the intravascular barrier, vascular endothelial barrier, extracellular barrier, cellular barrier and endosomal barrier. Copyright 2020. Reproduced with permission from the Elsevier.21 | ||
During long circulation, nanoparticles need to pass through the vascular endothelial barrier to reach the target organ and enrich at the treatment site.22 The integrity of the endothelial barrier varies widely across tissues, organs, and pathophysiological conditions.23 There is a large amount of fenestrated endothelium in the liver vessels, so the nanoparticles perfuse and accumulate in the liver.24 When tissue injury, infection, cancer and other inflammatory diseases occur in the body, the vascular endothelial cells in the lesion will become dysfunctional, resulting in the opening of blood vessels similar to the liver, forming a large number of relatively leaking blood vessels.25 This phenomenon, known as enhanced permeability and retention (EPR) effect, is particularly important for nanoparticle therapy at tumor and inflammatory sites.26
After the nanoparticles are enriched in the target organs, due to the combination of treatment sites, corresponding barriers need to be faced. Nanoparticles may need to pass through the extracellular matrix (ECM) to reach target cells in order to be effective, and some parts of the ECM are relatively dense, making it difficult for larger nanoparticles to pass through.21 When there is a disease, changes in ECM may create additional biological barriers, such as high pressure at the tumor site due to poor lymphatic reflux, thereby hindering the action of nanoparticles.4 Some nanoparticles need to cross the cell barrier to enter the cell. Drugs also need the endosome escape to overcome the danger of nanoparticle degradation through the endosome pathway.27 Nanoparticles that need to enter other organelles such as nucleus or mitochondria also need corresponding localization signals to help cross the barrier and enter the target site.1
3. Nanoparticle targeting mechanism
In this section, the targeting mechanisms of various targeted nanoparticles are discussed, providing guidance for the design of nanoparticles. Because nanoparticles need to pass through the above-mentioned series of barriers to play a role in the body, the number of nanoparticles that eventually reach the target site is very small.5 Therefore, it is very necessary to design nanoparticles that are combined with multiple targeting strategies in practical applications.3.1 Passive targeting
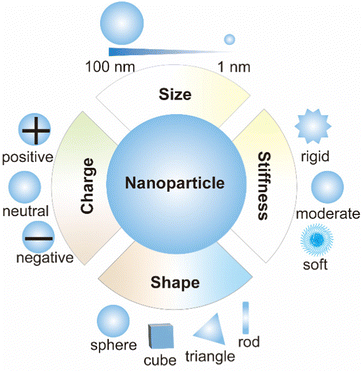
Passive targeting is mainly unmodified nanoparticles, which exploit the surface properties of nanoparticles, such as size, shape, charge, stiffness, etc. (Fig. 4).28 It is a targeting strategy to transfer nanoparticles to the target area through endocytosis and adsorption of various tissues and cells, capillary entrapment or exploiting the high permeability of blood vessels in diseased tissues.29 The study of passive targeting in tumor delivery is widely used in nanomedicine. Most of the tumor nanomedicines approved for marketing are passive targeting drug delivery. The theoretical basis of tumor passive targeting (EPR effect) has encountered many challenges in clinical trials.30 EPR effect varies among different tissues, organs and species. To address this challenge, a large number of literature studies have adopted various methods to enhance the EPR effects. Rodriguez et al. designed a minimal “self” peptide based on CD47 to prevent phagocytosis and enhance drug delivery.31 Deepagan et al. designed a nitric oxide (NO)-producing nanoparticle that releases NO at the target site. NO can relax vascular smooth muscle and increase local blood flow and vascular permeability, thereby enhancing passive targeting.32 In addition to cancer, passive targeting can also be used for organ targeting therapy for other diseases, mainly by utilizing its own surface properties, such as small-size nanoparticles for acute kidney injury treatment.33The size of nanoparticles is one of the distinguishing features of passive targeting. Nanoparticles in blood circulation accumulate in different organs due to their different sizes.34 Since the heart and brain are not sensitive to the size of nanoparticles, passive targeting nanoparticles rarely enter the heart and brain.34 Nanoparticles smaller than 10 nm are easily filtered and captured by kidneys, and nanoparticles larger than 200 nm can activate RES to remove nanoparticles and eventually accumulate in liver and spleen.35 Muscle-targeted drug delivery study showed that 11–32 nm poly(ethylene glycol) (PEG) graft copolymers increased permeability in Duchenne muscular dystrophy mice.36 When designing nanoparticles targeted to other organs, nanoparticles with a size of 50–200 nm are suitable for long-term circulation in the blood and can be more enriched in other targeted organs. For example, a study using poly(lactic-co-glycolic acid) (PLGA) nanoparticles showed that small nanoparticles (∼120 nm) could enhance uptake in lung and bone marrow.34
The shape of nanoparticles also plays an important role in organ-based passive targeting as nanoparticle shape affects their opsonization, internalization, migration, and circulation time.37 For example, the internalization of nanoparticles with a high aspect ratio in cells is more efficient than that of nanoparticles with a low aspect ratio.38 The shape of particles is important to avoid opsonization and promote long circulation.39 Compared with spherical nanoparticles, rod-shaped nanoparticles have a higher aspect ratio, less phagocytosis, and a longer circulation time.40 Therefore, the accumulation of rod-shaped nanoparticles in the liver is less than that of spherical nanoparticles because the elongated shape is better able to escape phagocytosis by Kupffer cells.39 Similar to rod-shaped nanoparticles, disk-shaped nanoparticles have a significantly longer circulation time than spherical nanoparticles and accumulated more in lung and heart.39 Spherical, elliptical, and cylindrical nanoparticles are more likely to be enriched in the liver, while irregular nanoparticles are more likely to be enriched in the spleen.39
Passive targeting also takes advantage of the stiffness and charge of nanoparticles.28 Macrophages tend to engulf stiffer nanoparticles, so less stiffness nanoparticles circulate in the body longer. For non-phagocytic cells, positively charged nanoparticles are taken up higher by cells due to the negatively charged cell membrane.39 The mobility of nanoparticles also affects the passage of drugs through the biological barriers. Wang et al. designed a nanoparticle formed by polymer-coated edible oil droplets, which is deformable and can effectively penetrate biological barriers such as BBB, deep brain tissues and mucus layers.41 However, only using the characteristics of nanoparticles, the passive targeting effect is not ideal, so people design nanoparticles and put forward the second generation of targeting nanoparticles.
3.2 Active targeting
Active targeting nanoparticles are the second generation of targeting nanoparticles, which are loaded with therapeutic molecules through surface modification, and transport therapeutic molecules to the target area so as to play a therapeutic role. Active targeting nanoparticles mainly include three parts, ligands, linker and cargo (Fig. 5). The cargo includes a variety of therapeutic molecules, such as small molecular drugs, nucleic acids, therapeutic peptides, etc. Ligands include targeting small molecules, aptamers, targeted peptides, antibodies or antibody fragments, cells and others.21 Therefore, active targeting is divided into the following strategies.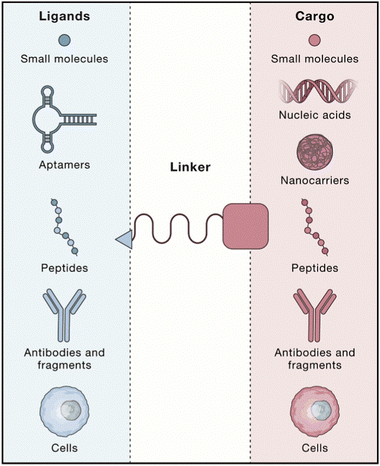 | ||
| Fig. 5 A schematic illustration of active targeting nanoparticles involving ligands, linker, and cargos. Copyright 2020. Reproduced with permission from the Elsevier.21 | ||
The targeting strategy based on small molecules uses small chemical molecules such as vitamins, monosaccharides and hyaluronic acid as warheads to accurately target the corresponding receptors. FA, one of the vitamins, is the most widely used small-molecule targeting agent, which is highly expressed in nearly half of tumors and activated macrophages.21 Thus, there are a large number of nanoparticles using the FA-targeting strategy. FA receptor-targeted two-dimensional palladium nanoparticles have been reported for multimode imaging of atherosclerosis using folic acid to target activated macrophages that are closely related to high-risk plaques.42 A similar tumor study showed that FA-modified lipid nanodisks avoided IgM-mediated opsonization and effectively targeted the FA receptor positive tumors in vivo.43 In addition to vitamins, monosaccharides are also excellent targeting ligands. For example, glucose can target the GLUT1 receptor, which is overexpressed in the BBB.44 Lectin-like receptors such as mannose-6-phosphate receptors respond to carbohydrates, and galactose can target asialoglycoprotein (ASGP) receptors.45 In addition, there are many small molecules that can be used for targeting. For example, urea derivatives such as glutamate urea can target prostate specific membrane antigen (PSMA), glycyrrhetinic acid (GA) and its derivatives can target hepatocyte surface protein kinase C (PKC), and bisphosphate can be used to target bone.21
Aptamers are also important molecules for potential targeting strategies. An aptamer is a structured oligonucleotide sequence (RNA or DNA) obtained using the in vitro screening technique: systematic evolution of ligands by exponential enrichment (SELEX). It can bind with high affinity and specificity to corresponding target molecules (proteins, metal ions, small molecules, peptides and even whole cells). Aptamers are easy to produce and have low immunogenicity, making them good candidates for organ targeted delivery.21 However, aptamer-based targeting strategies face enormous challenges and there are no drugs in the market. Among them, the degradation in organisms is the biggest problem,21 and some research groups have carried out research on this problem. Tan et al. designed and prepared a nucleic acid aptamer-multidrug conjugate, and the aptamer was placed on the shell of the nanomicelle to enhance its resistance to nuclease.46 Niu et al. designed a bone-targeting nanoparticle using PEGylated dendritic mesoporous silica nanoparticles to protect anti-sclerosing protein aptamers for the treatment of osteoporosis.47
The antibody-based targeting strategy is a promising clinical targeting strategy. FDA has approved a number of antibody–drug conjugates (ADCs), which are effective for tumor treatment.48 Ge et al. reported an antibody-modified DNA origami nanostructure for targeted therapy of prostate cancer.49 Current clinical studies on ADCs are mainly in the field of oncology, and antibodies against chronic inflammation and autoimmune diseases are being studied.50 Bertheloot et al. developed nanoantibodies and demonstrated their efficacy in disassembling post-pyroptotic inflammasome and treating inflammatory diseases in animal models.51
The peptide-based targeting strategy is another very representative targeting strategy, and a large amount of studies has focused on this field. The most commonly used method for the discovery of targeted peptides is through bioinformatics and biomolecule identification to find the corresponding target protein homologous sequence and synthesize the peptide library for screening.21In vivo or in vitro phage display is another common method for finding targeted peptides.21 Through the targeted combination of the random peptide sequences displayed on the phage surface, peptide sequences targeting specific tissues and organs can be screened out.52 For example, THRPPMWSPVWP peptide could target human transferrin receptor (TfR) positive cells, CMPRLRGC peptide could target low-density lipoprotein (LDL) receptors, and peptides with GFE motifs could target membrane dipeptides in alveolar capillaries.52 Homing peptides and cell penetration peptides were also screened for targeting delivery, such as TAT, NGR and RGD.53 Among them, RGD is the most commonly used targeting motif, which can bind to αvβ3 and αvβ5 integrin receptors, including cyclic RGD, bicyclic RGD and iRGD.53 Other peptides were also developed to target integrin receptors. Dan Peer et al. reported recombinant fusion protein-modified lipid nanoparticles that recognized the high-affinity conformation of integrin α4β7 for the treatment of colitis in mice.54
Cell-based targeting strategies take full advantage of the natural targeting process in the body. By utilizing a variety of cellular structures, including red blood cells, white blood cells, platelets, extracellular vesicles and even bacteria, nanoparticles or drugs can be delivered to specific organs.55 This strategy can be divided into cell membrane biomimetic delivery and living cell delivery. Cell membrane biomimetic delivery has a longer circulation time and better targeting ability, which mainly depends on homing mechanism and masking immune recognition.56 Cell membrane biomimetic delivery began in 2011. Zhang et al. reported for the first time that red blood cell membranes and PLGA were coextruded into a core–shell structure to facilitate drug delivery in vivo.57 Since then, a large number of studies have poured into this field, and cell membrane biomimetic delivery has sprung up in the fields of tumor therapy,58 inflammation targeting,59 antibacterial,60 detoxification61 and immunotherapy.62 Then, some research groups began to study its internal mechanism. Fan et al. constructed a membrane-derived plasma probe for targeted imaging. Experimental observations suggested that the integrin-mediated targeting is a universal mechanism independent of the core wrapped in the cell membrane.63 Lehto et al. used the fluorescence quenching method to detect the integrity of the cell membrane coating and proposed that the integrity of the coating affected the internalization of biomimetic nanoparticles. The more complete the membrane coating, the better the internalization.64 Bacterial cell membranes can also be used to coat nanoparticles, and related research is mainly focused on the field of immunotherapy. Nie et al. constructed a personalized tumor vaccine platform based on bacterial outer membrane vesicles (OMVs), which could quickly display a variety of tumor antigens and achieve “Plug-and-Display” of tumor vaccines.65 In addition, cell membrane biomimetics can also target specific organs, such as the ability of aging red blood cell membranes with high expression of phosphatidyl serine to be targeted to the spleen.66 Other immune cells such as macrophages and neutrophils can also target brain and other organs.21 Living cell delivery is also a good targeting strategy. Engineered stem cells can encode therapeutic proteins in the targeted area, and platelets can be used as the drug delivery system to target injured sites.21 Li et al. co-assembled doxorubicin with a cyclin-dependent kinase 5 inhibitor into platelet membrane fragments and delivered them to residual tumors after surgery. The nanosystem effectively disrupted the negative feedback loop between the recovery of interferon-γ and the upregulation of programmed death ligand-1 during postoperative adjuvant chemotherapy and resolved the problem of postoperative tumor progression.67 Zhang et al. used click chemistry to load biomimetic nanoparticles carrying drug cores onto microalgal cells and used the movement of microalgal cells to infiltrate deep tissues to treat acute bacterial pneumonia.68 In short, active targeting can equip drug missiles with automatic targeting and tracking systems to help target the interest site, which is of great significance in the design of targeting nanoparticles.
3.3 Endogenous targeting
Endogenous targeting refers to designing of the composition of nanoparticles to bind to different plasma proteins in the blood, thereby guiding them to target organs and promoting uptake by specific cells.69 In the past two years, the research on endogenous targeting has mainly focused on LNPs.9 As an FDA-approved drug delivery vehicle, LNP is well suited for therapeutic nucleic acid delivery.70 There have been a large number of literature studies on the active targeting of LNPs to adapt to various diseases, and its commercial prospects are very broad.71 The endogenous targeting of LNPs can be optimized by adjusting parameters such as structure, type, proportion, pH and others (Fig. 6).9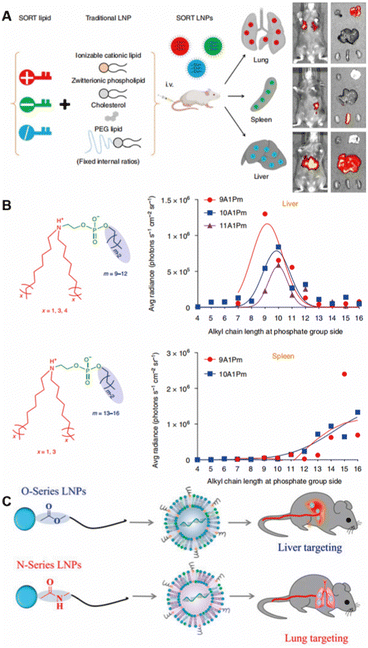 | ||
| Fig. 6 The composition of LNPs alters organ targeting. (A) “SORT” molecules with different charges added. Copyright 2020 The Authors. Reproduced with permission from Springer Nature.10 (B) Altered chemical structure of functional phospholipids. Copyright 2021 The Authors. Reproduced with permission from Springer Nature.72 (C) Structural changes of lipidoid tail groups. Copyright 2022 The Authors. Reproduced with permission from National Academy of Science.73 | ||
The surface composition of nanoparticles has a great influence on organ targeting, and surface lipids affect the fate of nanoparticles in vivo, especially in selective organ targeting.74 A research team found that when delivering plasmid DNA (pDNA), 3% Tween 20 (a surfactant connected to the short lipid tail by branched PEG) was highly selectively transfected in the lymph nodes compared with the traditional LNPs stabilized by linear PEG and two saturated lipid tails.75 The first siRNA drug patisiran (trade name: Onpattro) was approved by FDA in 2018.74 During the development of patisiran, the ionizable cationic lipids were optimized from the initial 1,2-dilinoleyl-N,N-dimethyl-3-aminopropane (DLinDMA) to 2,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane (DLin-KC2-DMA) and finally selected as heptatriaconta-6,9,28,31-tetraen-19-yl-4-(dimethylamino) butanoate (DLin-MC3-DMA). Recently, a research group has investigated the delivery of pDNA by LNPs composed of 1,2-dioleoyl-3-dimethylammoniumpropane (DODAP), DLin-KC2-DMA, or DLin-MC3-DMA.76 The results showed that the transfection efficiency of DLin-MC3-DMA, which worked best in patisiran, was lower than that of DLin-KC2-DMA, which may be due to the difference in the length of the negatively charged backbone of siRNA and pDNA.
Siegwart et al. were the first to report the modulation of organ targeting LNPs by adding components.10 They added supplementary “SORT” molecules to LNPs, adding different SORT molecules to target different organs (Fig. 6A). Ionizable cationic lipids enhanced targeting to the liver, anionic lipids could be delivered specifically to the spleen, and permanent cationic lipids with quaternary ammonium salt headgroups increased lung targeting. Different ratios of the same SORT molecules also affected the organ targeting of LNPs. As the molar percentage of ionizable cationic lipids increased, the organ targeting of LNPs shifted from the liver to the spleen and then to the lungs. Using changes in the ratio of ionizable cationic lipids, they constructed modified LNPs for selective organ targeting of the liver and lungs to deliver Cas9/sgRNA ribonucleoprotein complexes for gene editing.77 This strategy has become a well-established method, and the preparation protocol has been published.78
In addition to compositional changes affecting organ targeting, the chemical structure of functional phospholipids can also control efficacy and organ selectivity in vivo (Fig. 6B).72 The best ionizable phospholipids have two lipid tails for the tertiary amino group and one lipid tail for the phosphate group. Changing the length of the lipid tail on the phosphate group switched organ targeting from the liver to the spleen to the lungs.79 Using this ionizable phospholipid, Siegwart et al. reported a series of new phospholipids with strong endosome escape properties, in which the length of the lipid tail of the phosphate group determined its organ targeting. Ionizable phospholipids with 12 carbon chains selectively delivered mRNA to liver for expression, and ionizable phospholipids with 13 to 16 carbon chains could deliver mRNA to the spleen for expression.72 On the basis of the above studies, Siegwart et al. proposed the mechanism of LNP organ targeting by studying the liver targeting mediated by apolipoprotein E (ApoE).69 After LNPs enter the blood, specific plasma proteins adsorb and recognize exposed SORT molecules, and surface binding proteins interact with homologous receptors highly expressed in specific tissues and organs to achieve organ targeting.
Regulating the formation of protein corona on nanoparticles to control the organ targeting holds great promise. The formation of protein corona can be tuned by adjusting the surface properties of nanoparticles,80 including shape,81 charge,82 roughness,83 hydrophobicity84 and others. A recent study has found that LNPs tailed with ester bonds tended to target the liver, and LNPs tailed with amide bonds could adsorb coagulation-related proteins such as fibrinogen to selectively target the lung, and lipids based on imidazole gave priority to target the spleen (Fig. 6C).73
In addition to the endogenous targeting of LNPs, other nanomaterials can also use the endogenous targeting mechanism to adsorb other endogenous proteins to form protein corona for targeting.85 Lin et al. reported that gold nanoparticles might bind to activated platelet factor 4 through injured blood vessels and then interact with heparan sulfate proteoglycans on endothelial cells to promote uptake.86 Huang et al. designed and synthesized hemoglobin conjugated poly(ε-caprolactone) polymers and self-assembled into biomimetic nano red blood cells.87 Hemoglobin could bind to endogenous plasma haptoglobin to specifically target M2 macrophages through CD163 surface receptors; therefore, nano red blood cells specifically targeted M2 tumor-associated macrophages.
3.4 Stimuli-responsive targeting
Stimuli-responsive targeting is an intelligent targeting strategy that utilizes the properties of the nanomaterial itself, which can be switched under specific conditions. When stimulated by conditions such as light, ultrasound, temperature, magnetic field, chemical reaction, etc., the size, surface groups, and motion of the nanoparticles will change, allowing the nanoparticles to target the designed site.88,89This strategy has broad applications in tumors and inflammation and can target small molecule, tissue, cellular and organ levels. Gu et al. developed a glucose concentration-dependent insulin release LNPs (Fig. 7A).90 They formed an LNP complex from phenylboric acid-modified quaternary ammonium cationic lipids with negatively charged recombinant human insulin. When encountered with a high concentration of glucose, the decrease of positive charge of nanoparticles triggered insulin release. Ji et al. coupled the N-terminal of pH response penetrating peptide to the Fc segment of specific antibodies and injected it into the body to selectively assemble on the tumor cells in response to tumor acidity, thereby enhancing NK cell immunotherapy.91 Nie et al. designed PLGA nanoparticles modified with different phenolic compounds based on the property of phenolic groups to form free radicals and cross-linked polymers under the action of excess myeloperoxidase (MPO) and reactive oxygen species (ROS).6 Inflamed tissues can be targeted by in situ accumulation stimulated by excessive enzymes and ROS in the inflammatory pathological microenvironment. It was determined that tyramine modification could significantly enhance the target ability and aggregation of nanoparticles in the inflammatory region.6 Wang et al. reported a magnetically targeted nano-carrier that activated peroxisome proliferator-activated receptor gamma agonists, which can be targeted by a magnetic field and enriched in the cerebral hematomas after intravenous injection (Fig. 7B).92
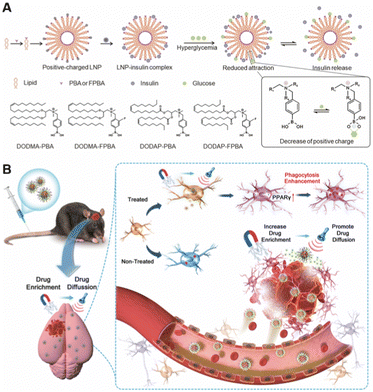 | ||
| Fig. 7 Stimuli-responsive targeting nanoparticles. (A) In vivo environment-responsive nanoparticles. Copyright 2023. Reproduced with permission from the Wiley.90 (B) External stimulus-responsive nanoparticles. Copyright 2023. Reproduced with permission from the Wiley.92 | ||
The above four strategies constitute the commonly used nanoparticle targeting strategies, and it has been reported that one or more of them are always used in the treatment of diseases. Similarly, in the design of organ targeting nanoparticles, the above-mentioned targeting strategies can be combined to exert a better organ targeting effect.
4. Nanoparticle targeting specific organs and their design
The design of nanoparticles targeting specific organs is important for drug delivery.1 Each organ has its own characteristics, and designing nanoparticles according to the characteristics of organs is the key to the design of organ targeting nanoparticles.5 In the design process, it is generally necessary to use a variety of targeting strategies comprehensively to achieve the best organ targeting effect as summarized in Table 1.4| Target organ | Targeting strategy | Targeted disease | Nanoparticle type | Ref. |
|---|---|---|---|---|
| Brain | Active targeting | Intracranial malignancies | Membrane biomimetic nanoparticle | 101 |
| Endogenous targeting | Alzheimer's disease | Liposome | 102 | |
| Stimuli-responsive targeting | Parkinson's disease | Piezoelectric nanoparticle | 103 | |
| Heart | Active targeting | Chronic heart failure | Porous silicon nanoparticle | 104 |
| Chronic heart failure | Extracellular vesicles | 105 | ||
| Myocardial Infarction | Liposomal nanoparticle | 106 | ||
| Stimuli-responsive targeting | Myocardial hypertrophy/ventricular fibrillation cardiac arrest | Polyphenol-based functional nanoparticle | 107 | |
| Liver | Passive targeting | Type 2 diabetes | Silicon nanoparticle | 108 |
| Active targeting | Diabetes | Polymer nanoparticle | 109 | |
| Endogenous targeting | Polyneuropathy | LNP | 74 | |
| Stimuli-responsive targeting | Acute liver injury | Phenol-functionalized nanoparticle | 6 | |
| Spleen | Endogenous targeting | Cancer | LNP | 110 |
| Lung | Passive targeting | Chronic lung disease | Tetra(piperazino)fullerene epoxide-based nanoparticle | 111 |
| Active targeting | Lung cancer | LNP | 112 | |
| Endogenous targeting | Pulmonary lymphangioleiomyomatosis | LNP | 73 | |
| Stimuli-responsive targeting | Acute lung injury | Phenol-functionalized nanoparticle | 6 | |
| Kidney | Passive targeting | Acute kidney injury | Ultra-small tungsten-based nanodot | 33 |
| Renal ischemia-reperfusion injury | Ceria nanoparticle | 113 | ||
| Active targeting | Renal fibrosis | Polymer nanoparticle | 114 | |
| Acute kidney injury | Membrane biomimetic nanoparticle | 115 | ||
| Eye | Active targeting | Inherited retinal degenerations | LNP | 116 |
| Bone | Active targeting | Osteoporosis | Mesoporous silica nanoparticle | 47 |
| Stimuli-responsive targeting | Upconversion nanoparticle | 117 | ||
| Bowel | Active targeting | Inflammatory bowel disease | LNP | 54 |
4.1 Brain
Due to the tight junction of the cerebrovascular BBB, brain-targeted drug delivery is relatively difficult to achieve.93 Most brain disease treatments are administered locally through invasive means, such as intracerebroventricular and intrathecal injections.94 Except for local injection, other intravascular administrations face the problem of the BBB.94The active targeting strategy is one of the most used strategies for brain-targeted drug delivery.94 Generally, transferrin, low-density lipoprotein, lactoferrin, rabies virus glycoprotein and others can be modified on the surface of the carrier to bind to specific receptors of BBB for enhancing the active transport of drugs.95 Zhong et al. designed a nanoparticle consisting of Angiopep-2 targeting peptide-linked polyamidoamine (PAMAM) for synergistic neuroprotection and oxidative stress treatment in Alzheimer's disease (AD).96 Nanoparticles leverage the ability of Angiopep-2 to target low-density lipoprotein receptor (LDLR)-associated protein-1 on the BBB and the ability of PAMAM to selectively target hyperactive microglia for delivery of Prussian blue. Prussian blue can efficiently scavenge ROS from microglia to treat AD.96 Also, using the Angiopep-2 targeting peptide that has a higher transcytosis ability than other proteins,97 Zhao et al. modified Angiopep-2 with liraglutide and assembled it with PEG into a nanostructure to target the brain and inhibit inflammatory pathways, thereby alleviating AD induced by Aβ.98 In this peptide-based active targeting strategy described above, the stability and affinity of the ligand play an important role in the BBB transport of nanoparticles. In another study, Li et al. demonstrated that nanoparticles with higher ligand stability and affinity had a significantly higher ability to cross the BBB than peptides with lower affinity.99 They used D-type amino acid ligands with a large number of interacting hydrogen bonds to compare with L-type ligands and found that the ability of D-type ligand-modified nanoparticles to cross the BBB was significantly improved compared with L-type ligand-modified nanoparticles.
In addition to peptide-based active targeting, cell-based active targeting can also be used for brain-targeted drug delivery.93 Li et al. designed a biomimetic nanoplatform camouflaged by red blood cell membranes to selectively deliver mRNA across the BBB to target glioblastoma (GBM).100 The delivery platform consists of mRNA, cationic polyethyleneimine (PEI), charge-switching citraconic anhydride-grafted poly-L-lysine (PLL-CA), and ApoE peptide-modified erythrocyte membranes. ApoE has high affinity for the LDLR family on the surface of endothelial cells in the BBB and GBM, so ApoE can mediate nanoparticles crossing the BBB and targeting GBM.100 The red blood cell membrane coating protects nanoparticles from opsonization in the blood, reduces immunogenicity and improves blood circulation time.58 PLL-CA can disrupt the red blood cell membrane and release mRNA under acidic conditions after the tumor internalization of nanoparticles. PEI is mainly used to effectively compress mRNA and achieve endosomal escape.100 Chen et al. constructed lipopolysaccharide-free biomimetic nanoparticles by exploiting the ability of Escherichia coli (E. coli) K1 to bind and invade BBB endothelial cells in native bacterial meningitis.101 Crossing the BBB using bacterial outer membrane protein A and gp96 has great potential for the treatment of gp96-overexpressing tumor cells.
Other targeted delivery strategies may also play a role in brain targeting.93 Zhang et al. modified the targeting ability of liposomes by manipulating the protein corona, modified a short peptide of Aβ1–42 to the surface of liposomes, and used it to adsorb apolipoproteins A1, E, and J to bind to the receptor binding domain of apolipoproteins for brain targeting.102 Chen et al. combined passive targeting with endogenous targeting, exploiting the ability of discoid nanoparticles to escape accelerated blood clearance.118 It was found that discoid LNPs could adsorb apolipoproteins and selectively target the brain. Whether it is active targeting or endogenous targeting, the corresponding targeting protein is required on the surface, so the target protein is very important in this process. By varying the type and density of nanoparticle surface modifications, Li et al. were able to bind nanoparticles to more targeted motifs, which maximized the permeability of nanoparticles through the BBB.119
Zhou et al. designed an enzyme-responsive nanoparticle, which used the ability of the nanoparticle to change size in the presence of an enzyme to improve the delivery efficiency of nanoparticles at the target site, thereby improving the therapeutic effect of stroke disease (Fig. 8A).120 Using the same strategy, they further optimized the nanoparticles with brain permeability and antioxidant activity for effective stroke treatment (Fig. 8B).121 Not only can the properties of nanoparticles be used to target the brain but also other auxiliary means can be used to help nanoparticles cross the BBB non-invasively.122 For example, ultrasound is used to temporarily open the BBB for therapeutic delivery,122,123 primarily through the paracellular pathway across the BBB.124 Rezai et al. reported that magnetic resonance-guided low-intensity focused ultrasound could reversibly open the BBB and facilitate brain-targeted delivery.125 Kim et al. also used ultrasound-assisted nanoparticles that triggered NO release, temporarily opening the tight junctions of the BBB and allowing nanoparticles to accumulate into the brain parenchyma (Fig. 8C).103
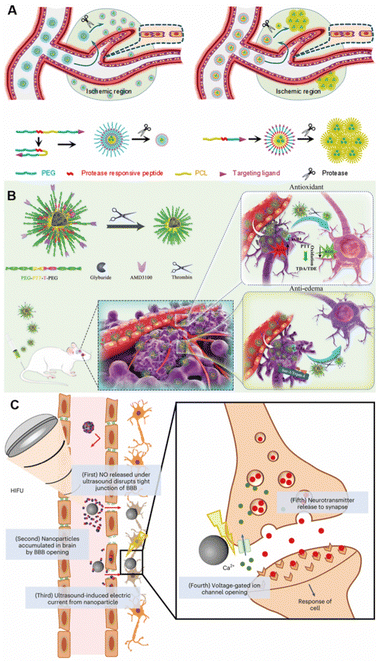 | ||
| Fig. 8 Stimuli-responsive targeting nanoparticles for brain targeting. (A) Protease-responsive size-variable nanoparticles. Copyright 2018. Reproduced with permission from American Chemical Society.120 (B) Thrombin-triggered shrinkable nanoparticles. Copyright 2022. Reproduced with permission from Wiley.121 (C) Ultrasound-responsive nanoparticles. Copyright 2023 The Authors. Reproduced with permission from Springer Nature.103 | ||
In addition to intravascular delivery, there are other alternative delivery strategies that can bypass the BBB, such as intranasal administration.94 Whether it is a pathway through the BBB or an alternative pathway, the design of these nanoparticles is based on the characteristics of the biological barrier that needs to be crossed, and a combination of methods can be used to achieve the best targeting effect.
4.2 Heart
It is reported that most cardiovascular diseases can be effectively treated by the use of nanotechnology, and the use of cardiac-targeted nanomedicines can increase drug efficacy and reduce the side effects.126 Cardiac targeting nanocarriers are an effective way to treat cardiovascular diseases such as myocardial infarction and heart failure.127Cardiac-targeted nanocarriers require overcoming the intravascular and endothelial barriers in vivo. Due to the high blood flow velocity in the heart, the stability and adhesion of nanoparticles in high-velocity fluids need to be considered; that is, the shape and size of nanoparticles need to be optimized.128 In addition, the heart tends to repel nanoparticles due to the nature of myocardial contraction, so the design of cardiac-targeted nanocarriers requires more active targeting strategies.128 Kim et al. extracted small extracellular vesicles from an engineered cell medium expressing high levels of cardiac targeting peptides (APWHLSSQYSRT) to deliver siRNA to reduce the inflammatory symptoms of myocarditis. The delivery efficiency was more than two times higher than that of normal small extracellular vesicles.129 Banik et al. designed dual-targeted nanoparticles for the treatment of atherosclerosis, mainly composed of a polymer-coated nuclear magnetic contrast agent.130 The surface modification of nanoparticles has three functional fragments, one targeting M2 macrophages that constitute atherosclerotic plaques and binding to mannose receptors overexpressed on their surface and the other enhancing cholesterol recognition and binding. The last fragment targets mitochondria to transport bound cholesterol to mitochondria to initiate cholesterol metabolism. Another type of nanoparticles integrating with diagnosis and treatment is delivered by neutrophils, which exploits their ability to target the site of ischemic inflammatory to deliver cargo for myocardial ischemic injury treatment.131 Neutrophils carry iron oxide nanoparticles modified with CD34 antibodies. After enrichment at inflammatory sites, neutrophils release nanoparticles and CD34 antibodies can capture endothelial progenitor cells and promote endothelial repair and neovascularization. Iron oxide nanoparticles can monitor this process in real time. It can be seen that nanocarriers play an important role in the process of heart repair and regeneration.132
However, with the contraction of the myocardium, the intracardiac pressure rises, the blood in the heart will be pumped out of the heart, and the nanoparticles will also be pumped out of the heart along with the blood. Therefore, nanoparticles are difficult to retain at the target site due to myocardial contraction. In order to solve this problem, Yang et al. designed a mixed injectable hydrogel containing microRNA for in situ injection; by modifying the penetrating peptide on the nanoparticles, the transfection efficiency can be significantly improved to improve myocardial repair and myocardial infarction symptoms.133 In another study, Liu et al. adopted a new approach to deliver nanoparticles for the treatment of heart failure.134 They used triphenylphosphine-modified ROS-scavenging nanoparticles for targeting mitochondria by non-invasive inhalation. Taking advantage of the ability of particles smaller than 100 nm to enter the pulmonary circulation and accumulate in the heart through the air-blood barrier, nanoparticles deposited in the lungs were continuously delivered to the heart through the pulmonary circulation, internalized to cardiomyocytes, and scavenged ROS to treat heart failure. Although there are still some difficulties to overcome,128 cardiac-targeted nanocarriers are still a promising direction worthy of investigation.
4.3 Liver
Generally, most nanoparticles accumulate passively in the liver after systemic administration.24 Therefore, nanoparticles are a natural liver targeted delivery system. The basic unit of liver structure is the hepatic lobule, which is composed of hepatocytes (HCs), hepatic stellate cells (HSCs), Kupffer cells (KCs), and liver sinusoidal endothelial cells (LSECs).135 After nanoparticles reach the liver, they can selectively target specific cells to play a therapeutic role.136 This requires nanoparticles to target specific receptors on different cells,136 and these receptors are shown in Table 2.| Cell type | Specific receptor |
|---|---|
| HC | Asialoglycoprotein receptor; |
| C–X–C chemokine receptor type 4; | |
| Folate receptor; | |
| Heparan sulfate proteoglycans; | |
| Low-density lipoprotein receptor; | |
| Scavenger receptor B1; | |
| Transferrin receptors. | |
| HSC | Collagen type VI receptor; |
| Mannose-6-phosphate receptor; | |
| Platelet-derived growth factor receptor-β; | |
| Retinol binding protein. | |
| KC | Fucose receptor; |
| Fc receptors; | |
| Galactose receptor; | |
| Mannose receptor; | |
| Scavenger receptors; | |
| LSEC | Hyaluronan receptor; |
| Low-density lipoprotein receptor; | |
| Mannose receptor; | |
| Scavenger receptor. | |
The asialoglycoprotein receptor is the most used receptor when targeting hepatocytes (Fig. 9A).45 It is specifically expressed in hepatocytes and can be targeted by a series of different ligands.137 After targeting, ligands can be quickly swallowed and recycled.137N-Acetylgalactosamine (GalNAc) is one of the targeting ligands for this receptor.137 A variety of GalNAc–nucleic acid conjugates have been introduced into clinical practice.9 GalNAc-based nucleic acid delivery has emerged as one of the most promising delivery systems.9
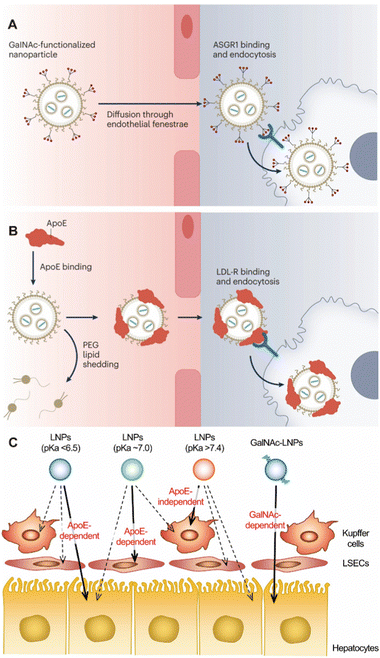 | ||
| Fig. 9 (A) GalNAc-based targeting nanoparticles and (B) endogenous targeting nanoparticles for liver. Copyright 2023. Reproduced with permission from Springer Nature.9 (C) A schematic illustration of the impact of pKa value and GalNAc modification on liver targeting nanoparticles. Copyright 2022. Reproduced with permission from the Elsevier.1 | ||
In addition to active targeted delivery, endogenous targeting also has a lot of room for development in the delivery of liver-targeted nanoparticles (Fig. 9B). Ahn et al. designed a DNA tetrahedral to deliver antisense oligonucleotides containing three cholesterol conjugates for the treatment of liver fibrosis.138 Cholesterol-modified nanoparticles could induce enhanced interactions with serum lipoproteins to selectively target hepatocytes.138 This design used endogenous targeting strategy to enhance the affinity of specific proteins and regulate the binding proteins on nanoparticles to control the targeting site. Siegwart et al. have long studied the endogenous targeting of LNPs and found that adding a small amount of cationic lipids can enhance the targeting to the liver.10 Ionizable phospholipids with 9–12 carbon chains in the phosphate tail can selectively deliver mRNA to the liver for expression.77 It can also target specific cells in the liver by adjusting the acid ionization constant (pKa).1,9 LNPs with pKa of 6.2–6.5 are the most suitable for targeting HC. With the increase of pKa, the targeting ability to LSEC is enhanced, and the targeting ability is the best in pKa = 7.1. When pKa > 7.4, it will be isolated by KC (Fig. 9C). There are still many problems to be studied in this strategy, such as what kind of molecular structure can specifically adsorb specific types of plasma proteins and then target specific organs or cells.9 Taking LNPs as an example, after the plasma protein ApoE is combined with LNPs, the composition of LNPs will be rearranged.1 It is still clear whether this composition rearrangement affects the accumulation in the liver.9 These issues still need a great deal of research so that nanoparticles can intelligently find the target sites in the body according to the design route and accurately target specific organs or cells.
4.4 Spleen
The spleen is one of the organs where nanoparticles naturally accumulate after entering the body.16 After entering the body, nanoparticles are easily swallowed by RES and accumulated in the liver and spleen, but the accumulation of nanoparticles in the liver can reach 80%.1 Therefore, the selective targeting of nanoparticles to the spleen is not as easy as one might think, and uptake by Kupffer cells needs to be avoided,1 so nanoparticles need to be modified to make them “invisible”. Jiang et al. delivered siRNA to the spleen using nanocapsules made of hydrophobic oil coated with arginine-modified gold nanoparticles.139 Arginine modification reduced the liver accumulation and increased the spleen accumulation of the nanoparticles. Nanoparticles larger than 200 nm have been reported to accumulate more in the spleen than small nanoparticles, and thus, spleen-targeted nanoparticles are large in size.1 Harris et al. added a peptide coating to nanoparticles to form 200 nm nanoparticles in serum, which could selectively deliver specific genes to the spleen.140The composition of nanoparticles also affects the ability to target the spleen.69 After adding anionic liposomes into LNPs, Siegwart et al. found that LNPs could selectively target spleen and a few other organs.10 In addition to anionic lipids, the addition of 10–15% permanent cationic lipids can also assist spleen targeting delivery.10 Ionizable phospholipids with lipid tails containing 13–16 carbon chains allow selective delivery of nucleic acids to the spleen for expression.72 This “SORT” strategy has been modified by changing the standard auxiliary phospholipids in LNPs to negatively charged phosphatidylserine, allowing more LNPs to be delivered to the spleen than the liver (Fig. 10).141 Kimura et al. greatly improved the ability of LNPs to target splenic immune cells through the complement C3 receptor by adjusting the ratio of ionizable lipid DODAP and helper phospholipid DOPE.110 Sinegra et al. modified DNA on the surface of LNPs and found that G-quadruplex-modified LNP could selectively target the spleen, which may be due to the change in protein corona.142 Zhao et al. screened a large number of liposomes and found a class of imidazolyl-based liposomes, which had the effect of spleen-targeted delivery of nucleic acid, but the transfection effect in other organs was low.143
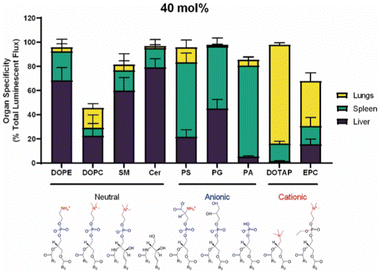 | ||
| Fig. 10 Organ targeting of nanoparticles containing 40 mol% of different kinds of helper lipids. Copyright 2022 The Authors. Reproduced with permission from the Elsevier.141 | ||
In addition, the stiffness and shape of nanoparticles also affect trapping in the spleen, which tends to capture rigid nanoparticles.1 Similarly, the spleen can remove the hard aging red blood cells in the body, and the aging red blood cells with high expression of phosphatidylserine can be used as delivery carriers to selectively target the spleen.21 In short, the design of spleen targeting nanoparticles also requires certain strategies, and it is not as simple as one might imagine.
4.5 Lung
After nanoparticles enter the blood vessels, some of them accumulate in the lungs.16 Nanoparticles that actively target the lungs often contain targeting peptides, such as GALA peptide.112 GALA peptide is a synthetic amphiphilic 30 peptide that can target the asialoglycosidic chain on lung endothelial cells.1 In addition to targeted peptides, monoclonal antibodies can also be used to deliver nanoparticles to the lung and bind to the corresponding overexpressed receptors, such as epidermal growth factor receptor (EGFR), epithelial cell adhesion molecule receptor, etc.144 Some monoclonal antibodies against lung cancer have been reviewed for selectively targeting the lung cancer after decorating on nanoparticles.144In addition to active targeting, endogenous targeting also strongly influences the accumulation of nanoparticles in the lung, where the composition and structure of nanoparticles significantly affect their endogenous targeting.9 Siegwart et al. found that the addition of 50% permanent cationic lipids to LNPs greatly increased the amount of LNPs in the lungs.10 LoPresti et al. replaced the standard auxiliary phospholipids in LNPs with the permanent cationic lipid DOTAP, thereby targeting the majority of nanoparticles in the body to the lungs than the liver (Fig. 10).141 Through library screening, Qiu et al. screened out that LNPs with amide bonds in the tails could adsorb coagulation-related proteins such as fibrinogen and promote selective targeting of the lungs (Fig. 11).73
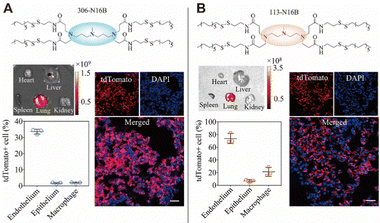 | ||
| Fig. 11 Different pulmonary cell types can be targeted by tuning the head structure of N-series LNPs. LNPs with a 113-N16B head structure were more capable of selectively targeting the lung. Copyright 2022 The Authors. Reproduced with permission from National Academy of Science.73 | ||
In selective lung-targeted nanoparticle delivery, the use of inhaled drug delivery enables local administration, maximizing the accumulation of nanoparticles in the lung.145 But there are also some biological barriers that prevent nanoparticles from working during pulmonary drug delivery.146 The mucus layer in the upper airway is the first mechanical barrier.146 The gas-blood barrier in the lung also hinders the treatment of nanoparticles, and the protein in the lung surfactant will form a protein corona on the surface of the nanoparticles, hindering its absorption.147 The physiochemical properties of nanoparticles, such as size, hydrophobicity, and charge, affect the ability of nanoparticles to pass through the biological barrier and the formation of protein corona.148 Zhang et al. used click chemistry to attach neutrophil membrane-coated polymer nanoparticles to the surface of natural microalgae and utilized the movement ability of natural microalgae to evenly distribute nanoparticles in deep lung tissues, providing a great opportunity for deep lung tissue targeting.68
4.6 Kidney
The kidney is mainly composed of renal corpuscles and associated tubules.150 Renal corpuscles are composed of glomeruli and mesangium, and glomerular filtration barrier (GFB) is located within the glomerulus.151 Glomerular filtration and tubular secretion are the main factors affecting the entry of nanoparticles into the kidney.152 The size, shape and material density of nanoparticles affect whether they enter and accumulate in the kidneys.150 In general, nanoparticles with a diameter of 1–160 nm can target the kidney and accumulate through certain mechanisms.150 Nanoparticles below 8–10 nm are generally filtered (Fig. 12A), but some accumulate in the glycocalyx on GFB, and nanoparticles of 10–20 nm can pass through GFB through slight deformation. Nanorods and nanodisks with one dimension smaller than 10 nm can also pass through GFB (Fig. 12C). Nanoparticles of 20–100 nm can enter GFB by decomposing into smaller particles and accumulate in the kidney through interaction with mesangial cells (Fig. 12D).149 Nanoparticles larger than 100 nm can enter the kidney through exocytosis of capillary epithelial cells around renal tubules and endocytosis of renal tubules (Fig. 12E).149 Han et al. used PEG–PLGA polymer nanoparticles of 300–400 nm to deliver Toll-like receptor 9 antagonists to alleviate ischemic acute renal injury.153 In addition, low-density nanoparticles flow faster and are easier to remove.150 Rod nanoparticles have a longer circulation time than spherical nanoparticles, resulting in more kidney accumulation.39 Positively charged nanoparticles easily enter the renal tubules through GFB (Fig. 12B), but are also easy cleared from the body, while negatively charged nanoparticles can be retained for a longer period of time.150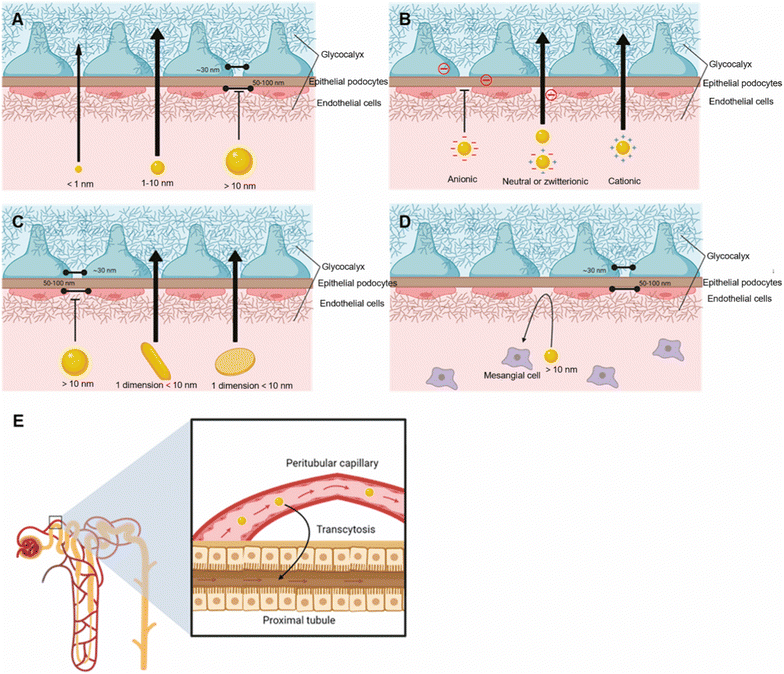 | ||
| Fig. 12 Effect of physical properties of nanoparticles on renal targeting. (A) The size of nanoparticles. (B) The charge of nanoparticles. (C) The shape of nanoparticles. The nanoparticles with > 10 nm enter the kidney through mesangial interaction (D) and exocytosis of peritubular capillary and endocytosis of proximal tubules (E). Copyright 2023. Reproduced with permission from the American Chemical Society.149 | ||
In addition to passive targeting, active targeting nanoparticles also play an important role in kidney targeting.150 A variety of targeting peptides and antibodies have been reported, such as CSAVPLC peptide, cyclo(RGDfC) peptide, PKNGSDP peptide, ELRGD(R/M)AX(W/L) peptide, and anti-CD-31 antibodies that bind to specific cells in the kidney.150 Among many targeting peptides, renal targeting peptide (KKEEE)3K has been widely studied, and it can bind to the multiligand binding receptor on the proximal tubule.154 Huang et al. studied in detail different variants of this kidney targeting peptide and investigated the effects of size, charge, and peptide length on kidney targeting.155
4.7 Other organs
In addition to the above-mentioned organs, there are many organs in the body, such as eyes, bones, stomach, small intestine, large intestine, pancreas, etc. These organs can also develop corresponding nanomedicines for selective targeted delivery. Here are a few examples to illustrate the design and application of other organ selective targeting nanoparticles.Drugs targeting the gastrointestinal tract are usually administered orally.156 Zhang et al. designed a micro-nano robot targeting gastrointestinal inflammation for oral delivery of nanomedicines.157 They loaded nanomedicines into yeast microcapsules, coupled glucose oxidase and catalase to half the surface area of yeast microcapsules, and prepared a micro-nano robot with an asymmetric structure. In the intestine, it generates energy by catalyzing high concentrations of glucose and product hydrogen peroxide, crosses through the intestinal epithelial barrier, and is phagocytosed by macrophages at Peyer's patches. Utilizing the inflammatory chemotaxis of macrophages, the drug can precisely target inflammatory sites of the gastrointestinal tract.
For bone metabolic diseases such as osteoporosis, it is usually necessary to design selectively targeted bone nanoparticles for treatment. Modification of aspartic acid-rich peptides and bisphosphonates on nanoparticles can selectively target bone tissue.158 Ye et al. designed a bone-targeting nanoparticle for the treatment of osteoporosis.117 They used upconversion materials to prepare nanoparticles, coated nanoparticles and NO donors with mesoporous silicon, and modified the outer layer with bisphosphonates for targeted delivery to bone. When irradiated with laser (808 nm), upconversion nanoparticles converted light into higher-energy light to stimulate NO production, promoted bone regeneration and reversed osteoporosis to some extent.
Drug delivery to the pancreas has been a challenge, especially for nanomedicines.159 Melamed et al. injected LNPs containing cationic lipids into the peritoneal cavity by intraperitoneal injection and used nanoparticles to interact with negatively charged mesothelial cells so that nanoparticles were retained in the peritoneal cavity.159 Then, nucleic acid drugs were enriched in pancreatic cells through extracellular vesicles of macrophages to engulf LNPs, achieving selective targeting of the pancreas.159
In addition to normal organs, nanoparticles can also be used to target diseased sites such as tumors and thrombus. As there are already many reviews summarizing cancer targeting strategies, this part will not be discussed in this review.29,160 Passive targeting nanoparticles can be used to treat thrombus, and it has been reported that disc-shaped nanoparticles similar to red blood cells can deliver drugs to thrombus.161 Active targeting nanoparticles are always designed to target the high concentrations of hydrogen peroxide and fibrin in blood clots.162 Lee et al. successively prepared two kinds of fibrin-targeting nanoparticles, which can respond to hydrogen peroxide and enhance the diagnostic and therapeutic effects.162,163 In addition, stimuli-responsive nanoparticles also play an important role in thrombus therapy. It has been reported that nanoparticles combined with high-intensity focused ultrasound can selectively destroy blood clots and avoid the formation of postoperative microembolism.164
In summary, when designing selective targeting nanoparticles, regardless of organs targeted, the choice of drug delivery modality requires an understanding of the biological barriers that need to be delivered to the corresponding organ. Depending on the corresponding biological barrier, a variety of targeting strategies can be selected to achieve ideal organ targeting.
5. Prospects and challenges of organ targeting nanoparticles
Nanoparticle-based drugs can not only act as therapeutic agents themselves but also serve as carriers to deliver therapeutic molecules to various organs. These nanomedicines can enhance the therapeutic effect of therapeutic molecules, selectively target specific organs, and reduce adverse reactions caused by non-specific distribution.165 Compared with traditional drugs, nanomedicines can increase the solubility of water-insoluble drugs, reduce systemic toxicity and the total dose of drugs, and exhibit better safety and effectiveness.166In the past two decades, drug research based on nanoparticles has become more and more popular, and research papers are increasing year by year. There are many papers being published to act as a reference for the clinical translation of nanomedicines, with a solid research foundation and broad prospects for clinical translation. FDA has approved about 100 nanomedicines;166 the market for nanomedicines is still very broad. In particular, the approval of LNP vaccines for COVID-19 pandemic has led to an upsurge in nanomedicine research. A large amount of capital has poured into this track, and many pharmaceutical companies are full of expectations for nanomedicine research and development. Up to now, there are 2313 clinical trials on liposomes, 57 on LNPs, 57 on micelles, 30 on nanocrystals, 14 on dendrimers, and 8 on carbon nanotubes. Overall, there is a great deal of scientific research on nanomedicine, but the clinical translation is still very limited.
Despite the ability of nanomedicines to selectively target specific organs, the laboratory scale of scientific research is disconnected from the need and production of clinical translation. At present, most of the approved nanomedicines target the liver or tumor tissue. The design of nanomedicines targeting other organs is complicated, and clinical translation has encountered many difficulties. Almost all active targeting nanoparticles have failed the clinical trials, which is largely related to the complexity of the design of active targeting nanoparticles. The clinical translation of nanomedicines selectively targeting to other organs is an unresolved issue that requires the participation of more researchers.
In the design of nanoparticles for disease treatment, it is necessary to understand what process the nanoparticles have experienced in the whole process of arriving at the disease site and which delivery method is more suitable. So far, the fate of nanomedicines in the body is still unknown,85 which requires more people to participate in the study of the fate of nanoparticles so that the design of nanoparticles becomes easier. In addition, the failure of many actively targeting nanoparticles to pass the clinic is also related to their failure to fully consider their journey in vivo. Protein corona plays an important role in determining the fate of nanoparticles. As soon as nanoparticles enter the body, a large amount of serum proteins are adsorbed on their surface. The types of these serum proteins vary according to the composition and structure of the nanoparticles, and different serum proteins will play different roles. As a result, nanoparticles are cleared or selectively targeted to different organs. Therefore, the role of protein corona is extremely important in the design of selective organ targeting nanoparticles. Knowing which structure of nanoparticle will adsorb which protein is the basis for clinical translation of selective organ targeting nanoparticles. In addition, a series of issues need to be considered, such as the cost and complexity of scale production, the stability and safety of nanoparticles and others.
In conclusion, the clinical translation prospects of selective organ targeting nanoparticles are very broad, but there are still a series of problems to be overcome, which are worth further studying. As long as the changes of nanoparticles in the body are discovered, people can regulate them, and the clinical translation of selective targeting nanoparticles based on nanoparticles can embark on a rapid development path.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the STI 2030-Major Projects (2021ZD0201602).References
- T. Nakamura, Y. Sato, Y. Yamada, M. M. Abd Elwakil, S. Kimura, M. A. Younis and H. Harashima, Adv. Drug Delivery Rev., 2022, 188, 114417 CrossRef CAS PubMed.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS PubMed.
- Z. Zhang, J. Qi, Y. Lu, W. Wu and H. Yuan, Med. Res. Rev., 2021, 41, 2590–2598 CrossRef CAS PubMed.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- W. Poon, B. R. Kingston, B. Ouyang, W. Ngo and W. C. W. Chan, Nat. Nanotechnol., 2020, 15, 819–829 CrossRef CAS PubMed.
- Q. Nie, C. Li, Y. Wang, Y. Hu, W. Pu, Q. Zhang, J. Cai, Y. Lin, G. Li, C. Wang, L. Li, Y. Dou and J. Zhang, Acta Pharm. Sin. B, 2023, 13, 390–409 CrossRef CAS PubMed.
- J. Zhang, Y. Yin, J. Zhang, J. Zhang, W. Su, H. Ma, F. Jia, G. Zhao and H. Wang, Nano Lett., 2022, 22, 2514–2520 CrossRef CAS PubMed.
- M. H. Zhu, X. D. Zhu, M. Long, X. Lai, Y. H. Yuan, Y. H. Huang, L. L. Zhang, Y. H. Gao, J. P. Shi, Q. Lu, P. Sun, J. F. Lovell, H. Z. Chen and C. Fang, Adv. Funct. Mater., 2023, 33, 2214842 CrossRef CAS.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed.
- J. Reinholz, K. Landfester and V. Mailänder, Drug Delivery, 2018, 25, 1694–1705 CrossRef CAS PubMed.
- H. He, Y. Lu, J. Qi, Q. Zhu, Z. Chen and W. Wu, Acta Pharm. Sin. B, 2019, 9, 36–48 CrossRef PubMed.
- D. M. Chopyk and A. Grakoui, Gastroenterology, 2020, 159, 849–863 CrossRef CAS PubMed.
- R. Juliano, J. Bauman, H. Kang and X. Ming, Mol. Pharm., 2009, 6, 686–695 CrossRef CAS PubMed.
- A. H. Okholm and J. Kjems, Adv. Drug Delivery Rev., 2016, 106, 183–191 CrossRef CAS PubMed.
- W. Poon, Y. N. Zhang, B. Ouyang, B. R. Kingston, J. L. Y. Wu, S. Wilhelm and W. C. W. Chan, ACS Nano, 2019, 13, 5785–5798 CrossRef CAS PubMed.
- J. J. Glass, Y. Li, R. De Rose, A. P. Johnston, E. I. Czuba, S. Y. Khor, J. F. Quinn, M. R. Whittaker, T. P. Davis and S. J. Kent, ACS Appl. Mater. Interfaces, 2017, 9, 12182–12194 CrossRef CAS PubMed.
- J. J. Glass, D. Yuen, J. Rae, A. P. R. Johnston, R. G. Parton, S. J. Kent and R. De Rose, Nanoscale, 2016, 8, 8255–8265 RSC.
- S. Y. Khor, M. N. Vu, E. H. Pilkington, A. P. R. Johnston, M. R. Whittaker, J. F. Quinn, N. P. Truong and T. P. Davis, Small, 2018, 14, 1801702 CrossRef PubMed.
- M. N. Vu, H. G. Kelly, A. K. Wheatley, S. Peng, E. H. Pilkington, N. A. Veldhuis, T. P. Davis, S. J. Kent and N. P. Truong, Small, 2020, 16, 2002861 CrossRef CAS PubMed.
- Z. Zhao, A. Ukidve, J. Kim and S. Mitragotri, Cell, 2020, 181, 151–167 CrossRef CAS PubMed.
- N. Wettschureck, B. Strilic and S. Offermanns, Physiol. Rev., 2019, 99, 1467–1525 CrossRef CAS.
- A. Jambusaria, Z. Hong, L. Zhang, S. Srivastava, A. Jana, P. T. Toth, Y. Dai, A. B. Malik and J. Rehman, eLife, 2020, 9, e51413 CrossRef CAS.
- Y. N. Zhang, W. Poon, A. J. Tavares, I. D. McGilvray and W. C. W. Chan, J. Controlled Release, 2016, 240, 332–348 CrossRef CAS PubMed.
- E. A. Azzopardi, E. L. Ferguson and D. W. Thomas, J. Antimicrob. Chemother., 2013, 68, 257–274 CrossRef CAS PubMed.
- J. Fang, W. Islam and H. Maeda, Adv. Drug Delivery Rev., 2020, 157, 142–160 CrossRef CAS.
- I. B. Kelly, 3rd, R. B. Fletcher, J. R. McBride, S. M. Weiss and C. L. Duvall, ACS Appl. Mater. Interfaces, 2020, 12, 39602–39611 CrossRef PubMed.
- J. M. Morachis, E. A. Mahmoud and A. Almutairi, Pharmacol. Rev., 2012, 64, 505–519 CrossRef CAS PubMed.
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Nat. Rev. Cancer, 2017, 17, 20–37 CrossRef CAS PubMed.
- S. Sindhwani, A. M. Syed, J. Ngai, B. R. Kingston, L. Maiorino, J. Rothschild, P. MacMillan, Y. Zhang, N. U. Rajesh, T. Hoang, J. L. Y. Wu, S. Wilhelm, A. Zilman, S. Gadde, A. Sulaiman, B. Ouyang, Z. Lin, L. Wang, M. Egeblad and W. C. W. Chan, Nat. Mater., 2020, 19, 566–575 CrossRef CAS PubMed.
- P. L. Rodriguez, T. Harada, D. A. Christian, D. A. Pantano, R. K. Tsai and D. E. Discher, Science, 2013, 339, 971–975 CrossRef CAS PubMed.
- V. G. Deepagan, H. Ko, S. Kwon, N. V. Rao, S. K. Kim, W. Um, S. Lee, J. Min, J. Lee, K. Y. Choi, S. Shin, M. Suh and J. H. Park, Nano Lett., 2018, 18, 2637–2644 CrossRef CAS PubMed.
- Q. Huang, Y. Yang, T. Zhao, Q. Chen, M. Liu, S. Ji, Y. Zhu, Y. Yang, J. Zhang, H. Zhao, Y. Nan and K. Ai, Bioact. Mater., 2023, 21, 381–393 CrossRef CAS PubMed.
- H. K. Mandl, E. Quijano, H. W. Suh, E. Sparago, S. Oeck, M. Grun, P. M. Glazer and W. M. Saltzman, J. Controlled Release, 2019, 314, 92–101 CrossRef CAS.
- X. Li, E. C. Montague, A. Pollinzi, A. Lofts and T. Hoare, Small, 2022, 18, 2104632 CrossRef CAS.
- M. Naito, Y. Watanuki, K. Toh, J. Yum, B. S. Kim, K. Taniwaki, S. Ogura, H. Ishida, M. Cho, H. Chaya, K. Miyajima, Y. Yamasaki, K. Osada, K. Minegishi, Y. Aoki and K. Miyata, J. Controlled Release, 2022, 347, 607–614 CrossRef CAS PubMed.
- S. Raj, S. Khurana, R. Choudhari, K. K. Kesari, M. A. Kamal, N. Garg, J. Ruokolainen, B. C. Das and D. Kumar, Semin. Cancer Biol., 2021, 69, 166–177 CrossRef CAS PubMed.
- J. A. Champion and S. Mitragotri, Pharm. Res., 2009, 26, 244–249 CrossRef CAS PubMed.
- S. Nejati, E. M. Vadeghani, S. Khorshidi and A. Karkhaneh, Eur. Polym. J., 2020, 122, 109353 CrossRef CAS.
- R. Agarwal, V. Singh, P. Jurney, L. Shi, S. V. Sreenivasan and K. Roy, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17247–17252 CrossRef CAS.
- C. Wang, J. Xiao, X. Hu, Q. Liu, Y. Zheng, Z. Kang, D. Guo, L. Shi and Y. Liu, Adv. Healthcare Mater., 2023, 12, e2201889 CrossRef PubMed.
- Z. Guo, L. Yang, M. Chen, X. Wen, H. Liu, J. Li, D. Xu, Y. An, C. Shi, J. Li, X. Su, Z. Li, T. Liu, R. Zhuang, N. Zheng, H. Zhu and X. Zhang, Nano Res., 2020, 13, 173–182 CrossRef.
- H. Wang, S. Lin, S. Wang, Z. Jiang, T. Ding, X. Wei, Y. Lu, F. Yang and C. Zhan, Nano Lett., 2022, 22, 6516–6522 CrossRef CAS PubMed.
- M.-R. Choi, R. Bardhan, K. J. Stanton-Maxey, S. Badve, H. Nakshatri, K. M. Stantz, N. Cao, N. J. Halas and S. E. Clare, Cancer Nanotechnol., 2012, 3, 47–54 CrossRef CAS PubMed.
- F. Chen and G. Huang, Eur. J. Med. Chem., 2019, 182, 111612 CrossRef CAS PubMed.
- Z. Geng, L. Wang, K. Liu, J. Liu and W. Tan, Angew. Chem., Int. Ed., 2021, 60, 15459–15465 CrossRef CAS PubMed.
- Y. Niu, Y. Yang, Z. Yang, X. Wang, P. Zhang, L. Lv, Y. Liu, Y. Liu and Y. Zhou, Nano Today, 2022, 45, 101529 CrossRef CAS.
- S. Zinn, R. Vazquez-Lombardi, C. Zimmermann, P. Sapra, L. Jermutus and D. Christ, Nat. Cancer, 2023, 4, 165–180 CrossRef PubMed.
- Z. Ge, L. Guo, G. Wu, J. Li, Y. Sun, Y. Hou, J. Shi, S. Song, L. Wang, C. Fan, H. Lu and Q. Li, Small, 2020, 16, e1904857 CrossRef PubMed.
- A. B. Silver, E. K. Leonard, J. R. Gould and J. B. Spangler, Trends Pharmacol. Sci., 2021, 42, 1064–1081 CrossRef CAS.
- D. Bertheloot, C. W. Wanderley, A. H. Schneider, L. D. Schiffelers, J. D. Wuerth, J. M. Tödtmann, S. Maasewerd, I. Hawwari, F. Duthie, C. Rohland, L. S. Ribeiro, L.-M. Jenster, N. Rosero, Y. M. Tesfamariam, F. Q. Cunha, F. I. Schmidt and B. S. Franklin, EMBO Mol. Med., 2022, 14, e15415 CrossRef CAS PubMed.
- J. Andrieu, F. Re, L. Russo and F. Nicotra, J. Drug Target., 2019, 27, 555–565 CrossRef CAS PubMed.
- N. Svensen, J. G. Walton and M. Bradley, Trends Pharmacol. Sci., 2012, 33, 186–192 CrossRef CAS PubMed.
- N. Dammes, M. Goldsmith, S. Ramishetti, J. L. J. Dearling, N. Veiga, A. B. Packard and D. Peer, Nat. Nanotechnol., 2021, 16, 1030–1038 CrossRef CAS PubMed.
- W. Li, Z. Su, M. Hao, C. Ju and C. Zhang, J. Controlled Release, 2020, 328, 313–324 CrossRef CAS PubMed.
- Y. Liu, J. Luo, X. Chen, W. Liu and T. Chen, Nanomicro Lett., 2019, 11, 100 CAS.
- C. M. Hu, L. Zhang, S. Aryal, C. Cheung, R. H. Fang and L. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10980–10985 CrossRef CAS PubMed.
- R. H. Fang, W. Gao and L. Zhang, Nat. Rev. Clin. Oncol., 2023, 20, 33–48 CrossRef PubMed.
- Q. Zhang, D. Dehaini, Y. Zhang, J. Zhou, X. Chen, L. Zhang, R. H. Fang, W. Gao and L. Zhang, Nat. Nanotechnol., 2018, 13, 1182–1190 CrossRef CAS PubMed.
- Y. Zhang, J. Zhang, W. Chen, P. Angsantikul, K. A. Spiekermann, R. H. Fang, W. Gao and L. Zhang, J. Controlled Release, 2017, 263, 185–191 CrossRef CAS PubMed.
- C.-M. J. Hu, R. H. Fang, J. Copp, B. T. Luk and L. Zhang, Nat. Nanotechnol., 2013, 8, 336–340 CrossRef CAS PubMed.
- A. V. Kroll, R. H. Fang, Y. Jiang, J. Zhou, X. Wei, C. L. Yu, J. Gao, B. T. Luk, D. Dehaini, W. Gao and L. Zhang, Adv. Mater., 2017, 29, 1703969 CrossRef PubMed.
- X. Xie, X. Hu, Q. Li, M. Yin, H. Song, J. Hu, L. Wang, C. Fan and N. Chen, Nano Lett., 2020, 20, 5228–5235 CrossRef CAS PubMed.
- L. Liu, X. Bai, M.-V. Martikainen, A. Kårlund, M. Roponen, W. Xu, G. Hu, E. Tasciotti and V.-P. Lehto, Nat. Commun., 2021, 12, 5726 CrossRef CAS PubMed.
- K. Cheng, R. Zhao, Y. Li, Y. Qi, Y. Wang, Y. Zhang, H. Qin, Y. Qin, L. Chen, C. Li, J. Liang, Y. Li, J. Xu, X. Han, G. J. Anderson, J. Shi, L. Ren, X. Zhao and G. Nie, Nat. Commun., 2021, 12, 2041 CrossRef CAS PubMed.
- J. S. Brenner, D. C. Pan, J. W. Myerson, O. A. Marcos-Contreras, C. H. Villa, P. Patel, H. Hekierski, S. Chatterjee, J.-Q. Tao, H. Parhiz, K. Bhamidipati, T. G. Uhler, E. D. Hood, R. Y. Kiseleva, V. S. Shuvaev, T. Shuvaeva, M. Khoshnejad, I. Johnston, J. V. Gregory, J. Lahann, T. Wang, E. Cantu, W. M. Armstead, S. Mitragotri and V. Muzykantov, Nat. Commun., 2018, 9, 2684 CrossRef PubMed.
- Q. Li, Y. Zhou, W. He, X. Ren, M. Zhang, Y. Jiang, Z. Zhou and Y. Luan, J. Controlled Release, 2021, 338, 33–45 CrossRef CAS PubMed.
- F. Zhang, J. Zhuang, Z. Li, H. Gong, B. E.-F. de Ávila, Y. Duan, Q. Zhang, J. Zhou, L. Yin, E. Karshalev, W. Gao, V. Nizet, R. H. Fang, L. Zhang and J. Wang, Nat. Mater., 2022, 21, 1324–1332 CrossRef CAS PubMed.
- S. A. Dilliard, Q. Cheng and D. J. Siegwart, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2109256118 CrossRef CAS PubMed.
- R. N. Kularatne, R. M. Crist and S. T. Stern, Pharmaceuticals, 2022, 15, 897 CrossRef CAS PubMed.
- I. Menon, M. Zaroudi, Y. Zhang, E. Aisenbrey and L. Hui, Mater. Today Adv., 2022, 16, 100299 CrossRef CAS.
- S. Liu, Q. Cheng, T. Wei, X. Yu, L. T. Johnson, L. Farbiak and D. J. Siegwart, Nat. Mater., 2021, 20, 701–710 CrossRef CAS.
- M. Qiu, Y. Tang, J. Chen, R. Muriph, Z. Ye, C. Huang, J. Evans, E. P. Henske and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2116271119 CrossRef CAS PubMed.
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope, T. D. Madden, B. L. Mui, S. C. Semple, Y. K. Tam, M. Ciufolini, D. Witzigmann, J. A. Kulkarni, R. van der Meel and P. R. Cullis, Nat. Nanotechnol., 2019, 14, 1084–1087 CrossRef CAS PubMed.
- A. Algarni, E. H. Pilkington, E. J. A. Suys, H. Al-Wassiti, C. W. Pouton and N. P. Truong, Biomater. Sci., 2022, 10, 2940–2952 RSC.
- D. Zukancic, E. J. A. Suys, E. H. Pilkington, A. Algarni, H. Al-Wassiti and N. P. Truong, Pharmaceutics, 2020, 12, 1068 CrossRef CAS PubMed.
- T. Wei, Q. Cheng, Y. L. Min, E. N. Olson and D. J. Siegwart, Nat. Commun., 2020, 11, 3232 CrossRef CAS PubMed.
- X. Wang, S. Liu, Y. Sun, X. Yu, S. M. Lee, Q. Cheng, T. Wei, J. Gong, J. Robinson, D. Zhang, X. Lian, P. Basak and D. J. Siegwart, Nat. Protoc., 2023, 18, 265–291 CrossRef CAS PubMed.
- S. G. Jadhav and S. F. Dowdy, Nat. Mater., 2021, 20, 575–577 CrossRef CAS PubMed.
- Q. Xiao, M. Zoulikha, M. Qiu, C. Teng, C. Lin, X. Li, M. A. Sallam, Q. Xu and W. He, Adv. Drug Delivery Rev., 2022, 186, 114356 CrossRef CAS PubMed.
- R. Madathiparambil Visalakshan, L. E. González García, M. R. Benzigar, A. Ghazaryan, J. Simon, A. Mierczynska-Vasilev, T. D. Michl, A. Vinu, V. Mailänder, S. Morsbach, K. Landfester and K. Vasilev, Small, 2020, 16, 2000285 CrossRef CAS PubMed.
- T. Bewersdorff, E. A. Glitscher, J. Bergueiro, M. Eravci, E. Miceli, A. Haase and M. Calderón, Mater. Sci. Eng., C, 2020, 117, 111270 CrossRef CAS PubMed.
- A. Piloni, C. K. Wong, F. Chen, M. Lord, A. Walther and M. H. Stenzel, Nanoscale, 2019, 11, 23259–23267 RSC.
- S. M. Pustulka, K. Ling, S. L. Pish and J. A. Champion, ACS Appl. Mater. Interfaces, 2020, 12, 48284–48295 CrossRef CAS PubMed.
- W. Kim, N. K. Ly, Y. He, Y. Li, Z. Yuan and Y. Yeo, Adv. Drug Delivery Rev., 2023, 192, 114635 CrossRef CAS PubMed.
- Z. P. Lin, W. Ngo, S. M. Mladjenovic, J. L. Y. Wu and W. C. W. Chan, Nano Lett., 2023, 23, 1003–1009 CrossRef CAS PubMed.
- Y. Wang, J. Yu, Z. Luo, Q. Shi, G. Liu, F. Wu, Z. Wang, Y. Huang and D. Zhou, Adv. Mater., 2021, 33, 2170304 CrossRef CAS.
- N. Krishnan, R. H. Fang and L. Zhang, Adv. Drug Delivery Rev., 2021, 179, 114006 CrossRef CAS PubMed.
- K. A. Hughes, B. Misra, M. Maghareh and S. Bobbala, Nano Res., 2023, 16, 6974–6990 CrossRef CAS PubMed.
- Y. Liu, Y. Wang, Y. Yao, J. Zhang, W. Liu, K. Ji, X. Wei, Y. Wang, X. Liu, S. Zhang, J. Wang and Z. Gu, Angew. Chem., Int. Ed., 2023, 62, e202303097 CrossRef CAS PubMed.
- T. Ji, J. Lang, B. Ning, F. Qi, H. Wang, Y. Zhang, R. Zhao, X. Yang, L. Zhang, W. Li, X. Shi, Z. Qin, Y. Zhao and G. Nie, Adv. Mater., 2019, 31, e1804395 Search PubMed.
- T. Wang, H. Lei, X. Li, N. Yang, C. Ma, G. Li, X. Gao, J. Ge, Z. Liu, L. Cheng and G. Chen, Small, 2023, 19, e2206982 CrossRef PubMed.
- G. C. Terstappen, A. H. Meyer, R. D. Bell and W. Zhang, Nat. Rev. Drug Discovery, 2021, 20, 362–383 CrossRef CAS PubMed.
- A. Duro-Castano, D. Moreira Leite, J. Forth, Y. Deng, D. Matias, C. Noble Jesus and G. Battaglia, Adv. Drug Delivery Rev., 2020, 160, 52–77 CrossRef CAS PubMed.
- M. Agrawal, S. Saraf, S. Saraf, S. K. Dubey, A. Puri, R. J. Patel, Ajazuddin, V. Ravichandiran, U. S. Murty and A. Alexander, J. Controlled Release, 2020, 321, 372–415 CrossRef CAS PubMed.
- G. Zhong, H. Long, T. Zhou, Y. Liu, J. Zhao, J. Han, X. Yang, Y. Yu, F. Chen and S. Shi, Biomaterials, 2022, 288, 121690 CrossRef CAS PubMed.
- Y. Guo, R. Liu, L. Zhou, H. Zhao, F. Lv, L. Liu, Y. Huang, H.-W. Zhang, C. Yu and S. Wang, Nano Today, 2020, 35, 100969 CrossRef CAS.
- Y. Zhao, S. Tian, J. Zhang, X. Cheng, W. Huang, G. Cao, Y.-Z. Chang, H. Wang, G. Nie and W. Qiu, Nano Today, 2022, 44, 101457 CrossRef CAS.
- Y. Li, Y. Pan, Y. Wang, Z. Jiang, O. U. Akakuru, M. Li, X. Zhang, B. Yuan, J. Xing, L. Luo, D. Larhammar, A. Wu and J. Li, Nano Today, 2022, 44, 101465 CrossRef CAS.
- Y. Liu, D. Zhang, Y. An, Y. Sun, J. Li, M. Zheng, Y. Zou and B. Shi, Nano Today, 2023, 49, 101790 CrossRef CAS.
- H. Chen, M. Zhou, Y. Zeng, T. Miao, H. Luo, Y. Tong, M. Zhao, R. Mu, J. Gu, S. Yang and L. Han, Adv. Sci., 2022, 9, e2105854 CrossRef PubMed.
- Z. Zhang, J. Guan, Z. Jiang, Y. Yang, J. Liu, W. Hua, Y. Mao, C. Li, W. Lu, J. Qian and C. Zhan, Nat. Commun., 2019, 10, 3561 CrossRef PubMed.
- T. Kim, H. J. Kim, W. Choi, Y. M. Lee, J. H. Pyo, J. Lee, J. Kim, J. Kim, J.-H. Kim, C. Kim and W. J. Kim, Nat. Biomed. Eng., 2023, 7, 149–163 CrossRef CAS PubMed.
- M. P. A. Ferreira, S. Ranjan, A. M. R. Correia, E. M. Mäkilä, S. M. Kinnunen, H. Zhang, M.-A. Shahbazi, P. V. Almeida, J. J. Salonen, H. J. Ruskoaho, A. J. Airaksinen, J. T. Hirvonen and H. A. Santos, Biomaterials, 2016, 94, 93–104 CrossRef CAS PubMed.
- N. El Harane, A. Kervadec, V. Bellamy, L. Pidial, H. J. Neametalla, M.-C. Perier, B. Lima Correa, L. Thiébault, N. Cagnard, A. Duché, C. Brunaud, M. Lemitre, J. Gauthier, A. T. Bourdillon, M. P. Renault, Y. Hovhannisyan, S. Paiva, A. R. Colas, O. Agbulut, A. Hagège, J.-S. Silvestre, P. Menasché and N. K. E. Renault, Eur. Heart J., 2018, 39, 1835–1847 CrossRef CAS PubMed.
- S. P. Kwon, B.-H. Hwang, E.-H. Park, H. Y. Kim, J.-R. Lee, M. Kang, S. Y. Song, M. Jung, H. S. Sohn, E. Kim, C. W. Kim, K. Y. Lee, G. C. Oh, E. Choo, S. Lim, Y. Chung, K. Chang and B.-S. Kim, Small, 2021, 17, 2101207 CrossRef CAS PubMed.
- Y. Qi, J. Li, Q. Nie, M. Gao, Q. Yang, Z. Li, Q. Li, S. Han, J. Ding, Y. Li and J. Zhang, Biomaterials, 2021, 275, 120952 CrossRef CAS PubMed.
- Z. Zhang, D. Zhou, X. Luan, X. Wang, Z. Zhu, W. Luo, J. Yang, S. Tang and Y. Song, ACS Nano, 2023, 17, 9313–9325 CrossRef CAS PubMed.
- T. Yang, A. Wang, D. Nie, W. Fan, X. Jiang, M. Yu, S. Guo, C. Zhu, G. Wei and Y. Gan, Nat. Commun., 2022, 13, 6649 CrossRef CAS PubMed.
- S. Kimura, I. A. Khalil, Y. H. A. Elewa and H. Harashima, J. Controlled Release, 2021, 330, 753–764 CrossRef CAS PubMed.
- D. Chen, S. Liu, D. Chen, J. Liu, J. Wu, H. Wang, Y. Su, G. Kwak, X. Zuo, D. Rao, H. Cui, C. Shu and J. S. Suk, Angew. Chem., Int. Ed., 2021, 60, 15225–15229 CrossRef CAS PubMed.
- M. M. Abd Elwakil, I. A. Khalil, Y. H. A. Elewa, K. Kusumoto, Y. Sato, N. Shobaki, Y. Kon and H. Harashima, Adv. Funct. Mater., 2019, 29, 1807677 CrossRef.
- L. Zhou, S. Tang, F. Li, Y. Wu, S. Li, L. Cui, J. Luo, L. Yang, Z. Ren, J. Zhang, D. Zhou, J. Jiang, X. Yang, X. Zhou and Y. Wu, Biomaterials, 2022, 287, 121686 CrossRef CAS PubMed.
- H.-T. Cheng, H.-C. Huang, T.-Y. Lee, Y.-H. Liao, Y.-H. Sheng, P.-R. Jin, K.-W. Huang, L.-H. Chen, Y.-T. Chen, Z.-Y. Liu, T.-C. Lin, H.-C. Wang, C.-H. Chao, I. P. Juang, C.-T. Su, K.-H. Huang, S.-L. Lin, J. Wang, Y.-C. Sung and Y. Chen, J. Controlled Release, 2022, 346, 169–179 CrossRef CAS PubMed.
- Z. Huang, C. Chun and X. Li, J. Controlled Release, 2023, 358, 368–381 CrossRef CAS PubMed.
- M. Herrera-Barrera, R. C. Ryals, M. Gautam, A. Jozic, M. Landry, T. Korzun, M. Gupta, C. Acosta, J. Stoddard, R. Reynaga, W. Tschetter, N. Jacomino, O. Taratula, C. Sun, A. K. Lauer, M. Neuringer and G. Sahay, Sci. Adv., 2023, 9, eadd4623 CrossRef PubMed.
- J. Ye, J. Jiang, Z. Zhou, Z. Weng, Y. Xu, L. Liu, W. Zhang, Y. Yang, J. Luo and X. Wang, ACS Nano, 2021, 15, 13692–13702 CrossRef CAS PubMed.
- T. Chen, F. Pan, G. Luo, K. Jiang, H. Wang, T. Ding, W. Li, C. Zhan and X. Wei, Nano Today, 2022, 46, 101609 CrossRef CAS.
- W. Li, J. H. Qiu, X. L. Li, S. Aday, J. D. Zhang, G. Conley, J. Xu, J. Joseph, H. Y. Lan, R. Langer, R. Mannix, J. M. Karp and N. Joshi, Sci. Adv., 2021, 7, 16 Search PubMed.
- X. Guo, G. Deng, J. Liu, P. Zou, F. Du, F. Liu, A. T. Chen, R. Hu, M. Li, S. Zhang, Z. Tang, L. Han, J. Liu, K. N. Sheth, Q. Chen, X. Gou and J. Zhou, ACS Nano, 2018, 12, 8723–8732 CrossRef CAS PubMed.
- H. Wu, B. Peng, F. S. Mohammed, X. Gao, Z. Qin, K. N. Sheth, J. Zhou and Z. Jiang, Small, 2022, 18, 2107126 CrossRef CAS PubMed.
- C. Y. Chien, L. Xu, C. P. Pacia, Y. Yue and H. Chen, Sci. Rep., 2022, 12, 16147 CrossRef CAS PubMed.
- Y. Guo, H. Lee, Z. Fang, A. Velalopoulou, J. Kim, M. B. Thomas, J. Liu, R. G. Abramowitz, Y. Kim, A. F. Coskun, D. P. Krummel, S. Sengupta, T. J. MacDonald and C. Arvanitis, Sci. Adv., 2021, 7, eabf7390 CrossRef CAS PubMed.
- J. Wu, Z. Zhu, W. Liu, Y. Zhang, Y. Kang, J. Liu, C. Hu, R. Wang, M. Zhang, L. Chen and L. Shao, ACS Nano, 2022, 16, 15627–15652 CrossRef CAS PubMed.
- A. R. Rezai, M. Ranjan, P.-F. D’Haese, M. W. Haut, J. Carpenter, U. Najib, R. I. Mehta, J. L. Chazen, Z. Zibly, J. R. Yates, S. L. Hodder and M. Kaplitt, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 9180–9182 CrossRef CAS PubMed.
- R. Pala, V. T. Anju, M. Dyavaiah, S. Busi and S. M. Nauli, Int. J. Nanomed., 2020, 15, 3741–3769 CrossRef CAS PubMed.
- R. S. Soumya and K. G. Raghu, J. Cardiol., 2023, 81, 10–18 CrossRef PubMed.
- F. Passaro, C. G. Tocchetti, G. Spinetti, F. Paudice, L. Ambrosone, C. Costagliola, F. Cacciatore, P. Abete and G. Testa, Adv. Drug Delivery Rev., 2021, 174, 461–481 CrossRef CAS PubMed.
- H. Kim, D. Mun, J. Y. Kang, S. H. Lee, N. Yun and B. Joung, Mol. Ther. Nucleic. Acids, 2021, 24, 1024–1032 CrossRef CAS PubMed.
- B. Banik, B. Surnar, B. W. Askins, M. Banerjee and S. Dhar, ACS Appl. Mater. Interfaces, 2020, 12, 6852–6862 CrossRef CAS PubMed.
- R. Sun, X. Wang, Y. Nie, A. Hu, H. Liu, K. Zhang, L. Zhang, Q. Wu, K. Li, C. Liu, H. Zhang, B. Zheng, H. Li, H. Xu, R. Xu, H. Fu, L. Dai, R. Jin and Y. Guo, Acta Biomater., 2022, 146, 421–433 CrossRef CAS PubMed.
- J. Qiu, X.-J. Liu, B.-A. You, N. Ren and H. Liu, Small, 2023, 19, 2206487 CrossRef CAS PubMed.
- H. Yang, X. Qin, H. Wang, X. Zhao, Y. Liu, H. T. Wo, C. Liu, M. Nishiga, H. Chen, J. Ge, N. Sayed, O. J. Abilez, D. Ding, S. C. Heilshorn and K. Li, ACS Nano, 2019, 13, 9880–9894 CrossRef CAS PubMed.
- C. Liu, L. Chen, Y. Ma, K. Hu, P. Wu, L. Pan, H. Chen, L. Li, H. Hu and J. Zhang, Theranostics, 2021, 11, 8550–8569 CrossRef CAS PubMed.
- E. Trefts, M. Gannon and D. H. Wasserman, Curr. Biol., 2017, 27, R1147–r1151 CrossRef CAS PubMed.
- R. Bottger, G. Pauli, P. H. Chao, N. Al Fayez, L. Hohenwarter and S. D. Li, Adv. Drug Delivery Rev., 2020, 154–155, 79–101 CrossRef CAS PubMed.
- Y. Sato, Y. Kinami, K. Hashiba and H. Harashima, J. Controlled Release, 2020, 322, 217–226 CrossRef CAS PubMed.
- K. R. Kim, J. Kim, J. H. Back, J. E. Lee and D. R. Ahn, ACS Nano, 2022, 16, 7331–7343 CrossRef CAS PubMed.
- Y. Jiang, J. Hardie, Y. Liu, M. Ray, X. Luo, R. Das, R. F. Landis, M. E. Farkas and V. M. Rotello, J. Controlled Release, 2018, 283, 235–240 CrossRef CAS PubMed.
- T. J. Harris, J. J. Green, P. W. Fung, R. Langer, D. G. Anderson and S. N. Bhatia, Biomaterials, 2010, 31, 998–1006 CrossRef CAS PubMed.
- S. T. LoPresti, M. L. Arral, N. Chaudhary and K. A. Whitehead, J. Controlled Release, 2022, 345, 819–831 CrossRef CAS PubMed.
- A. J. Sinegra, M. Evangelopoulos, J. Park, Z. Huang and C. A. Mirkin, Nano Lett., 2021, 21, 6584–6591 CrossRef CAS PubMed.
- X. Zhao, J. Chen, M. Qiu, Y. Li, Z. Glass and Q. Xu, Angew. Chem., Int. Ed., 2020, 59, 20083–20089 CrossRef CAS PubMed.
- N. Wathoni, L. E. Puluhulawa, I. M. Joni, M. Muchtaridi, A. F. A. Mohammed, K. M. Elamin, T. Milanda and D. Gozali, Drug Delivery, 2022, 29, 2959–2970 CrossRef CAS PubMed.
- R. Y. K. Chang and H.-K. Chan, Nat. Biomed. Eng., 2021, 5, 949–950 CrossRef CAS PubMed.
- A. Garcia-Fernandez, F. Sancenon and R. Martinez-Manez, Adv. Drug Delivery Rev., 2021, 177, 113953 CrossRef CAS PubMed.
- A. Hidalgo, A. Cruz and J. Pérez-Gil, Eur. J. Pharm. Biopharm., 2015, 95, 117–127 CrossRef CAS PubMed.
- Q. Liu, J. Guan, R. Song, X. Zhang and S. Mao, Acta Biomater., 2022, 140, 76–87 CrossRef CAS PubMed.
- N. Trac, A. Ashraf, J. Giblin, S. Prakash, S. Mitragotri and E. J. Chung, ACS Nano, 2023, 17, 6165–6177 CrossRef CAS PubMed.
- Y. Huang, J. Wang, K. Jiang and E. J. Chung, J. Controlled Release, 2021, 334, 127–137 CrossRef CAS PubMed.
- E. Arif and D. Nihalani, Postdoc J., 2013, 1, 33–45 Search PubMed.
- M. A. Felmlee, R. A. Dave and M. E. Morris, AAPS J., 2013, 15, 278–287 CrossRef CAS PubMed.
- S. J. Han, R. M. Williams, V. D'Agati, E. A. Jaimes, D. A. Heller and H. T. Lee, Kidney Int., 2020, 98, 76–87 CrossRef CAS PubMed.
- A. Wischnjow, D. Sarko, M. Janzer, C. Kaufman, B. Beijer, S. Brings, U. Haberkorn, G. Larbig, A. Kübelbeck and W. Mier, Bioconjugate Chem., 2016, 27, 1050–1057 CrossRef CAS PubMed.
- Y. Huang, K. Jiang, X. Zhang and E. J. Chung, Bioeng. Transl. Med., 2020, 5, e10173 CAS.
- J. N. Chu and G. Traverso, Nat. Rev. Gastroenterol. Hepatol., 2022, 19, 219–238 CrossRef CAS PubMed.
- B. Zhang, H. Pan, Z. Chen, T. Yin, M. Zheng and L. Cai, Sci. Adv., 2023, 9, eadc8978 CrossRef CAS PubMed.
- X. Gao, L. Li, X. Cai, Q. Huang, J. Xiao and Y. Cheng, Biomaterials, 2021, 265, 120404 CrossRef CAS PubMed.
- J. R. Melamed, S. S. Yerneni, M. L. Arral, S. T. LoPresti, N. Chaudhary, A. Sehrawat, H. Muramatsu, M. G. Alameh, N. Pardi, D. Weissman, G. K. Gittes and K. A. Whitehead, Sci. Adv., 2023, 9, eade1444 CrossRef CAS PubMed.
- J. Wang, Y. Li and G. Nie, Nat. Rev. Mater., 2021, 6, 766–783 CrossRef CAS PubMed.
- M. Colasuonno, A. L. Palange, R. Aid, M. Ferreira, H. Mollica, R. Palomba, M. Emdin, M. Del Sette, C. Chauvierre, D. Letourneur and P. Decuzzi, ACS Nano, 2018, 12, 12224–12237 CrossRef CAS PubMed.
- C. Kang, S. Gwon, C. Song, P. M. Kang, S.-C. Park, J. Jeon, D. W. Hwang and D. Lee, ACS Nano, 2017, 11, 6194–6203 CrossRef CAS PubMed.
- E. Jung, C. Kang, J. Lee, D. Yoo, D. W. Hwang, D. Kim, S.-C. Park, S. K. Lim, C. Song and D. Lee, ACS Nano, 2018, 12, 392–401 CrossRef CAS PubMed.
- N. T. Blum, C. M. Gyorkos, S. J. Narowetz, E. N. Mueller and A. P. Goodwin, ACS Appl. Mater. Interfaces, 2019, 11, 36324–36332 CrossRef CAS PubMed.
- H. Havel, G. Finch, P. Strode, M. Wolfgang, S. Zale, I. Bobe, H. Youssoufian, M. Peterson and M. Liu, AAPS J., 2016, 18, 1373–1378 CrossRef CAS PubMed.
- F. Farjadian, A. Ghasemi, O. Gohari, A. Roointan, M. Karimi and M. R. Hamblin, Nanomedicine, 2019, 14, 93–126 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |