Recent progress in nucleic acid detection with CRISPR
Frank X.
Liu
a,
Johnson Q.
Cui
a,
Zhihao
Wu
 b and
Shuhuai
Yao
b and
Shuhuai
Yao
 *ac
*ac
aDepartment of Mechanical and Aerospace Engineering, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong. E-mail: meshyao@ust.hk
bIIP-Advanced Materials, Interdisciplinary Program Office (IPO), Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong
cDepartment of Chemical and Biological Engineering, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong
First published on 24th January 2023
Abstract
Recent advances in CRISPR-based biotechnologies have greatly expanded our capabilities to repurpose CRISPR for the development of molecular diagnostic systems. The key attribute that allows CRISPR to be widely utilized is its programmable and highly specific nature. In this review, we first illustrate the principle of the class 2 CRISPR nucleases for molecular diagnostics which originates from their immunologic defence systems. Next, we present the CRISPR-based schemes in the application of diagnostics with amplification-assisted or amplification-free strategies. By highlighting some of the recent advances we interpret how general bioengineering methodologies can be integrated with CRISPR. Finally, we discuss the challenges and exciting prospects for future CRISPR-based biosensing development. We hope that this review will guide the reader to systematically learn the start-of-the-art development of CRISPR-mediated nucleic acid detection and understand how to apply the CRISPR nucleases with different design concepts to more general applications in diagnostics and beyond.
1 Introduction
Detection of nucleic acids is of great importance for the identification of genetic diseases and pathogenic diagnosis, as well as its use in therapeutics. In recent decades, large-scale recurring disease outbreaks caused by emerging viruses, such as SARS, H1N1, Ebola, Zika, and SARS-CoV-2,1 highlight that the current gold standard approach polymerase chain reaction (PCR), which requires centralized diagnostic facilities, is unable to respond quickly to these pandemic or epidemic crises. The response time is significantly delayed by sample collection, transport, preparation, and slow reaction time. As guided by the World Health Organisation (WHO), the ideal pathogen detection should be affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-users (ASSURED). Therefore, there is an urgent need for deployable and miniaturized point-of-care testing (POCT) platforms to address viral detection.Over the past few years, the emerging clustered regularly interspaced short palindromic repeats (CRISPR) technology has revolutionized the field of gene editing and molecular diagnostics.2 Originally, the CRISPR–Cas system was a prokaryotic adaptive immune system used against viruses and plasmids in most archaea and bacteria.3 In the immune system, the CRISPR array inserts a portion of the target DNA sequence next to the protospacer adjacent motif (PAM) sequence to become a spacer where the CRISPR array is transcribed as a single transcript containing the spacer sequence and parts of the flanking repeats.4 The CRISPR RNA (crRNA) coupled with Cas nucleases recognizes the protospacer sequence in the invading objects, then the invading viruses or plasmids are cleaved and deactivated by the Cas nucleases.5–9 Although this was first discovered in the 1980s,10 it was not until 2012 that the first in vitro demonstration of an engineered Cas9 system was used to specifically cut the targeted sequence by fusing RNA molecules into a single-guide RNA (sgRNA).11 By programmable design of the spacer of the sgRNA, the CRISPR coupled with Cas effectors offer unparalleled potential for genome-editing applications such as sequencing and molecular diagnostics.11–16 The subsequent discovery of the cis and trans-cleavage ability of Cas12 and Cas13 has shown more values in molecular diagnostics where Cas proteins trans-cleave the surrounding single-stranded reporters after the cis base-pair matching between the guided RNA spacer and targeted protospacer sequences. The applications of the CRISPR toolbox in molecular diagnostics imply its merits as follows: (i) programmable and easy to design, (ii) near-ambient operational temperature, and (iii) high sensitivity to single mutation variants.
The first diagnostic application of CRISPR was reported in 2016. With the preamplification of NASBA, the Cas9 achieved superior specific and sensitive detection of the Zika virus at a single nucleotide polymorphism (SNP).12 In the same year, the collateral activity of Cas13a was reported,17 and employed for direct RNA detection without amplification.18 The amplification-associated and amplification-free detection ability of trace amounts of nucleic acids by the CRISPR–Cas system has since been harnessed for the next generation of nucleic acid detection. The CRISPR-driven diagnostic tools have been intensively developed as an alternative to the PCR test. Several of these tools including LAMP-DETECTR and LAMP-SHERLOCK have been approved by the US Food Drug Administration (FDA).19
CRISPR-mediated molecular diagnostics has been a hot topic in recent reviews.20–29 These review papers provide different aspects of recent advances in CRISPR diagnostic technologies including understanding the cellular biology of CRISPR and principles for diagnostics,20,22,24 the transition of CRISPR technology to molecular diagnostics from tube to device for POCT,27,29 strategies and technologies that can be coupled with CRISPR such as microfluidics and nanotechnologies,23,25 and the application for COVID-19 diagnostics to meet the urgent pandemic emergency.21,28 Nevertheless, the design principles of how the CRISPR effectors work with different strategies and technologies to meet the sensing and quantification demands have not been elaborated and clearly outlined. More importantly, with the continuous expansion of CRISPR-based tools in the past few years, it is important to summarize and update new working principles and trends of this field. In this review, we tend to focus on the selection preference of Cas effectors, amplification methods, and reporting schemes. We summarize the development of CRISPR-based diagnostics in two main categories in coupling with nucleic acid amplification-assisted and amplification-free (signal amplification) techniques, respectively. With the aid of theory and examples, we present a synthesized overview from the design principles to the detection techniques in detail to illustrate how different CRISPR effectors can be integrated with nucleic acid detection protocols.
The review is structured as follows: first, we provide a short summary of the mechanisms and characteristics of class 2 CRISPR–Cas systems including conjugation strategies and cleavage purposes that guide the design concepts. Second, we summarize the isothermal preamplification schemes associated with CRISPR, illustrating the advantages and disadvantages of these methods in different scenarios (Tables 3–5). Third, we review the amplification-free (i.e., without polymerase adoption) assays in combination with CRISPR indicating that the CRISPR systems are promising for facile detection without target molecule amplification (Table 6). Lastly, from CRISPR implementation theory to diagnostic applications, we highlight advanced functions of CRISPR for quantitative, multiplexed, and all-in-one diagnostic purposes, and present our views on the outstanding issues and potential solutions regarding how to make the detection schemes fit for specific scenarios and how to simplify the sample-in and results-out procedures toward POCT. We hope that this review will guide the reader to choose their adventure of CRISPR-based diagnostics with either amplification-assisted or amplification-free methods for specific applications.
2 CRISPR classification and diagnostic mechanisms
As various CRISPR–Cas systems have been discovered, they have been categorized into two classes (class 1 and class 2) depending on whether it is a single protein effector during the interference process. While the class 1 nucleases use multiple proteins as an effector complex for the interference function, the class 2 enzymes rely on a single protein effector during the interference process such as the families of Cas9, Cas12, and Cas13. The details of the hierarchical categorization can be found in ref. 9. To date, most of the nucleic acid and associated molecular diagnostics are based on the class 2 CRISPR family with a few exceptions.30,31 Moreover, different variants have shown notable differences in terms of PAM sequences, sensitivity, and cleavage loci, offering more options for the CRISPR diagnostic toolbox. Therefore, we summarize the commonly used Cas proteins according to their features, such as origins, PAM/protospacer flanking site (PFS) sequences, target type, and sensitivity (the sensitivity here was measured by previous research using fluorescence without amplification) in Table 1. Santiago's group has investigated the sensitivity of some of those listed enzymes using Michaelis–Menten enzyme kinetics theory systematically,32–34 showing that the sensitivity of those enzymes is governed by enzymatic kinetics. Among all the identified enzymes, the reaction temperature of the enzymes was found to be around 37–42 °C, except for the thermostable AsCas12a, AapCas12b, and BrCas12b which can work up to 70 °C.35–37| Type | Variant | Origin | Target | Spacer length | PAM/FPS (5′ to 3′) | Cleavage loci | Sensitivity | Ref. |
|---|---|---|---|---|---|---|---|---|
| Cas9 | spCas9 | Streptococcus pyogenes | dsDNA | 20 | NGG | 4th | — | 11 |
| Cas12 | Lbcas12a | Lachnospiraceae bacterium | ssDNA, dsDNA | 22 | TTTV | 22nd | 41.5 pM | 45 |
| Ascas12a | Acidaminococcus sp. | ssDNA, dsDNA | 20 | TTTV | 24th | 50 nM | 46, 47 | |
| AacCas12b | Alicyclobacillus acidoterrestris | ssDNA, dsDNA | 20 | TTTV | 24th | — | 37, 48 | |
| BhCas12b | Bacillus hisashii | ssDNA, dsDNA | 22 | TTN | — | — | 49 | |
| AapCas12b | Alicyclobacillus acidiphilus | dsDNA | 20 | TTN | 23rd | 1.6 nM (ssDNA) | 50, 51 | |
| 8 nM (dsDNA) | ||||||||
| Un1Cas12f1 (Cas14) | Uncultivated archaea | ssDNA | 20 | NA | 23rd | — | 52 | |
| Un1Cas12f1 (Cas14) | Uncultivated archaea | dsDNA | 20 | TTTA | 20–24 bp | — | 53 | |
| Cas13 | LshCas13a | Leptotrichia shahii | RNA | 28 | FPS, H | Multiple | — | 54 |
| LwaCas13a | Leptotrichia wadei | RNA | 28 | NA | Multiple | 50 pM | 46 | |
| LbuCas13a | Leptotrichia buccalis | RNA | 20 | NA | Multiple | 10 fM | 18, 55 | |
| LbaCas13a | L. bacterium NK4A179 | RNA | 28 | NA | Multiple | 1 nM | 46 | |
| CcaCas13b | C. canimorsus | RNA | 30 | NA | Multiple | 50 pM | 46 | |
| PsmCas13b | P. sp. A2016 | RNA | 30 | NA | Multiple | 5 nM | 46 |
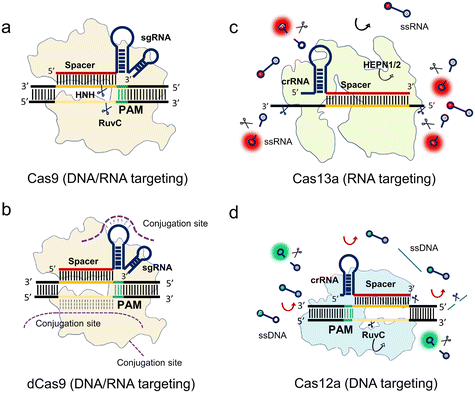
To elucidate the fundamental working principles of the class 2 nucleases, we select class 2 representative proteins including spCas9, catalytically inactivate spCas9 (dCas9), LbCas12a, and LwCas13a to explain their working mechanisms for the nucleic acid-targeting diagnostics in Fig. 1.
The spCas9 variant isolated from Streptococcus pyogenes was the first CRISPR system discovered to be programmable and engineered for sequence cleavage.11 The Cas9 ternary as shown in Fig. 1a consists of Cas9 protein, programmable sgRNA, and target sequence. The DNase activity of Cas9 is directed by a dual crRNA-trans-activating CRISPR RNA (tracrRNA) complex to break the target dsDNA at specific loci near the PAM, where the chimeric sgRNA is fused by the crRNA and tracrRNA. After the programmable sgRNA spacer binds to the target protospacer sequence, the HNH and RuvC nuclease domains of the enzyme are activated to cleave both the targeting and non-targeting sequences, respectively (Fig. 1a). In addition, Cas9 can be guided to cleave ssRNA with the presence of PAM-presenting oligonucleotides (PAMmers).38 Recently, it was found that in the case of the Cas9 derived from Campylobacter jejuni, the tracrRNA hybridizes to cellular RNAs which leads to the formation of “noncanonical” crRNAs capable of guiding DNA targeting by Cas9, where the guided RNA is unassociated with viral defence.39 This discovery inspired the reprograming of tracrRNA that can be linked to the presence of any RNA of interest for DNA cutting. Therefore, this method expands the programmable designs of the Cas9 system within the dual RNAs complex from the conventional crRNA spacer region to tracrRNA.
After mapping the locations of both HNH and RuvC nuclease domains, either of these two nuclease domains can be silenced by point mutation to create a programmable Cas9 nickase (Cas9n) that generates a nick at the targeted sequence. Furthermore, spCas9 can be switched to catalytically inactive Cas9 (dCas9) without any nuclease activities.40 As shown in Fig. 1b, the dCas9 possesses the same Cas9 ternary including dCas9, sgRNA, and target sequence. This feature allows the dCas9 system to recognize any sites in the genome near the PAM, similar to the function of antibody and antigen conjugation. To extend this powerful labeling ability, extended functions of the dCas9 system are developed via conjugations from different sites (Fig. 1b). Oligonucleotide probes with different tags such as nanoparticles and fluorophores can be conjugated at the nucleic acid stem-loop and non-target sequence sites. Comparably, the dCas9 protein is another conjugation site for other functional proteins and oligonucleotide probes.
Cas9 guided by sgRNA only cleaves the target strand within the spacer region. In contrast, Cas13 has been reported as an RNA-guided RNA targeting system that not only cleaves the target strand but also the surrounding RNAs. This RNase activity of trans-cleaving the surrounding ssRNAs is named collateral activity or trans-cleavage (Fig. 1c).17 Specifically, the crRNA guides LwCas13a to detect the target RNA. LwCas13a can trans-cleave the surrounding ssRNA, after binding to the target RNA.17,18,41,42 Unlike the Cas9 family, most reported variants of the Cas13 family have no PAM requirements except for LshCas13a from Leptotrichia shahii, which can be constrained by a sequence constraint termed PFS restriction where no G base is required.17
Similar to the Cas13 system, the Cas12a family has a trans-cleavage ability of ssDNA.43,44 As shown in Fig. 1d, LbCas12a is guided by its crRNA to target the dsDNA sequence near the PAM. After binding to the target sequence, Cas12a cleaves both the targeting strand and the non-targeting strand at a specific position. At the same time, the trans-cleavage ability is activated to cleave the surrounding ssDNA. Most of the Cas12 family has PAM sequence constraints like Cas9.
In summary, the mechanisms of CRISPR-based detection can be broadly classified into two categories, non-collateral cleavages (Cas9 and dCas9) and collateral cleavages (Cas12 and Cas13). For the non-collateral detection methods, Cas9 is a programmable endonuclease. In addition, dCas9, in which the sequence nicking ability is muted, can be utilized for specific bio-recognition, or target enrichment via different links including the sgRNA, protospacer, and protein conjugation. Comparably, the collateral effect of Cas12 and Cas13 harnesses the trans-cleavage ability for enhanced bio-reporting. Thus, some of these Cas enzymes exhibit high sensitivity, ranging from femtomolar to picomolar, allowing amplification-free detection in certain scenarios.
3 Isothermal amplification-assisted detection
3.1. Isothermal amplification schemes in CRISPR systems
The CRISPR-based diagnostic tool is programmable and specific, but its sensitivity would be still limited to the picomolar level without any amplification. For ultrasensitive detection, preamplification is required for increasing the limit of detection (LOD). Among the nucleic acid amplification schemes, isothermal alternatives require less complicated setups compared to PCR thermal cycling, which is promising for meeting the ASSURED standard. The isothermal assays, including loop-mediated isothermal amplification (LAMP),56 recombinase polymerase amplification (RPA),57 strand displacement amplification (SDA),58 nucleic acid sequence-based amplification (NASBA),59 exponential amplification reaction (EXPAR),60 and rolling circle amplification (RCA),61 have all been demonstrated in combination with CRISPR assays (Table 2). More recently, some of these methods were developed to combat the SARS-CoV-2 pandemic in different formats.62–69 Compared to the gold standard PCR which has a ∼109 amplification efficiency and SNP detection ability,70 isothermal methods such as HRCA, LAMP, and RPA have shown equal amplification efficiency to PCR, while NASBA and EXPAR are less efficient. Overall, isothermal amplification methods tend to generate non-specific results because of their relatively lower annealing temperature. Augmentation of those assays always requires additional special designs (e.g., probes, and exonuclease) to enhance specificity.71–73 Therefore, the recent integrated methods of isothermal amplification and CRISPR detection have shown unprecedented specificity at extremely low concentrations (e.g., down to the attomolar level), where the signals are only triggered in the presence of the target objects.| Method | Enzymes | No. of primers | Temperature (°C) | Reaction time (min) | Efficiency | Ref. |
|---|---|---|---|---|---|---|
| NASBA | Reverse transcriptase and RNA polymerase (RNase H) | 2 | ∼41 | 90–120 | 106–109 | 59, 74 |
| E-SDA | DNA polymerase and NEase | 2 or 4 | 37 | 120 | 107 | 58, 75 |
| HRCA | Ligase and DNA polymerase | 2 | 37 | 90 | 109 | 61, 76 |
| LAMP | DNA polymerase | 4 or 6 | 60–65 | 30–60 | 109 | 56, 77 |
| RPA | DNA polymerase and recombinase | 2 | 37–42 | 10–30 | 109 | 57 |
| EXPAR | DNA polymerase and NEase | 0 | 50–60 | 30 | 106–108 | 60, 78 |
| Variant | Name | Amplification methods | Time (min) | Target | LOD (aM) | LOD (cp mL−1) | Visualization methods | Ref. |
|---|---|---|---|---|---|---|---|---|
| spCas9 | NASBACC | NASBA | 180 | RNA | 1 × 103 | 6 × 105 | Fluorescence | 12 |
| spCas9 | CAS-EXPAR | EXPAR | 60 | DNA | 0.82 | 4.9 × 102 | Fluorescence | 80 |
| spCas9 | CRISDA | SDA | 190 | DNA | 6.7 | 4 × 103 | Fluorescence | 79 |
| dCas9 | RPA | 20 | DNA RNA | 0.54 | 3 × 102 | Silicon microring resonator | 90 | |
| dcas9 | RCH | RCA | 240 | miRNA | 35.4 | 2.1 × 104 | Colorimetry | 87 |
| spCas9 | Cas9nAR | Nickase-based amplification reaction | 60 | DNA | 0.167 | 1 × 102 | Fluorescence | 82 |
| spCas9 | RACE | RCA | 240 | microRNA | 9 × 104 | 5.4 × 107 | Fluorescence | 91 |
| spCas9 | CUT-LAMP | LAMP | 60 | DNA | 5 | 3 × 106 | Fluorescence | 92 |
| spCas9 | CASLFA | RPA | 60 | DNA | 12.5 | 7.5 × 103 | LFA | 85 |
| spCas9 | Nickase-triggered amplification | 80 | DNA | 16 | 9.6 × 103 | LFA | 83 | |
| spCas9 | SDA–RCA | 150 | DNA | 1.87 × 103 | 1.1 × 106 | Fluorescence | 93 | |
| dCas9 | RT-RPA | 37 | RNA | 6.7 | 4 × 103 | LFA | 86 | |
| spCas9 | SDA | 65 | DNA | 1.3 × 107 | 7.8 × 109 | Electrochemical | 94 | |
| FnCas9 | FELUDA | RPA | 60 | RNA | 33 | 2 × 103 | LFA | 95 |
| dCas9 | Vigilant | RPA | 35 | RNA | 4.2 | 2.5 × 103 | LFA | 96 |
| dCas9 | Bio-SCAN | RPA | 24 | RNA | 6.67 | 4 × 103 | LFA | 89 |
| dCas9 | RPA–RCA | — | DNA | 20 | 1.2 × 104 | LFA | 97 | |
| spCas9 | Cas9 nAR-v2 | SDA | 60 | RNA | 0.83 | 5 × 102 | Fluorescence/LFA | 98 |
| spCas9 | DAMPR | LAMP | 50 | RNA | ∼1 | 6 × 102 | Fluorescence | 99 |
| Variant | Name | Amplification methods | Time (min) | Target | LOD (aM) | LOD (cp mL−1) | Visualization | Ref. |
|---|---|---|---|---|---|---|---|---|
| Lwcas13a | SHERLOCK | RPA | 60 | NA | 2 | 1 × 103 | Fluorescence | 41 |
| Lwcas13a, LbCas13b, Ascas12a | SHERLOCKv2 | RPA | 60 | NA | 8 × 10−3 | 4.8 | Fluorescence | 46 |
| LFA | ||||||||
| Lwcas13a | HUDSON–SHERLOCK | RPA | 60 | RNA | 2 | 1 × 103 | Fluorescence | 100 |
| LFA | ||||||||
| Lwcas13a | RPA | 46 | NA | 16.7 | 1 × 104 | Colorimetric | 115 | |
| LbuCas13a | DISCoVER | LAMP | 35 | RNA | 16.7 | 1 × 104 | Fluorescence | 109 |
| Lwcas13a | CARMEN–Cas13 | RPA | 60 | NA | 2 | 1 × 103 | Digital fluorescence (multiplexed) | 104 |
| Lwcas13a | RPA | 105 | DNA | 0.6 | 4 × 102 | LFA | 116 | |
| LwCas13a | RPA | 60 | RNA | 1.4 | 8.4 × 102 | LFA/fluorescence | 117 | |
| Lwcas13a | RPA | 60 | RNA | 16.7 | 1 × 104 | LFA/fluorescence | 101 | |
| LwCas13a | SHINE | RPA | 40 | RNA | 16.7 | 1 × 104 | LFA | 102 |
| LwCas13a | MEDICA | RPA | 25 | DNA | 2 | 1 × 103 | Digital fluorescence | 108 |
| LwCas13a | PADLOCK | RPA | 30 | DNA | 2 | 1 × 103 | Digital fluorescence | 73 |
| LbuCas13a | RCA | 120 | miRNA | 5 × 102 | 3 × 106 | Fluorescence | 111 | |
| LwCas13a | SHINEv2 | RPA | 90 | RNA | 1.7 × 102 | 1 × 106 | LFA (POCT) | 118 |
| LwCas13a | RPA | 60 | RNA | 0.3 | 1.8 × 102 | Fluorescence (multiplexed) | 119 | |
| PsmCas13b | ||||||||
| LwCas13a | ADESSO | RPA | 60 | RNA | 4.2 | 2.5 × 103 | Fluorescence | 120 |
| TccCas13a | OPTIMA-dx | LAMP | 45 | RNA | 16.7 | 1 × 104 | Fluorescence | 121 |
| Variant | Name | Amplification methods | Time (min) | Target | LOD (aM) | LOD (cp mL−1) | Visualization methods | Ref. |
|---|---|---|---|---|---|---|---|---|
| LbCas12a | DETECTR | RPA | 70 | DNA | 2 | 1 × 103 | Fluorescence | 44 |
| LbCas12a | ULC | LAMP | 45 | DNA | 16.7 | 1 × 104 | Fluorescence | 139 |
| AacCas12b | HOLMESv2 | LAMP | 60 | NA | 10 | 6 × 103 | Fluorescence | 37 |
| LbCas12a | Cas12a-SCR | RCT | 420 | miRNA | 1.7 × 105 | 1 × 108 | Fluorescence | 135 |
| LbCas12a | RPA | 75 | DNA | 16.7 | 1 × 104 | Fluorescence | 140 | |
| LbCas12a | CIA | LAMP | 50 | DNA | 1 | 6 × 102 | LFA | 141 |
| LbCas12a | LAMP | 40 | RNA | 1.3 | 7.8 × 102 | Fluorescence | 142 | |
| LbCas12a | LAMP | 35 | DNA | — | — | Colorimetry | 143 | |
| LbCas12a | SHERLOCK-Cas12a | RPA | 60 | DNA RNA | 50 | 3 × 104 | LFA | 144 |
| LbCas12a | LAMP | 160 | DNA | 10 | 6 × 103 | Colorimetry | 145 | |
| LbCas12a | HRCA | 420 | miRNA | 2 × 103 | 1 × 106 | Fluorescence | 136 | |
| LbCas12a | LAMP | 40 | RNA | 2 | 1 × 103 | LFA | 129 | |
| LbCas12a | LAMP–RPA | 60 | RNA | 0.1 | 60 | LFA | 47 | |
| LbCas12a | AIOD-CRISPR | RPA | 40 | RNA | 0.33 | 2 × 102 | Fluorescence | 122 |
| AapCas12b | STOP | LAMP | 45 | RNA | 0.055 | 3.3 × 101 | Fluorescence | 50 |
| AsCas12a | CIRI | RCA | — | DNA | 1 | 6 × 102 | Optomagnetic | 134 |
| LbCas12a | ITP–CRISPR | LAMP | 35 | RNA | 16.7 | 1 × 104 | Fluorescence | 130 |
| LbCas12a | CIALFB | LAMP | 50 | DNA | 3.1 | 1.8 × 103 | LFA | 146 |
| LbCas12a | LAMP | 60 | DNA | — | — | Fluorescence | 147 | |
| LbCas12a | LAMP | 40 | RNA | 50 | 3 × 104 | Fluorescence | 148 | |
| LbCas12a AacCas12b AapCas12b | iSCAN | LAMP | 60 | RNA | 0.33 | 2 × 102 | Fluorescence | 149 |
| LbCas12a | RPA | 30 | RNA | 3.3 × 10−2 | 20 | Surface plasmon resonance colorimetric | 150 | |
| LbCas12a | MCDC | RPA | 55 | RNA | 1.67 | 1 × 103 | Colorimetry | 151 |
| LbCas12a | LAMP | 40 | DNA | 800 CFU ml−1 | — | Fluorescence | 152 | |
| LbCas12a | opvCRISPR | LAMP | 45 | RNA | 0.21 | 1.3 × 102 | Fluorescence | 153 |
| LbCas12a | RPA | 40 | RNA | 0.63 | 3.8 × 102 | Fluorescence | 113 | |
| LbCas12a | WS-CRISPR | LAMP | 90 | RNA | 83.3 | 5 × 104 | Digital fluorescence | 133 |
| LbCas12a | LAMP | 60 | DNA | 1.67 | 1 × 103 | Fluorescence–LFA | 154 | |
| LbCas12a | DECOVID | RPA | 30 | RNA | 1.67 | 1 × 103 | Digital fluorescence | 126 |
| LbCas12a | RADICA | RPA | 60 | DNA | 1.5 | 9 × 102 | Digital fluorescence | 155 |
| AsCas12a | VaNGuard | LAMP | 52 | RNA | 3.33 | 2 × 103 | Fluorescence | 156 |
| LFA | ||||||||
| Lbcas12a | CAL-LAMP | LAMP | — | miRNA | 1 × 104 | 6 × 105 | Fluorescence | 157 |
| LbCas12a | CLAP | LAMP | 40 | RNA | 6.67 | 4 × 103 | Colorimetry | 158 |
| LbCas12a | RPA | 60 | RNA | 6.67 | 4 × 103 | Fluorescence (POCT) | 159 | |
| LbCas12a | POC-CRISPR | RPA | 60 | RNA | 1 GE μL−1 | Fluorescence (POCT) | 160 | |
| Aapcas12b | WS-RADICA | LAMP | 60 | RNA | 2 | 1.2 × 103 | Digital fluorescence | 161 |
| BrCas12b | LAMP | 30 | RNA | 10 | 6 × 103 | Fluorescence | 132 | |
| LbCas12a | sPAMC | RPA | 20 | RNA | 2 | 1.2 × 103 | Fluorescence | 123 |
| LbCas12a | LAMP | 45–75 | RNA | 166.7 | 1 × 105 | Fluorescence | 162 | |
| LbCas12a | RPA | 40 | DNA | 0.16 | 96 | Fluorescence | 163 | |
| LbCas12a | DropCRISPR | LAMP | >60 | DNA | 102 cfu mL−1 | — | Fluorescence | 164 |
| LbCas12a | FAST | RPA | 50 | RNA | 1 × 104 | 6 × 106 | Fluorescence | 165 |
| AsCas12a | UCAD | RPA | >40 | Antibodies | 10 | 6 × 104 | Fluorescence | 166 |
| LbCas12a | MiCaR | RPA | 40 | DNA | 0.26 | 1.6 × 102 | Fluorescence (multiplexed) | 167 |
| LbCas12a | RPA | 30 | RNA | 1.67 | 1 × 103 | Fluorescence | 168 | |
| LbCas12a | MAPnavi | RPA | 40 | RNA | 0.33 | 2 × 102 | Fluorescence | 169 |
| Type | Variant | Name | Time (min) | Target | LOD (aM) | LOD (cp mL−1) | Visualization | Ref. |
|---|---|---|---|---|---|---|---|---|
| Cas9 | dcas9 | CRISPR DNA-FISH | 30 | DNA | 10 cfu ml−1 | Fluorescence | 180 | |
| dcas9 | Paired dCas9 (PC) reporter | 30 | DNA | 5 × 104 | 3 × 107 | Luminescence | 201 | |
| dcas9 | 30 | DNA | — | — | Colorimetry | 202 | ||
| dcas9 | CRISPR chip | 15 | DNA | 1.7 × 103 | 1 × 106 | FET-electricity | 176 | |
| dcas9 | 90 | RNA | 1.4 × 108 | 8.4 × 1010 | Colorimetry | 203 | ||
| dcas9 | CRISPR-SERS | 60 | DNA | 8.1 × 103 | 4.8 × 106 | Surface-enhanced Raman | 181 | |
| dcas9 | CRISPR-SNP-Chip | 60 | DNA | — | — | FET-electricity | 182 | |
| dCas9 | 60 | DNA | 3 × 102 | 1.8 × 105 | LFA | 187 | ||
| Cas12 | Lbcas12a | E-CRISPR | 40 | DNA | 5 × 107 | 3 × 1010 | Electrochemical | 183 |
| Lbcas12a | IMPACT | 120 | DNA | 1 × 108 | 6 × 1010 | Fluorescence | 204 | |
| LbCas12a | E-DNA | 30 | DNA | 1 × 104 | 6 × 106 | Electrochemical | 205 | |
| Lbcas12a | 1440 | DNA | 1 × 104 | 6 × 106 | Fluorescence | 206 | ||
| Ascas12a | 20 | DNA | 1 × 105 | 1 × 107 | Electrochemical | 186 | ||
| Lbcas12a | CONAN | 30 | DNA | 5 | 3 × 102 | Fluorescence | 175 | |
| LbCas12a | CSMBA | 10 | DNA | 3 | 1.8 × 103 | Fluorescence | 207 | |
| LbCas12a | 30 | DNA | 1 × 106 | 6 × 108 | Fluorescence (tandem crRNA) | 208 | ||
| LbCas12a | 5 | miRNA | 1 × 104 | 6 × 106 | Fluorescence (gold nanoparticles) | 209 | ||
| LbCas12a LwCas13a | >30 | DNA RNA | 16.67 | 1 × 104 | Quantum dot-based molecular beacons | 210 | ||
| LbCas12a | MOPCS | 38 | RNA | 1.5 × 104 | 9 × 106 | Plasmon resonance | 211 | |
| Cas13 | LbuCas13a | 30 | RNA | 1 × 106 | 6 × 108 | Fluorescence | 18 | |
| LbuCas13a | 30 | miRNA | 4.5 × 105 | 2.7 × 108 | Fluorescence | 110 | ||
| LwCas13a | 5 | RNA | 9 × 104 | 5.4 × 107 | Fluorescence | 212 | ||
| LwCas13a | 240 | miRNA | 1 × 107 | 6 × 109 | Electrochemical | 194 | ||
| LbuCas13a | CLISA | 300 | Protein | 8.1 × 103 | 4.8 × 106 | Fluorescence | 213 | |
| LbuCas13a | PECL-CRISPR | 70 | miRNA | 1 × 103 | 6 × 105 | Electricity | 195 | |
| LwCas13a | CRISPR-Biosensor X | 180 | miRNA | 2 × 106 | 1.2 × 109 | Electrochemical | 196 | |
| LwaCas13a | CRISPR-FET | 20 | RNA | 1.56 | 9.4 × 102 | FET-electricity | 214 | |
| LbuCas13a | SHARK | 90 | RNA | 1 × 104 | 6 × 106 | LFA/digital bioluminescence | 215 | |
| LwaCas13a | CRISPR Cas13agFET biosensor | 30 | RNA | 1 | 6 × 102 | gFET-electricity | 177 | |
| LtrCas13a | opn-SATORI | 9 | RNA | 6.5 | 3.9 × 103 | Digital fluorescence | 216 | |
| LbuCas13a LwaCas13a | CrisprZyme | 270 | RNA | 8.3 × 104 | 5 × 107 | Colorimetric | 217 | |
| LwaCas13a | 30 | RNA | 1 | 6 × 102 | Electrochemical | 218 |
The selection of the nucleic acid amplification scheme is largely dependent on the type of target and CRISPR effectors. DNA and RNA targets can be commonly amplified by RPA, LAMP, or NASBA, while SDA, EXPAR, and RCA are more frequently applied to miRNAs because the length of miRNAs restricts the primer design.70 When it comes to the influence of CRISPR effectors, non-collateral detection of Cas9 requires additional design for signal reporting, which would significantly affect the options of the amplification methods. In comparison, the final activators of Cas12 and Cas13 for collateral cleavage are DNAs and RNAs, respectively. Since most of the amplification methods would generate DNA amplicons using DNA polymerase, the specific design of T7 transcription is required to produce RNA amplicons. Therefore, the Cas12 family can be easily integrated with most of these methods. In contrast, the Cas13 family requires an extra T7 transcription step to generate RNA activators. To date, RPA and LAMP are regarded as the most popular candidates to be integrated with CRISPR owing to their fast reaction and high amplification efficiency.
3.2. Cas9 detection systems with isothermal amplification
Cas9 is a powerful CRISPR tool that can be used as a programmable endonuclease for specific sequence cleavage with switchable nuclease activities. In addition to the nuclease function, the engineered dCas9 with different conjugations has been applied to molecular diagnostics, in situ nucleic acid imaging, etc. In this section, we summarize the strategies using Cas9 for diagnostics in different scenarios.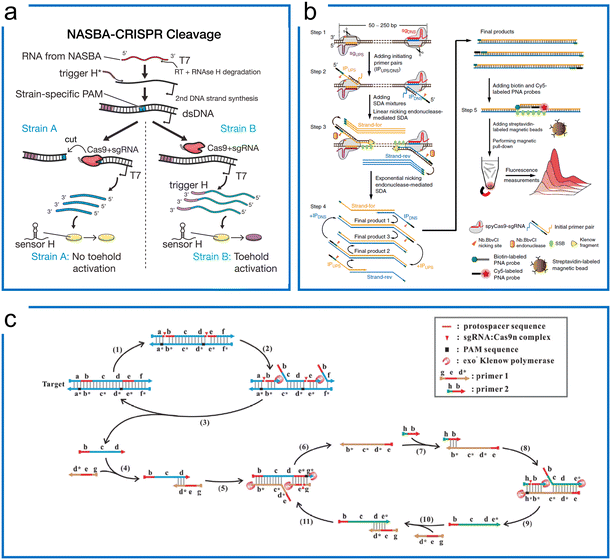 | ||
| Fig. 2 Isothermal amplification enhanced Cas9 nucleic acid detection. (a) The Zika virus detection with Cas9 for SNP visualization. The target RNA was firstly amplified by NASBA, followed by the Cas9 cleavage to differentiate the SNP difference due to the PAM requirement. Finally, the synthetic biology of the toehold sensor enables colorimetric visualization. The figure was reproduced from ref. 12 with permission from Elsevier, copyright 2016. (b) The CRISDA method uses SDA and Cas9, where programmable cas9n nicks the binding region for SDA primer binding and amplification. The amplicons were visualized by additional fluorescence. The figure was reproduced from ref. 79 under Creative Commons Attribution License from Springer Nature, copyright 2018. (c) The Cas9nAR design for universal dsDNA detection includes primers, DNA polymerase, and sgRNA/Cas9n complex. The Cas9n serves as the programmable nicking enzyme in the SDA. The figure was reproduced from ref. 82 with permission from John Wiley and Sons, copyright 2019. | ||
A similar design of EXPAR in combination with Cas9 was used for mRNA detection.80 Since EXPAR is widely adopted for short nucleic acid detection such as microRNAs,81 the desired RNA sequence is cut off by Cas9 directed by a sgRNA in the presence of the PAMmer. Subsequently, the cleaved fragments hybridize with the EXPAR template and are amplified accordingly. This method can detect any sites of single-strand targets by the programmable sgRNA with the sequence selection of the Cas9 and EXPAR system.
However, even though CRISPR–Cas9 can be applied to specifically cut off the target sequence, these two aforementioned methods still rely on extra nicking endonucleases in their design even though Cas9 already has a nicking ability. The Cas9 nickase-based amplification reaction (Cas9nAR) was developed for genomic DNA upon the precise recognition ability of the target object site (Fig. 2c),82 whereby the 11-step reaction in a one-pot assay can be designed as a universal method containing cycles of DNA replication through priming, extension, nicking, and displacement reaction steps. This method harnesses an sgRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cas9n complex with a single-strand nicking property, a strand displacing DNA polymerase, and two primers bearing the cleavage sequence of Cas9n. The Cas9 ribonucleoprotein has been utilized for selecting the target of interest from the raw genomic DNA as well as for SDA amplification as the nicking enzymes. This one-pot reaction has achieved 0.1 copies per μL sensitivity within 60 min, highlighting the potential applications of Cas9-based SDA methods for universal detection. With the lateral flow assay (LFA) visualization introduced later, this method achieved 16.7 aM sensitivity in detecting food-borne pathogens.83 However, this approach is challenging to implement due to the constraint of the two PAM sequence designs.
Cas9n complex with a single-strand nicking property, a strand displacing DNA polymerase, and two primers bearing the cleavage sequence of Cas9n. The Cas9 ribonucleoprotein has been utilized for selecting the target of interest from the raw genomic DNA as well as for SDA amplification as the nicking enzymes. This one-pot reaction has achieved 0.1 copies per μL sensitivity within 60 min, highlighting the potential applications of Cas9-based SDA methods for universal detection. With the lateral flow assay (LFA) visualization introduced later, this method achieved 16.7 aM sensitivity in detecting food-borne pathogens.83 However, this approach is challenging to implement due to the constraint of the two PAM sequence designs.
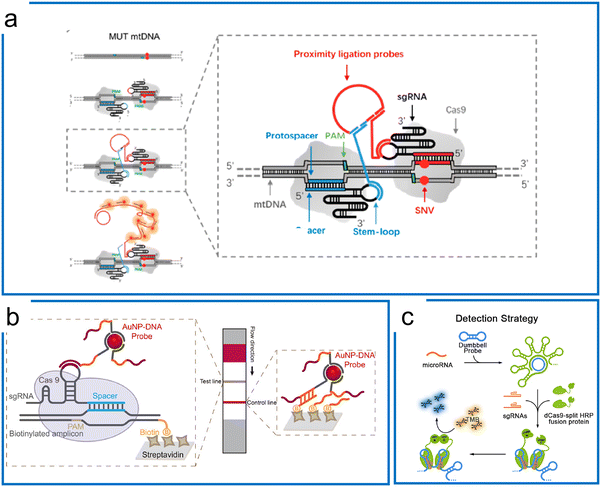 | ||
| Fig. 3 Isothermal amplification assisted dCas9 molecular diagnosis. (a) The dCas9 system with a proximity ligation assay coupled with RCA for SNV detection. The two Cas9 ribonucleoproteins differentiate the mutations of the sequence in the protospacer region. The in situ difference is visualized by the proximity ligation assay using two DNA probes binding to stem-loops of these two sgRNAs. Adapted from ref. 84. Copyright 2018 American Chemical Society. (b) The dCas9-based detection using AuNPs–DNA probes hybridizing to either the stem-loop of sgRNA or non-targeting sequence. The RPA preamplification tags the biotin for hybridization with the streptavidin on the surface of the LFA strip. Adapted from ref. 85. Copyright 2020 American Chemical Society. (c) The microRNA detection using dCas9-splint HRP fusion protein for colorimetric readout. The dumbbell amplicons are synthesized by RCA amplification using microRNA and padlock probe ligation. After dCas9 systems are bound to the dsDNA region of the amplicons, the TMB is introduced to react with HRP for colorimetric readout. Adapted from ref. 87. Copyright 2018 American Chemical Society. | ||
3.3. Cas13 detection systems with isothermal amplification
The significant difference between Cas9 and Cas13 is that Cas13 is an RNA targeting system with both cis and trans-cleavage ability. Therefore, the nucleic acid diagnostics with the Cas13a system utilize the specific cis targeting ability of the Cas13 system to enable trans-cleavage of engineered ssRNAs (e.g., ssRNA probes). To achieve a comparable LOD as PCR, the Cas13-based diagnostics involve preamplification and Cas13 detection including T7 transcription and Cas13 visualization. Here, we summarize the representative Cas13 systems according to the different isothermal amplification methods.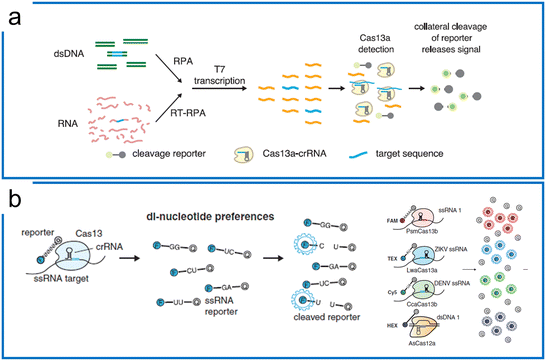 | ||
| Fig. 4 Theory of Cas13-based diagnostics. (a) SHERLOCK nucleic acid detection platform includes RPA preamplification, T7 transcription, and Cas13a detection. The figure was reproduced from ref. 41 with permission from the American Association for the Advancement of Science, copyright 2017. (b) The di-nucleotide preferences of the Cas13 family and AsCas12a combination enable 4 channel multiplexed diagnostics with different fluorescence probes including ssRNA probes (Cas13 family) and ssDNA probe (Cas12). The figure was reproduced from ref. 46 with permission from the American Association for the Advancement of Science, copyright 2018. | ||
Subsequently, the SHERLOCK assay was coupled with the HUDSON (unextracted author manuscript diagnostic samples to obliterate nucleases) protocol to detect the Zika virus and dengue fever virus in patient samples with the LOD of a single copy per microliter (Fig. 5a).100 This protocol was then further applied to detect Ebola and SARS-CoV-2.101,102 Recently, machine learning was applied to simplify the sequence design process and optimize the primers and crRNA of the SHERLOCK assay.103 The SHERLOCK assay has been combined with droplet array technology to realize massively multiplexed pathogen detection. This method is termed combinatorial arrayed reactions for multiplexed evaluation of nucleic acids with Cas13 (CARMEN–Cas13) (Fig. 5b).104 The high multiplexable ability was achieved by the preparation of the color code droplets of 4 fluorescent colors with different concentrations individually to make 1050 color codes.105 Each RPA preamplified sample or Cas13a detection mix was emulsified into droplets with a distinct color code. All droplets were pooled and loaded on a microarray chip where the droplets self-organized into pairs and the positions of each sample and crRNA were identified by their color codes. The droplet pairs were then merged for the initiation of CRISPR assays to obtain the positive detection results. Later, Myhrvold et al. upgraded the CARMEN with Fluidigm microfluidics to reduce the operation time.106 This isothermal massive detection has the advantage of decreasing reagent costs per test by more than 300 times compared to the conventional bulk reaction. Quantitative measurement is of great importance for biological studies, and in order to increase the accuracy of quantification and reduce false-positive events, a microfluidic-enabled digital isothermal Cas13a assay (termed MEDICA) has been developed (Fig. 5c). In traditional setups, the RPA preamplification starts as soon as the magnesium initiator is added even at ambient temperature, causing premature amplification prior to droplet partitioning.107 In this method, a Y-shaped microfluidic chip was designed to separate the reaction initiator from the target-included master mix.108 Moreover, a droplet picoinjector was applied to address this RPA premature amplification, allowing complete premature amplification-free quantification.73
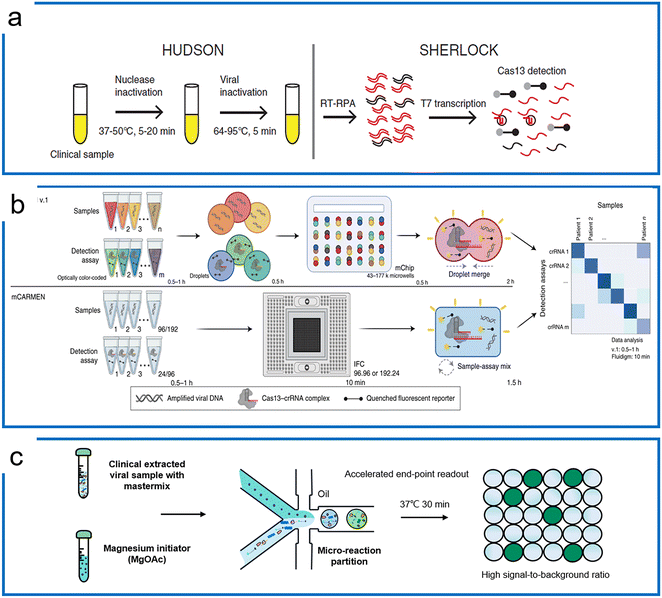 | ||
| Fig. 5 RPA-based Cas13 diagnostics. (a) The HUDSON protocol that pairs with SHERLOCK for viral detection directly from body fluids; the figure was reproduced from ref. 100 with permission from the American Association for the Advancement of Science, copyright 2018. (b) The massively multiplexed viral detection using Cas13a with microfluidics. The combinatorial droplet merge technique enables amplified targets with droplet color barcodes to merge for multiplexed visualization. The figure was reproduced from ref. 112 under Creative Commons Attribution License from Springer Nature, copyright 2022. (c) Droplet digital quantification with one-pot SHERLOCK. Droplet microfluidics allows the RPA reaction to be triggered after droplets are formed, and enables background-free binarization of droplets where the signals are only triggered by the trans reporting of Cas13a in the presence of the desired targets. Adapted from ref. 108 with the permission from American Chemical Society, copyright 2022. | ||
3.4. Cas12 detection systems with isothermal amplification
Unlike Cas13 which is an RNA targeting system, Cas12 is a DNA targeting system with a trans-cleavage ability of ssDNA. Upon the specific binding to the target sequence, Cas12 not only cleaves on the target strand but is activated for trans-cleavage of the surrounding ssDNA. When bound to dsDNA, the turnover rate of Lbcas12a trans-cleavage is over 5-times faster than ssDNA targeting. We summarize the representative Cas12 systems according to the combination of different isothermal amplification methods (see Table 5).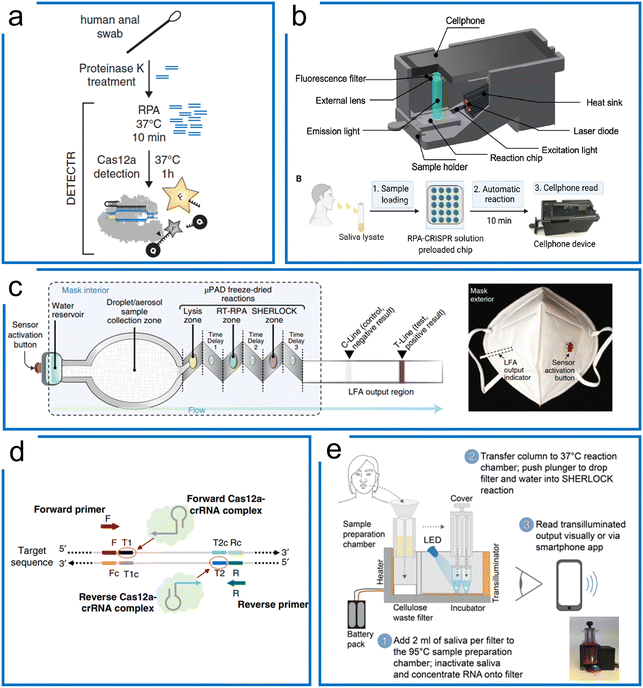 | ||
| Fig. 6 The RPA-based Cas12a systems for nucleic acid detection. (a) The DETECTR system of Cas12a diagnostics for HPV viral detection, the sample was treated with proteinase K, followed by RPA, and then the targets were detected by Cas12a ribonucleoprotein for trans-cleavage of the ssDNA reporters. The figure was reproduced from ref. 44 with permission from the American Association for the Advancement of Science, copyright 2018. (b) A miniaturized system with cell phone readout using the RPA–Cas12a system that allows sensitive and quantitative readout of saliva CRISPR-FDS assays at sites. Sample preparation, amplification, and detection are all combined in one device. The figure was reproduced from ref. 113 under Creative Commons Attribution License from the American Association for the Advancement of Science, copyright 2021. (c) The RPA–Cas12a diagnostics with paper microfluidics in a mask. Once the water reservoir was opened, the target was driven by the capillary force of paper microfluidics through different folded layers for three purposes including sample lysis, RT-RPA amplification, and Cas12a-based detection. All these chemicals associated with these purposes were lyophilized reagents in the mask. The figure was reproduced from ref. 114 with permission from Springer Nature, copyright 2021. (d) A one-pot RPA–Cas12a assay with two targeting sites of crRNAs for enhanced sensitivity of Cas12a diagnosis of COVID-19. After RPA amplification, the amplicons can be targeted using two crRNAs to boost the sensitivity higher compared to the conventional one-crRNA method. The figure was reproduced from ref. 122 under Creative Commons Attribution License from Springer Nature, copyright 2020. (e) A portable device with enhanced one-pot Cas12a and RPA reaction for SARS-CoV-2 and related variants' detection. The target sample was firstly treated with a high temperature for sample preparation, following liquid transfer to another lyophilized power tube to perform the one-pot RPA–Cas12a reaction. Sample preparation, RPA amplification, and Cas12a detection steps were integrated into a small device with a mobile phone readout. The figure was reproduced from ref. 125 under Creative Commons Attribution License from the American Association for the Advancement of Science, copyright 2021. | ||
Although the RPA and Cas12a combination can achieve high sensitivity and specificity, the multistep process may cause higher cross-contamination risks, alongside the requirement for extra expertise and equipment. Therefore, a one-pot assay of RPA and LbCas12a was developed to fuse the RPA preamplification and CRISPR visualization into a single step. To increase the reported sensitivity, Liu et al. designed an approach using two crRNAs for detecting both strands of RPA amplicons (Fig. 6d). They demonstrated that the two-crRNA combination can achieve a higher sensitivity compared to the one-crRNA method. This method achieved 0.33 aM in a one-pot RPA and aCas12a reaction to detect the SARS-Cov-2 N gene within 20 minutes.122 Besides, Yin et al. found that the suboptimal PAM of Cas12a accelerated the reaction with high specificity and sensitivity. Thus, the one-pot RPA and Cas12a reaction uses the suboptimal PAM of Cas12a (e.g., NTTV and TTNT) rather than the canonical PAM (TTTV). This method achieved an LOD of 1 cp μL−1 in SARS-COV-2 diagnostics.123 Comparably, Collins et al. optimized reaction factors such as the reaction temperature, reporter concentration, and for RT commercial brands, crRNA and RPA reaction primers, resulting in the improvement of the LOD to 50 aM for Plasmodium species in the case of symptomatic and asymptomatic malaria.124 In another work (Fig. 6e), they further optimized the reaction buffer and enzymes, as well as adding RNase H to improve the RT efficiency for SARS-Cov-2 and related variants. In order to deliver a deployable system, they further developed a minimally instrumented SHERLOCK (miSHERLOCK) POC diagnostic platform, which integrates instrument-free, built-in RNA paper-capture from saliva, room temperature stable reagents, battery-powered incubation, and mobile phone-enabled readout with an LOD of 1000 copies per milliliter.125
The one-pot assay has also been extended to digital quantification. Wang et al. have applied the nano-well chip for absolute quantification of COVID-19 (ref. 126) and have demonstrated fewer false-positive results. They were able to show that digital detection can be accomplished within 30 minutes to a single-molecule level sensitivity. Yu et al. were able to apply yet another commercial digital PCR system for Cas12a and RPA one-pot quantification.127 This system partitioned the one-pot master mix inside a tube for thousands of partitions similar to nano-well technology. They compared the performance of digital PCR, qPCR, and digital Cas12a and RPA results to demonstrate that this novel technology can be applied to the digital quantification of viral target objects. However, these two technologies have a serious drawback in that the RPA reaction can start at ambient temperature as soon as the magnesium initiator is added,107,128 which can lead to inaccurate results because of premature amplification during partition.
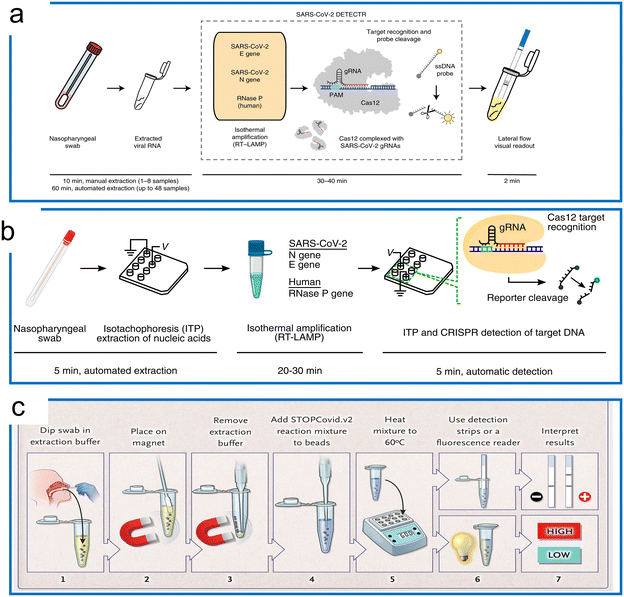 | ||
| Fig. 7 The LAMP-based Cas12a detection. (a) The DETECTR with the LAMP system for SARS-CoV-2 detection. The traditional RNA extraction method was combined with the LAMP–Cas12a detection scheme. The test results were visualized by either fluorescence or LFA assay. The figure was reproduced from ref. 129 with permission from Springer Nature, copyright 2020. (b) The microfluidic ITP enables COVID-19 detection with LAMP preamplification and Cas12a detection. A streamlined workflow was established with ITP extraction of nucleic acids, off-chip incubation with LAMP, and visualization with Cas12a on an ITP chip. The figure was reproduced from ref. 130 under Creative Commons Attribution License from National Academy of Sciences, copyright 2020. (c) The STOPCovid system with one-pot LAMP and Cas12b for COVID-19 detection. The sample was first collected and treated with extraction buffer assisted by a magnet. Then the lysed target was transferred to perform a LAMP–Cas12b one-pot reaction at 60 °C. Lastly, the readout was conducted using an LFA or fluorescence assay to inspect the results. The figure was reproduced from ref. 50 with permission from Massachusetts Medical Society, copyright 2020. | ||
Because different reaction temperatures are required for LAMP (∼65 °C) and Cas12a (∼37 °C), a combined one-pot assay requires the system to be compatible with a wider range of temperatures. Through the screening of the type V enzymes, AacCas12b from Alicyclobacillus acidoterrestris, a thermophilic RNA-guided endonuclease that can work around 55 °C, was reported to be first combined with LAMP for a one-pot reaction.37 However, the sensitivity of this one-pot LAMP and AaCas12b assay is lower compared to the two-step reaction (10 aM). AapCas12b from Alicyclobacillus acidiphilus was used by Zhang's group in a one-pot assay working at 60 °C.50 The guide RNA of AmCas12b was used to guide AapCas12b because the AapCas12b locus does not contain an identifiable CRISPR array.45 This protocol, termed STOPCovid2 (SHERLOCK Testing in One Pot), has been applied for COVID-19 detection with a 55 zM sensitivity, which is more sensitive compared to the standard PCR test (Fig. 7c). It is notable that whilst the standard LAMP method can generate a nonspecific signal, the STOPCovid only produced a signal in the presence of the target object, which further indicates that CRISPR detection can enhance the test specificity. Similar work of LAMP using CRISPR–Cas9 to trigger signals only in the presence of the target has also been reported.131 In addition to the AapCas12b, which has shown excellent performance at a high operation temperature, Nguyen et al. recently found that the BrCas12b from Brevibacillus sp. exhibited phenomenal stability at high temperatures (e.g., up to 70 °C in the optimal buffers). In combination with LAMP, this thermostable Cas enzyme has been adopted for diagnostics of SARS-COV-2 variants including alpha, beta, gamma, delta, and omicron in a one-pot reaction.132 The LOD for detection of these variants ranges from 20 aM to 833 aM. Apart from the one-pot reaction that can be utilized for quantification using a standard curve, a LAMP-based LbCas12a reaction has also been developed for digital quantification.133 The reaction temperature range of LbCas12a can work up to 55 °C. Thus, a dual-priming isothermal amplification (DMAP), a variant of the LAMP method that works at a lower temperature, was designed to combine with LbCas12a for hot-start initiation. The digital quantification using a commercial nano-well chip achieved a 5 cp μL−1 sensitivity, which was 10 times higher compared to reactions in bulk.137
4 Amplification-free CRISPR detection systems
Although preamplification greatly enhances the sensitivity of the CRISPR diagnostic assays, it inevitably poses limitations to the assay design since additional steps and assay compatibility are required in a more complex system. The amplification-free CRISPR methods present enormous value as testing methods due to their simplicity and rapidity as direct detection platforms. As aforementioned, CRISPR-based diagnostics have a natural sensitivity around the picomolar level using fluorescence measurement.170 To further increase the sensitivity of amplification-free CRISPR methods, advanced sensing technologies such as surface-enhanced Raman spectroscopy (SERS),171 electrochemistry,172 Förster resonance energy transfer (FRET),173,174 molecular circuits of self-amplification,175 field-effect transistor (FET),176,177 tandem crRNA or enzymes,178,179etc. have been utilized (see a summary of these methods in Table 6). The ability to deploy in resource-limited settings will meet a great need for public health monitoring and general pathogen detection.4.1. dCas9-based amplification-free assays
The Cas9 system, especially dCas9, is a powerful CRISPR tool for target recognition. The dCas9-based amplification-free schemes apply the programmable recognition ability to identify and enrich the targets before measurement. In general, the dCas9 system can be conjugated with different substrates to immobilize the target oligonucleotide, which can be visualized using different methods including fluorescence,180 Raman,181 gFET electrical detection,176,182etc.Although the fluorescence in situ hybridization (FISH) method enables high-resolution detection of specific DNA in single cells, there are some major drawbacks, such as complex and time-consuming operation, and the need to denature the target DNA. Jung et al. have designed a DNA-FISH-based method with dCas9 to detect the dsDNA target within 30 minutes.180 The hypothesis is that the dCas9–sgRNA complex functions as a targeting material and SYBR Green I (SG I) as a label probe. In this design, the dCas9–sgRNA complex conjugated to a magnetic bead can specifically bind to the target DNA in un-purified cell lysates, and after simple magnet enrichment, SG I staining and washing steps, the LOD of methicillin-resistant Staphylococcus aureus (MRSA) detection can reach 10 CFU ml−1.
SERS is a phenomenon where the molecular Raman signal increases near the SERS active surface, which has been applied for nucleic acid detection at single-molecule resolution.171 Kang et al. have been able to introduce SERS into the dCas9 system for the nucleic acid detection of multidrug-resistant bacteria (Fig. 8a). This work combines the specificity of dCas9 and SERS Raman sensitivity. The dCas9–sgRNA complex was first immobilized on gold-coated magnetic nanoparticles (Au MNPs). The dCas9–AuNP complex reacted with the superbugs to recognize the genomic DNA. Then, methylene blue (MB), a Raman reporter, was added to this mixture, intercalating into the genomic DNA.181 By applying an external magnet to collect the dCas9 ternary, the Raman intensity was then measured for target detection. Thus, the SERS coupled with dCas9 is an ultra-sensitive detection method, having an LOD of 8.1fM without amplification.
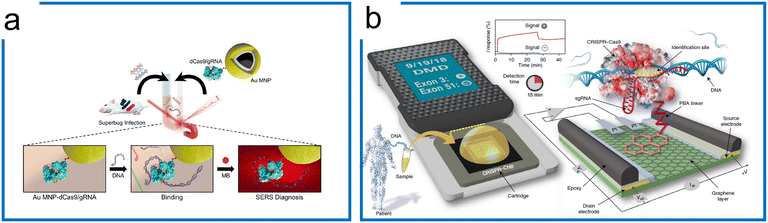 | ||
| Fig. 8 The amplification-free system with dCas9. (a) A SERS detection strategy with dCas9. The Au MNP-dCas9/gRNA probe can label the genomic DNA of a superbug-infected mouse. MB was introduced to interact with the genomic DNA. After the external magnetic enrichment, the SERS measurement was conducted. Adapted from ref. 181. Copyright 2020 American Chemical Society. (b) A CRISPR-Chip for dsDNA detection. The dCas9 complexed with a target-specific sgRNA (referred to as dRNP) is immobilized on the surface of the graphene within a three-terminal gFET. The dRNP scans the whole genomic DNA and unzips the double helix until it identifies and binds to its target sequence (complementary to the 5′ end of sgRNA). The binding of the target DNA to the dRNP complex can sensitively modulate the electrical characteristics of the gFET, resulting in an electrical signal output within 15 min. The figure was reproduced from ref. 176 with permission from Springer Nature, copyright 2019. | ||
Apart from the sensitive SERS, a CRISPR-Chip using a graphene-based field-effect transistor (gFET) system was developed (Fig. 8b).176 Using the gFET as a detection method, CRISPR-Chip can achieve an LOD of 1.7 fM without amplification within 15 min. In this work, the dCas9–sgRNA complex has been linked to the graphene layer with a 1-pyrenebutanoic acid (PBA) linker. The target sequence was bound to the dCas9–sgRNA complex specifically, which enhances the output signal relative to samples lacking the target sequence. To further expand the application of the CRISPR-Chip for SNP detection, two optimizations based on this design were accomplished by (1) monitoring multiple electrical signals including I, C, and V responses, and (2) applying multiple Cas9 orthologs to improve test discrimination.182
4.2. Cas12a-based amplification-free assays
Different from tdCas9, which has been widely adopted for sequence labeling and enrichment purposes, the Cas12a system harnesses the trans-cleavage ability for amplification-free diagnostics mainly focusing on augmentation of the signal output. Therefore, a series of visualization techniques including electrochemistry,183–186 LFA,187 nanoparticle reporter,188 piezoresistive effect,189 surface plasmon resonance,188 and molecular circuits175,190 have been applied to amplify the trans-cleavage for enhanced diagnostics.Liu et al. developed an electrochemical biosensor based on CRISPR–Cas12a (E-CRISPR) (Fig. 9a).183 The ssDNA tagged MBs were first immobilized on the gold electrode as the state condition, whereby the Cas12a ribonucleoprotein binding to the target sequence region then trans-cleaves the immobilized ssDNA. The electrical current difference was recorded for diagnostic visualization and an LOD of 50 pM was achieved without amplification.
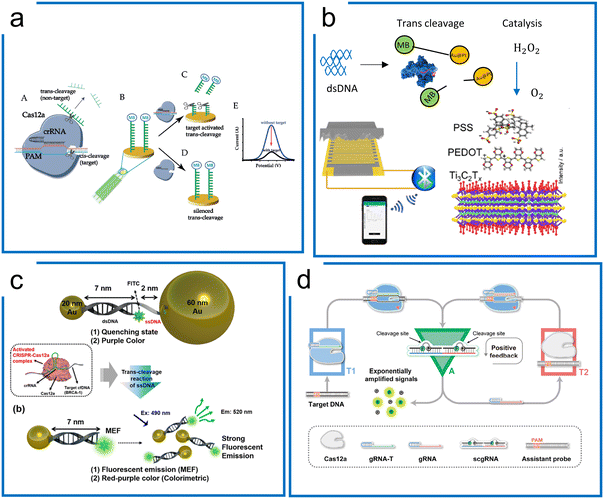 | ||
| Fig. 9 Amplification-free diagnostics with Cas12. (a) A Cas12a system for dsDNA detection using electrochemistry. MB conjugated ssDNA was immobilized on the electrochemical sensor surface to form the detection machinery. The signal can be measured once Cas12a trans-cleaves the ssDNA reporter on the surface of the electrode. The figure was reproduced from ref. 183 with permission from John Wiley and Sons, copyright 2019. (b) The piezoresistive biosensor with Cas12a for HPV-16 detection. When the Cas12a system recognizes the target sequence to activate the trans-cleavage, Cas12a enables the separation of AuPt and magnetic beads, which is conjugated via ssDNA. The released AuPt catalyzes H2O2 into O2, compressing the piezoresistive sensor to cause the current difference. The figure was reproduced from ref. 189 with permission from Elsevier, copyright 2021. (c) A metal-enhanced fluorescence and colorimetric system with Cas12a. Two different-sized AuNP (20 nm and 60 nm) pairs are connected by DNA hybridization with an ssDNA region to achieve the induction of metal-enhanced fluorescence (MEF) with target DNA and fluorescence quenching in the absence of target DNA. The presence of target DNA enables the trans-cleavage of Cas12a via the ssDNA region to switch from fluorescence quenching to MEF. Adapted from ref. 188. Copyright 2021 American Chemical Society. (d) A self-amplified system (CONAN) for dsDNA detection, in which a cycling loop is developed after Cas12a recognizes the target sequence. The trans-cleavage ability of Cas12a triggers the separation of the RNA–DNA probe to emit fluorescence and a new sgRNA. The released sgRNA guides Cas12a in targeting the assistant probe to release more fluorescence and sgRNA. The figure was reproduced from ref. 175 under Creative Commons Attribution License from the American Association for the Advancement of Science, copyright 2021. | ||
Tang et al. combined the piezoresistive wireless biosensor with Cas12a for HPV diagnosis (Fig. 9b).189 The trans-cleavage ability of Cas12a enables the separation of MB-ssDNA-AuPt reporters upon recognizing the target HPV 16. Subsequently, the separated AuPt particles were enriched by magnetism and then catalyzed H2O2 into O2, which compresses the sensor for increased current, enabling a LOD of 15.22 pM.
Unique surface plasmon resonance properties have also been identified with noble metal nanomaterials (NMNs),190 which can be regarded as the vibrational motion of an electromagnetic wave propagating in parallel along with the precious metal and dielectric interface. Fluorescence enhancement or fluorescence quenching is exhibited when noble metals are at different distances from the fluorescent dye. Choi et al. were able to design a AuNP assisted metal-enhanced fluorescence method for cell-free DNA detection using a Cas12a trans-cleavage (in Fig. 9c).188 A pair of AuNPs of two different sizes (20 nm and 60 nm) is linked through 7 nm long dsDNA and 2 nm long ssDNA, showing fluorescence quenching in the initial state. In the presence of the target DNA, the trans-cleavage ability of Cas12a cleaves the non-hybridized ssDNA part to release the 60 nm AuNPs. Therefore, the 20 nm AuNP can induce a strong signal from the fluorophore.
An internal autocatalysis-driven feedback amplification network was designed utilizing Cas12a by Nie et al.175 In this method, the CRISPR–Cas-only amplification network (CONAN) achieves a 5 aM sensitivity without DNA amplification (Fig. 9d). Here, they designed a probe comprising ssDNA reporters binding to the crRNA. When the target was bound to the Cas12a ribonucleoprotein, the probe was cleaved, releasing a fluorescent signal and new crRNA. The released crRNA assembled a new active Cas12a protein and participated in the reaction to achieve autocatalytic signal amplification. Moreover, the entropy driven catalysis DNA networks,191 as a self-driven amplification method without DNA polymerase, have been integrated with Cas12a for diagnostics and shown to reach picomolar sensitivity.190
4.3. Cas13-based amplification-free assays
Cas13 has been widely adopted for direct detection of RNA including mRNAs and microRNAs at picomolar concentrations without amplification. Signal augmentation methods, such as electrochemical biosensors,194–196 droplet confinement,193,197,198 and specifically molecular designs, have been combined with Cas13 to increase the LOD.The confinement of single molecules in cell-like-sized droplets/reactors can enhance local concentrations of the target and reporter by 3 orders of magnitude, and thus may break the detection limit of the most conventional biochemical assays or detectors. Zhou et al. achieved >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold enhancement in sensitivity compared to the bulk Cas13a assay using droplet microfluidics without nucleic acid amplification (Fig. 10a).192 This work has been further applied to detect SARS-CoV-2 without amplification. Similar work has been demonstrated using a microchamber array to improve the sensitivity of LwCas13a diagnostics down to 5 fM, which is 3 orders of magnitude more sensitive than the bulk test.198
000-fold enhancement in sensitivity compared to the bulk Cas13a assay using droplet microfluidics without nucleic acid amplification (Fig. 10a).192 This work has been further applied to detect SARS-CoV-2 without amplification. Similar work has been demonstrated using a microchamber array to improve the sensitivity of LwCas13a diagnostics down to 5 fM, which is 3 orders of magnitude more sensitive than the bulk test.198
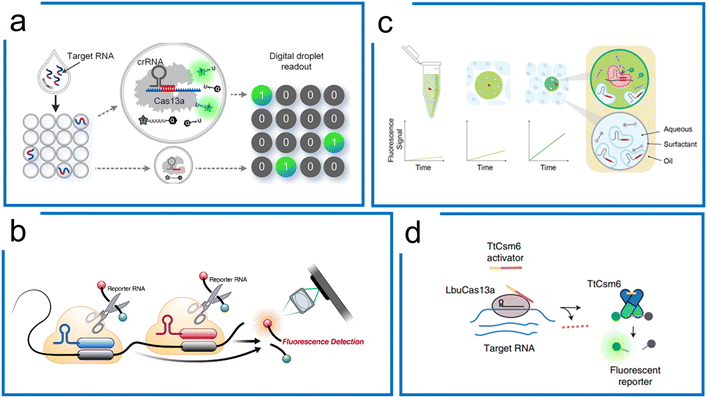 | ||
| Fig. 10 Cas13a-based amplification-free diagnostics. (a) Droplet microfluidics for Cas13a direct detection of RNA samples. By droplet compartment, local concentrations of the target and reporter are enhanced simultaneously, compared to the bulk Cas13a assay. It enables absolute digital single-molecule RNA quantitation. Adapted from ref. 192. Copyright 2021 American Chemical Society. (b) The tandem crRNAs for SARS-CoV-2 detection. Compared to the single-crRNA guiding Cas13 system, the multiple crRNA for the same target sequence improves the kinetic activity of trans-cleavage to higher sensitivity. The figure was reproduced from ref. 178 with permission from Elsevier, copyright 2021. (c) Multiplexed detection using Cas13a kinetic barcodes in droplets. The kinetic difference of Cas13a governed by both target RNA and crRNA is harnessed to achieve multiplexed diagnostics in droplet partition, where the fluorescence trajectory of different micro reactions within the droplets is recorded in real-time to differentiate the different targets. Adapted from ref. 193 under Creative Commons Attribution License from medRxiv. (d) Accelerated RNA detection using tandem CRISPR nucleases. Combining RNA-guided Cas13 and Csm6, a tandem nuclease assay is created with a chemically stabilized activator to achieve both high sensitivity and fast signal generation without requiring a preceding target amplification step. The figure was reproduced from ref. 179 with permission from Springer Nature, copyright 2021. | ||
Subsequently, Ott et al. demonstrated that the combination of multiple crRNAs in the LbuCas13a system can be used to directly detect viral RNA in the attomolar range without amplification. This method integrated with a cell phone readout was used for SARS-CoV-2 detection with a sensitivity of 167 aM within 30 minutes (Fig. 10b).178 The Cas enzymatic kinetics were found to be associated with the crRNA. Accordingly, Fletcher et al. proposed a multiplexed diagnostic method with 2 crRNA that has been used in combination with droplet microfluidics. Based on the stochastic nuclease activities, they analysed the kinetic features of each droplet over time for multiplexing (Fig. 10c).193
In addition to the tandem crRNA strategy, the tandem use of unrelated CRISPR nucleases can increase the SHERLOCK sensitivity.179 Doudna et al. engineered the Csm6 with a chemically stabilized activator. Once LbuCas13a guided by multiple crRNAs was activated, the Csm6 would trans-cleave extra reporters, creating a one-step amplification-free SARS-CoV-2 detection assay. This method, termed the Fast Integrated Nuclease Detection In Tandem (FIND-IT) approach, enabled an efficient detection (50 aM) within 20 minutes, highlighting the potential for amplification-free RNA detection ability using LbuCas13a (Fig. 10d).
Besides fluorescence visualization, an electrochemical biosensor combined with Cas13a has been reported to directly detect microRNA 19b.194 In this design, Urban et al. were able to immobilize the anti-fluorescein antibodies attached to glucose oxidase (GOx) via the FAM and biotin-tagged ssRNA probes on the streptavidin-coated substrate surface. The electrochemical detection was accomplished using GOx to catalyze the substrate, producing H2O2, where the current measurement was detected. In the presence of the target microRNA, the activated Cas13a trans-cleaved the ssRNA, interrupting the catalytic process. To expand the detection ability, they then further developed a multi-channel microfluidic chip to enable 4 multiple detections.196 To increase the signal output, a hairpin-based enzyme-free amplification was applied to increase the electrochemical signal via a catalytic chain reaction.199,200
5 Perspectives
5.1. Target quantification
Accurate quantification of biomarker molecule concentrations is of great importance in order to respond to treatment in companion diagnostic and precision medicine development. Similar to PCR, quantification using a CRISPR-based diagnostics system can be obtained by relative quantification using a standard curve or digital quantification using Poisson statistics.219 The requirement for quantification through comparison with a standard curve reference and continuous measurement of a signal hampers the feasibility of the CRISPR-based diagnostics system as a POCT tool.Digital quantification has been reported to greatly reduce uncertainty at low concentrations.220 By employing nano-well-based221 or droplet-based222,223 technologies for sample partitioning, the digital format of CRISPR-based assays enables absolute quantification of nucleic acids with no dependency on endogenous references, as it measures the number of target molecules directly by observing the end-point fluorescence in individual partitions.
To date, most CRISPR effectors in diagnostics operate at ambient temperature. In combination with isothermal amplification, two concerns may arise:224 target overestimation associated with the premature amplification induced by the near ambient temperature reaction (e.g., RPA-based preamplification) and target loss in the multistep procedures. The possible solutions to address these issues can be classified into three aspects. First, sample preparation at a lower temperature such as in a cold room or on ice and rapid transfer or operation during experiments are demanded. Second, digital quantification reforms from an ambient reaction to a hot-start reaction such as LAMP. With the expansion of the CRISPR Cas family, the thermostability of Cas proteins is discovered.36,121 Those thermostable proteins of Cas12 and Cas13 families allow the CRISPR diagnostics along with LAMP.161,225 Finally, the preamplification issue can be avoided by the creation of a reaction initiator (e.g., Mg initiator or DNA polymerase). For instance, the mixing of the MgOAc initiator into the RPA-CRISPR reaction before partitioning can solve the premature amplification using microfluidic technologies such as Y-shaped droplet generator,108 picoinjector,226 droplet emerging,105 droplet array,104 Slipchip,227etc. Nevertheless, all the quantification methods require sophisticated ancillary equipment from droplet partition to visualization. The miniaturized and operation-free digital quantification might be another chapter in diagnostics, such as high-throughput droplet partition,228–230 vortex droplet partition,231 advanced pore materials,232 and self-digitalization systems.107,233,234
5.2. Multiplexed detection
Simultaneous diagnosis of multiple targets in one reaction is desirable in order to reduce the overall testing cost and improve the throughput in certain testing demands. With more and more unique properties of Cas enzymes uncovered in recent years, those special features of CRISPR have been leveraged to expand the multiplexing ability of CRISPR diagnostics such as (1) programmable crRNA-based Cas9 system (LEOPARD);39 (2) orthogonal cleavage properties of Cas enzymes (SHERLOCKv2);46 (3) droplet barcoding and pairing enabled massively multiplexed detection (CARMEN);104 (4) heterogeneous nuclease kinetics in a confined volume (droplet kinetic barcoding).193The LEOPARD platform is based on the discovery of the RNA guidance of CjeCas9 derived from cellular RNAs unrelated to viral defense. Thus, the RNA guidance can be reprogrammed to any RNA to affect the activation of CjeCas9 nuclease. This discovery extended the programmability of the spacer region of crRNA to tracrRNA. The sensing of RNA is enabled by creating fluorescent DNA sensors that would be cleaved at a specific locus by Cas9. The current proof-of-principle demonstration of LEOPARD allows the detection of multiple different RNA sequences using gel-based readouts. Alternative methods such as lateral flow and colorimetric readouts would be beneficial for point-of-care detection.
In the Cas12 and Cas13-based detection systems, the signals are normally derived from nonspecific trans-cleavage with universal probes, making the multiplexing of various targets a challenging task. In SHERLOCKv2, the Cas13 family has been discovered with the di-nucleotide preference to cleave different sequences. Four Cas13 effectors were discovered to have specific cleavage preferences on di-nucleotide reporters (AU, UC, AC, and GA), and thus their orthogonal collateral-cleavage activities (including Cas13 and Cas12a enzymes) have been harnessed to achieve up to 5 channel multiplexing using distinct fluorescent dyes.46 Additional multiplexing in a single reaction is possible as newer Cas effectors that possess the di-nucleotide preference are still being explored. However, the combination of multiple nuclease-crRNAs with different probes would be a crowding reaction that may result in high background and low sensitivity.
CARMEN–Cas13 combines the specific sensing ability of the CRISPR-based detection technology SHERLOCK with the throughput capabilities of the combinatorial droplet array technology to enable parallel, scalable, and massively multiplexed diagnostics.104 To enable high multiplexing with high sample throughput, a set of 1050 color codes was generated using ratios of 4 different fluorescent dyes. Each amplified sample or Cas13 detection mix was emulsified into color-coded droplets. The droplets of all samples and detection mixes were pooled and loaded into a droplet pairing array for registration of the droplet identity with a distinct color code. After the droplet pairs were merged, the Cas13a-triggered fluorescence reported a positive reaction. The high sensitivity and specificity of Cas13a allowed the simultaneous detection of 169 human-associated viruses in a single microchip. An upgraded version of CARMEN (mCARMEN) by leveraging the Fluidigm microfluidic platform has decreased the processing time.106 Unfortunately, a two-step process is required which compromises its quantification abilities. Future development of one-pot assays or CRISPR-based amplification-free techniques may further simplify mCARMEN. CARMEN or mCARMEN methods have the advantage of extremely high-throughput and multiplex detection capabilities. Further improvements can be focused on the integration and optimization of the multiplexed platforms with advanced microfluidic systems. Similarly, ideas of distributing crRNAs into different microchannels or wells have been developed for the detection of multiple targets using microchip-based space coding.167
Droplet microfluidics offers the ability to isolate single molecules and reduce inhibition or interference by confinement as well as accelerated reaction and improved sensitivity in a low reaction volume. Rapid and diverse kinetic activities of the Cas13 reaction were observed when the reaction volume shrank to even a single target in a confined droplet.196 Thus, this feature is leveraged for multiplexed diagnostics at single-molecule resolution using droplet microfluidics. The kinetic behavior depends on a specific combination of crRNA and its target; thus it can be harnessed for CRISPR-based multiplexed diagnostics by tracking the real-time fluorescence trajectory of individual droplets with single-molecule level sensitivity. As a result, this droplet-based CRISPR/Cas13a assay can realize absolute quantification and multiplexed detection simultaneously. However, this method has only been demonstrated in two-channel multiplexed detection. The multiplexibility might be restricted by the differentiation of the kinetics difference.
5.3. Streamlined point-of-care diagnostics
To date, the majority of efforts in the development of CRISPR-based diagnostic systems have been focused on amplification and detection techniques. For a POCT system, streamlined steps from sample preparation, amplification or amplification-free methods, and signal visualization are all equally important. The nucleic acid preparation can be divided into three steps including target lysis, purification, and elusion. Target lysis then relies on mechanical, chemical, and physical approaches to break the cell membrane and wall to expose the cytoplasm.235 Some methods have subsequently claimed purification-free nucleic acid detection after lysis.236–238 However, purification is highly sought after, followed by the elusion step in order to reduce the inhabitation and increase the sensitivity.239 More recently, the promising thermal lysis method HUDSON has been developed, which combines the sample preparation and CRISPR isothermal diagnostics for Zika virus and DENV detection at 1 copy per microliter level.100 Similarly, a chemical-based direct sampling method coupled with LAMP and Cas13 diagnostics has been reported to achieve 25 cp μL−1 around 30 min.240 These methods do not require cumbersome experimental operations such as centrifugal separation used in standard purification protocols, and thus reduce the need for expensive labor force and centralized facilities. These merits imply that the potential POCT with CRISPR could be massively simplified and miniaturized, overcoming the limitations of the PCR system including thermal cycling requirements and inhibitors to DNA polymerase.Hence, the possible streamlined diagnostics would be via either a multistep test kit with easy operations,241 or an automatic all-in-one system.109,125 Inspired by the rapid test kit for COVID-19 used massively worldwide, the new detection style is accepted by several with simple liquid handing from the collection of the non-invasive sample into reactions with lyophilized or liquid chemicals. Besides, the ultra-miniaturized system with a simple heating function for sample lysis and incubation and fluorescence reader or LFA visualization are new pathways to further reduce the cost of a POCT device with a more sensitive readout. In addition, a facile and robust molecular assay is highly desired. However, the current one-pot assays have shown less sensitivity compared to the two-step reactions. To address this problem, dedicated optimization of the current molecular reaction schemes and characterization of new Cas effectors are in urgent need for the advancement of POCT.
5.4. Beyond nucleic acid detection
Other than nucleic acids, various biomarkers such as proteins/peptides, small molecules, exosomes, etc. are also important sensing targets in environmental protection, microbiome monitoring, food safety, drug testing, and antimicrobial resistance diagnosis. Although CRISPR-based detection schemes cannot be directly applied to non-nucleic acids, the modularity of the CRISPR–Cas system offers a great many opportunities for the development of CRISPR-mediated strategies for sensing non-nucleic-acid targets. The general principle is to convert the non-nucleic-acid targets into pre-designed DNA or RNA intermediates for use of CRISPR–Cas as target-induced ssDNases or RNases. CRISPR–Cas systems coupled with transduction elements including oligonucleotide–protein conjugates,242–245 aptamers,246–251 DNAzymes,252–254 allosteric transcriptional factors,255,256 riboswitches,257 and enzymatic reactions258,259 have been developed for detecting non-nucleic-acid targets. Such developments have expanded the applications of the CRISPR–Cas toolbox to meet diverse purposes such as control of gene expression, nanopore sequencing, live cell imaging, environmental monitoring, etc.However, due to the lack of inherent mechanisms for the amplification of non-nucleic-acid targets, the detection limit of non-nucleic-acid targets by CRISPR systems is still comparably inferior, pending more innovative coupling and integration techniques with advanced sensing instruments. Moreover, simultaneous detection of multiple non-nucleic-acid targets remains a challenge and the development of such schemes is still in its infancy. Various approaches, such as assorted labeling and barcoding strategies, super-resolution imaging, microfluidics, microarrays, etc., should be explored for multi-target sensing or fit-for-purpose applications.
6 Conclusions
In this review, we discuss the developments of CRISPR-based detection strategies utilizing different Cas subsystems, the mechanisms of CRISPR/Cas as elegant molecular scissors that recognize and cut nucleic acid targets, and the engineering of such molecular scissors for a broader detection of biomolecules with flexibility, sensitivity, and specificity. The coupling of CRISPR–Cas enzymes with either nuclei acid amplification or signal amplification methods has provided us flexible choices of schemes to meet the detection requirements in terms of the types of targets and sensitivity needed. Besides, the development of direct targeting using CRISPR with new signal transformation schemes has shown a blooming prosperity of this field to avoid enzyme-based amplification methods. Moreover, the advancement of emerging technologies such as nanotechnologies, microfluidics, and wearable electronics will continuously propel the CRISPR-based technologies towards rapid, high-throughput, integrated, multiplexed, and digital detection. However, the practical applications have to wait for a little bit longer till the chemical stability (e.g., crRNAs, enzymes), storage, and overall design of easy-to-use schemes can be fully achieved. Combining microfluidics, wearable technology, flexible materials, and nanotechnology, CRISPR-based technologies will lead to many powerful POCT devices meeting the growing necessity for quick, accurate and resource-limited medical tests to help quickly respond to outbreaks of viral and other diseases.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Research Grants Council of Hong Kong under the General Research Fund (16205619) and the Collaborative Research Fund (C6107-20GF), and the Innovation and Technology Commission under Midstream Research Programme for Universities (MRP/077/20).Notes and references
- C. Wang, P. W. Horby, F. G. Hayden and G. F. Gao, Lancet, 2020, 395, 470–473 CrossRef CAS PubMed.
- D. S. Chertow, Science, 2018, 360, 381–382 CrossRef PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf, O. S. Alkhnbashi, F. Costa, S. A. Shah, S. J. Saunders, R. Barrangou, S. J. Brouns, E. Charpentier and D. H. Haft, Nat. Rev. Microbiol., 2015, 13, 722–736 CrossRef CAS PubMed.
- P. Mohanraju, K. S. Makarova, B. Zetsche, F. Zhang, E. V. Koonin and J. Van der Oost, Science, 2016, 353(6299), aad5147 CrossRef PubMed.
- S. A. Jackson, R. E. McKenzie, R. D. Fagerlund, S. N. Kieper, P. C. Fineran and S. J. Brouns, Science, 2017, 356, eaal5056 CrossRef PubMed.
- R. Barrangou and P. Horvath, Nat. Microbiol., 2017, 2, 1–9 Search PubMed.
- J. McGinn and L. A. Marraffini, Nat. Rev. Microbiol., 2019, 17, 7–12 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf, J. Iranzo, S. A. Shmakov, O. S. Alkhnbashi, S. J. Brouns, E. Charpentier, D. Cheng, D. H. Haft and P. Horvath, Nat. Rev. Microbiol., 2020, 18, 67–83 CrossRef CAS PubMed.
- Y. Ishino, H. Shinagawa, K. Makino, M. Amemura and A. Nakata, J. Bacteriol., 1987, 169, 5429–5433 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan and J. J. Collins, Cell, 2016, 165, 1255–1266 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang and L. A. Marraffini, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- J. Quan, C. Langelier, A. Kuchta, J. Batson, N. Teyssier, A. Lyden, S. Caldera, A. McGeever, B. Dimitrov and R. King, Nucleic Acids Res., 2019, 47, e83 CrossRef CAS PubMed.
- P. Giesselmann, B. Brändl, E. Raimondeau, R. Bowen, C. Rohrandt, R. Tandon, H. Kretzmer, G. Assum, C. Galonska and R. Siebert, Nat. Biotechnol., 2019, 37, 1478–1481 CrossRef CAS PubMed.
- T. Gilpatrick, I. Lee, J. E. Graham, E. Raimondeau, R. Bowen, A. Heron, B. Downs, S. Sukumar, F. J. Sedlazeck and W. Timp, Nat. Biotechnol., 2020, 38, 433–438 CrossRef CAS PubMed.
- O. O. Abudayyeh, J. S. Gootenberg, S. Konermann, J. Joung, I. M. Slaymaker, D. B. Cox, S. Shmakov, K. S. Makarova, E. Semenova and L. Minakhin, Science, 2016, 353(6299), aaf5573 CrossRef PubMed.
- A. East-Seletsky, M. R. O'Connell, S. C. Knight, D. Burstein, J. H. Cate, R. Tjian and J. A. Doudna, Nature, 2016, 538, 270–273 CrossRef CAS PubMed.
- B. Perez-Lopez and M. Mir, Talanta, 2021, 225, 121898 CrossRef CAS PubMed.
- Y. Tang, L. Gao, W. Feng, C. Guo, Q. Yang, F. Li and X. C. Le, Chem. Soc. Rev., 2021, 50, 11844 RSC.
- H. Rahimi, M. Salehiabar, M. Barsbay, M. Ghaffarlou, T. Kavetskyy, A. Sharafi, S. Davaran, S. C. Chauhan, H. Danafar and S. Kaboli, ACS Sens., 2021, 6, 1430–1445 CrossRef CAS PubMed.
- M. M. Kaminski, O. O. Abudayyeh, J. S. Gootenberg, F. Zhang and J. J. Collins, Nat. Biomed. Eng., 2021, 5, 643–656 CrossRef CAS PubMed.
- P. Li, H. Xiong, B. Yang, X. Jiang, J. Kong and X. Fang, TrAC, Trends Anal. Chem., 2022, 116812 CrossRef CAS.
- Y. Dai, Y. Wu, G. Liu and J. J. Gooding, Angew. Chem., Int. Ed., 2020, 59, 20754–20766 CrossRef CAS PubMed.
- Y. Chen, S. Qian, X. Yu, J. Wu and J. Xu, Trends Biotechnol., 2022 DOI:10.1016/j.tibtech.2022.07.015.
- J. E. van Dongen, J. T. Berendsen, R. D. Steenbergen, R. M. Wolthuis, J. C. Eijkel and L. I. Segerink, Biosens. Bioelectron., 2020, 166, 112445 CrossRef CAS PubMed.
- H. Yue, M. Huang, T. Tian, E. Xiong and X. Zhou, ACS Nano, 2021, 15, 7848–7859 CrossRef CAS PubMed.
- R. Nouri, Z. Tang, M. Dong, T. Liu, A. Kshirsagar and W. Guan, Biosens. Bioelectron., 2021, 178, 113012 CrossRef CAS PubMed.
- A. Lau, C. Ren and L. P. Lee, Shengwu Yixue Gongchengxue Jinzhan, 2020, 3, 012001 Search PubMed.
- J. A. Steens, Y. Zhu, D. W. Taylor, J. P. Bravo, S. H. Prinsen, C. D. Schoen, B. J. Keijser, M. Ossendrijver, L. M. Hofstra and S. J. Brouns, Nat. Commun., 2021, 12, 1–12 CrossRef PubMed.
- K. Yoshimi, K. Takeshita, S. Yamayoshi, S. Shibumura, Y. Yamauchi, M. Yamamoto, H. Yotsuyanagi, Y. Kawaoka and T. Mashimo, iScience, 2022, 25, 103830 CrossRef CAS PubMed.
- D. A. Huyke, A. Ramachandran, V. I. Bashkirov, E. K. Kotseroglou, T. Kotseroglou and J. G. Santiago, Anal. Chem., 2022, 94, 9826–9834 CrossRef CAS PubMed.
- A. S. Avaro and J. G. Santiago, Angew. Chem., Int. Ed., 2022, 61, e202209527 CrossRef CAS PubMed.
- A. Ramachandran and J. G. Santiago, Anal. Chem., 2021, 93, 7456–7464 CrossRef CAS PubMed.
- F. Teng, T. Cui, G. Feng, L. Guo, K. Xu, Q. Gao, T. Li, J. Li, Q. Zhou and W. Li, Cell Discovery, 2018, 4, 1–15 CrossRef CAS PubMed.
- L. T. Nguyen, N. C. Macaluso, B. L. Pizzano, M. N. Cash, J. Spacek, J. Karasek, M. R. Miller, J. A. Lednicky, R. R. Dinglasan and M. Salemi, EBioMedicine, 2022, 77, 103926 CrossRef CAS PubMed.
- L. Li, S. Li, N. Wu, J. Wu, G. Wang, G. Zhao and J. Wang, ACS Synth. Biol., 2019, 8, 2228–2237 CrossRef CAS PubMed.
- M. R. O'Connell, B. L. Oakes, S. H. Sternberg, A. East-Seletsky, M. Kaplan and J. A. Doudna, Nature, 2014, 516, 263–266 CrossRef PubMed.
- C. Jiao, S. Sharma, G. Dugar, N. L. Peeck, T. Bischler, F. Wimmer, Y. Yu, L. Barquist, C. Schoen and O. Kurzai, Science, 2021, 372, 941–948 CrossRef CAS PubMed.
- L. S. Qi, M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173–1183 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer and C. A. Freije, Science, 2017, 356, 438–442 CrossRef CAS PubMed.
- M. J. Kellner, J. G. Koob, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, Nat. Protoc., 2019, 14, 2986–3012 CrossRef CAS PubMed.
- S. Li, Q. Cheng, J. Wang, X. Li, Z. Zhang, S. Gao, R. Cao, G. Zhao and J. Wang, Cell Discovery, 2018, 4, 1–4 CAS.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- P. Ma, Q. Meng, B. Sun, B. Zhao, L. Dang, M. Zhong, S. Liu, H. Xu, H. Mei and J. Liu, Adv. Sci., 2020, 7, 2001300 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed.
- L. T. Nguyen, B. M. Smith and P. K. Jain, Nat. Commun., 2020, 11, 1–13 CrossRef PubMed.
- H. Yang, P. Gao, K. R. Rajashankar and D. J. Patel, Cell, 2016, 167, 1814–1828 CrossRef CAS PubMed , e12.
- J. Strecker, S. Jones, B. Koopal, J. Schmid-Burgk, B. Zetsche, L. Gao, K. S. Makarova, E. V. Koonin and F. Zhang, Nat. Commun., 2019, 10, 1–8 CrossRef PubMed.
- J. Joung, A. Ladha, M. Saito, N. Kim, A. E. Woolley, M. Segel, R. P. Barretto, A. Ranu, R. K. Macrae, G. Faure and F. Zhang, N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed.
- F. Teng, L. Guo, T. Cui, X. Wang, K. Xu, Q. Gao, Q. Zhou and W. Li, Genome Biol., 2019, 20, 1–7 CrossRef CAS PubMed.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Science, 2018, 362, 839–842 CrossRef CAS PubMed.
- T. Karvelis, G. Bigelyte, J. K. Young, Z. Hou, R. Zedaveinyte, K. Budre, S. Paulraj, V. Djukanovic, S. Gasior and A. Silanskas, Nucleic Acids Res., 2020, 48, 5016–5023 CrossRef CAS PubMed.
- O. O. Abudayyeh, J. S. Gootenberg, P. Essletzbichler, S. Han, J. Joung, J. J. Belanto, V. Verdine, D. B. Cox, M. J. Kellner and A. Regev, Nature, 2017, 550, 280–284 CrossRef PubMed.
- A. East-Seletsky, M. R. O'Connell, D. Burstein, G. J. Knott and J. A. Doudna, Mol. Cell, 2017, 66, 373–383 CrossRef CAS PubMed , e3.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- O. Piepenburg, C. H. Williams, D. L. Stemple and N. A. Armes, PLoS Biol., 2006, 4, e204 CrossRef PubMed.
- G. T. Walker, M. S. Fraiser, J. L. Schram, M. C. Little, J. G. Nadeau and D. P. Malinowski, Nucleic Acids Res., 1992, 20, 1691–1696 CrossRef CAS PubMed.
- J. Compton, Nature, 1991, 350, 91–92 CrossRef CAS PubMed.
- J. Van Ness, L. K. Van Ness and D. J. Galas, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4504–4509 CrossRef CAS PubMed.
- P. M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas and D. C. Ward, Nat. Genet., 1998, 19, 225–232 CrossRef CAS PubMed.
- A. Ganguli, A. Mostafa, J. Berger, M. Y. Aydin, F. Sun, S. A. Stewart de Ramirez, E. Valera, B. T. Cunningham, W. P. King and R. Bashir, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 22727–22735 CrossRef CAS PubMed.
- V. L. D. Thi, K. Herbst, K. Boerner, M. Meurer, L. P. Kremer, D. Kirrmaier, A. Freistaedter, D. Papagiannidis, C. Galmozzi and M. L. Stanifer, Sci. Transl. Med., 2020, 12, eabc7075 CrossRef PubMed.
- B. A. Rabe and C. Cepko, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24450–24458 CrossRef CAS PubMed.
- M. El-Tholoth, H. H. Bau and J. Song, ChemRxiv, 2020, preprint, DOI:10.26434/chemrxiv.11860137.v1.
- T. Chaibun, J. Puenpa, T. Ngamdee, N. Boonapatcharoen, P. Athamanolap, A. P. O'Mullane, S. Vongpunsawad, Y. Poovorawan, S. Y. Lee and B. Lertanantawong, Nat. Commun., 2021, 12, 1–10 CrossRef PubMed.
- B. Tian, F. Gao, J. Fock, M. Dufva and M. F. Hansen, Biosens. Bioelectron., 2020, 165, 112356 CrossRef CAS PubMed.
- Q. Wu, C. Suo, T. Brown, T. Wang, S. A. Teichmann and A. R. Bassett, Sci. Adv., 2021, 7, eabe5054 CrossRef CAS PubMed.
- J. G. Carter, L. O. Iturbe, J. H. Duprey, I. R. Carter, C. D. Southern, M. Rana, C. M. Whalley, A. Bosworth, A. D. Beggs, M. R. Hicks, J. H. Tucker and T. R. Dafforn, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(35), e2100347118 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- M. L. Powell, F. R. Bowler, A. J. Martinez, C. J. Greenwood, N. Armes and O. Piepenburg, Anal. Biochem., 2018, 543, 108–115 CrossRef CAS PubMed.
- X. Lu, H. L. Yu, H. Lin, Y. Cao and I. Hsing, Sens. Actuators, B, 2022, 131385 CrossRef CAS.
- J. Q. Cui, F. X. Liu, H. Park, K. W. Chan, T. Leung, B. Z. Tang and S. Yao, Biosens. Bioelectron., 2022, 114019 CrossRef CAS PubMed.
- J. C. Guatelli, K. M. Whitfield, D. Y. Kwoh, K. J. Barringer, D. D. Richman and T. R. Gingeras, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 1874–1878 CrossRef CAS PubMed.
- G. T. Walker, M. C. Little, J. G. Nadeau and D. D. Shank, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 392–396 CrossRef CAS PubMed.
- F. B. Dean, J. R. Nelson, T. L. Giesler and R. S. Lasken, Genome Res., 2001, 11, 1095–1099 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- H. Jia, Z. Li, C. Liu and Y. Cheng, Angew. Chem., Int. Ed., 2010, 49, 5498–5501 CrossRef CAS PubMed.
- W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu and X. Yu, Nat. Commun., 2018, 9, 1–11 CrossRef PubMed.
- M. Huang, X. Zhou, H. Wang and D. Xing, Anal. Chem., 2018, 90, 2193–2200 CrossRef CAS PubMed.
- M. S. Reid, X. C. Le and H. Zhang, Angew. Chem., Int. Ed., 2018, 57, 11856–11866 CrossRef CAS PubMed.
- T. Wang, Y. Liu, H. Sun, B. Yin and B. Ye, Angew. Chem., 2019, 131, 5436–5440 CrossRef.
- L. Wang, X. Shen, T. Wang, P. Chen, N. Qi, B. Yin and B. Ye, Biosens. Bioelectron., 2020, 165, 112364 CrossRef CAS PubMed.
- K. Zhang, R. Deng, X. Teng, Y. Li, Y. Sun, X. Ren and J. Li, J. Am. Chem. Soc., 2018, 140, 11293–11301 CrossRef CAS PubMed.
- X. Wang, E. Xiong, T. Tian, M. Cheng, W. Lin, H. Wang, G. Zhang, J. Sun and X. Zhou, ACS Nano, 2020, 14, 2497–2508 CrossRef CAS PubMed.
- E. Xiong, L. Jiang, T. Tian, M. Hu, H. Yue, M. Huang, W. Lin, Y. Jiang, D. Zhu and X. Zhou, Angew. Chem., 2021, 133, 5367–5375 CrossRef.
- X. Qiu, L. Zhu, C. Zhu, J. Ma, T. Hou, X. Wu, S. Xie, L. Min, D. Tan and D. Zhang, ACS Synth. Biol., 2018, 7, 807–813 CrossRef CAS PubMed.
- T. Marsic, Z. Ali, M. Tehseen, A. Mahas, S. Hamdan and M. Mahfouz, Nano Lett., 2021, 21, 3596–3603 CrossRef CAS PubMed.
- Z. Ali, E. Sánchez, M. Tehseen, A. Mahas, T. Marsic, R. Aman, G. Sivakrishna Rao, F. S. Alhamlan, M. S. Alsanea and A. A. Al-Qahtani, ACS Synth. Biol., 2022, 11, 406–419 CrossRef CAS PubMed.
- B. Koo, D. Kim, J. Kweon, C. E. Jin, S. Kim, Y. Kim and Y. Shin, Sens. Actuators, B, 2018, 273, 316–321 CrossRef CAS PubMed.
- R. Wang, X. Zhao, X. Chen, X. Qiu, G. Qing, H. Zhang, L. Zhang, X. Hu, Z. He and D. Zhong, Anal. Chem., 2019, 92, 2176–2185 CrossRef PubMed.
- Y. Bao, Y. Jiang, E. Xiong, T. Tian, Z. Zhang, J. Lv, Y. Li and X. Zhou, ACS Sens., 2020, 5, 1082–1091 CrossRef CAS PubMed.
- X. Sun, Y. Wang, L. Zhang, S. Liu, M. Zhang, J. Wang, B. Ning, Y. Peng, J. He and Y. Hu, Anal. Chem., 2020, 92, 3032–3041 CrossRef CAS PubMed.
- M. Chen, D. Wu, S. Tu, C. Yang, D. Chen and Y. Xu, Biosens. Bioelectron., 2021, 173, 112821 CrossRef CAS PubMed.
- M. Azhar, R. Phutela, M. Kumar, A. H. Ansari, R. Rauthan, S. Gulati, N. Sharma, D. Sinha, S. Sharma and S. Singh, Biosens. Bioelectron., 2021, 183, 113207 CrossRef CAS PubMed.
- T. Marsic, Z. Ali, M. Tehseen, A. Mahas, S. Hamdan and M. Mahfouz, Nano Lett., 2021, 21, 3596–3603 CrossRef CAS PubMed.
- M. Bengtson, M. Bharadwaj, O. Franch, J. van der Torre, V. Meerdink, H. Schallig and C. Dekker, Nanoscale, 2022, 14, 1885–1895 RSC.
- T. Wang, Y. Wang, P. Chen, B. Yin and B. Ye, Anal. Chem., 2022, 94, 12461–12471 CrossRef CAS PubMed.
- J. Song, B. Cha, J. Moon, H. Jang, S. Kim, J. Jang, D. Yong, H. Kwon, I. Lee and E. Lim, ACS Nano, 2022, 16, 11300–11314 CrossRef CAS PubMed.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul and L. A. Parham, Science, 2018, 360, 444–448 CrossRef CAS PubMed.
- K. G. Barnes, A. E. Lachenauer, A. Nitido, S. Siddiqui, R. Gross, B. Beitzel, K. J. Siddle, C. A. Freije, B. Dighero-Kemp and S. B. Mehta, Nat. Commun., 2020, 11, 1–10 CrossRef PubMed.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm, S. Siddiqui, B. M. Shaw, G. Adams, T. F. Kosoko-Thoroddsen and M. E. Kemball, Nat. Commun., 2020, 11, 1–9 CrossRef PubMed.
- H. C. Metsky, N. L. Welch, P. P. Pillai, N. J. Haradhvala, L. Rumker, S. Mantena, Y. B. Zhang, D. K. Yang, C. M. Ackerman and J. Weller, Nat. Biotechnol., 2022, 1–9 Search PubMed.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky, D. K. Yang, H. Y. Simon, C. K. Boehm, T. F. Kosoko-Thoroddsen and J. Kehe, Nature, 2020, 582, 277–282 CrossRef CAS PubMed.
- A. Kulesa, J. Kehe, J. E. Hurtado, P. Tawde and P. C. Blainey, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 6685–6690 CrossRef PubMed.
- N. L. Welch, M. Zhu, C. Hua, J. Weller, M. E. Mirhashemi, T. G. Nguyen, S. Mantena, M. R. Bauer, B. M. Shaw, C. M. Ackerman and C. Myhrvold, Nat. Med., 2022, 1 Search PubMed.
- E. Yeh, C. Fu, L. Hu, R. Thakur, J. Feng and L. P. Lee, Sci. Adv., 2017, 3, e1501645 CrossRef PubMed.
- F. X. Liu, J. Q. Cui, H. Park, K. W. Chan, T. Leung, B. Z. Tang and S. Yao, Anal. Chem., 2022, 94, 5883–5892 CrossRef CAS PubMed.
- S. S. Chandrasekaran, S. Agrawal, A. Fanton, A. R. Jangid, B. Charrez, A. M. Escajeda, S. Son, R. Mcintosh, H. Tran, A. Bhuiya and P. D. Hsu, Nat. Biomed. Eng., 2022, 1–13 Search PubMed.
- Y. Shan, X. Zhou, R. Huang and D. Xing, Anal. Chem., 2019, 91, 5278–5285 CrossRef CAS PubMed.
- M. Huang, R. Huang, H. Yue, Y. Shan and D. Xing, Sens. Actuators, B, 2020, 325, 128799 CrossRef CAS.
- N. L. Welch, M. Zhu, C. Hua, J. Weller, M. E. Mirhashemi, T. G. Nguyen, S. Mantena, M. R. Bauer, B. M. Shaw and C. M. Ackerman, Nat. Med., 2022, 1–12 Search PubMed.
- B. Ning, T. Yu, S. Zhang, Z. Huang, D. Tian, Z. Lin, A. Niu, N. Golden, K. Hensley and B. Threeton, Sci. Adv., 2021, 7, eabe3703 CrossRef CAS PubMed.
- P. Q. Nguyen, L. R. Soenksen, N. M. Donghia, N. M. Angenent-Mari, H. de Puig, A. Huang, R. Lee, S. Slomovic, T. Galbersanini, G. Lansberry and J. J. Collins, Nat. Biotechnol., 2021, 39, 1366–1374 CrossRef CAS PubMed.
- C. Yuan, T. Tian, J. Sun, M. Hu, X. Wang, E. Xiong, M. Cheng, Y. Bao, W. Lin and J. Jiang, Anal. Chem., 2020, 92, 4029–4037 CrossRef CAS PubMed.
- M. M. Kaminski, M. A. Alcantar, I. T. Lape, R. Greensmith, A. C. Huske, J. A. Valeri, F. M. Marty, V. Klämbt, J. Azzi and E. Akalin, Nat. Biomed. Eng., 2020, 4, 601–609 CrossRef CAS PubMed.
- M. Patchsung, K. Jantarug, A. Pattama, K. Aphicho, S. Suraritdechachai, P. Meesawat, K. Sappakhaw, N. Leelahakorn, T. Ruenkam and T. Wongsatit, Nat. Biomed. Eng., 2020, 4, 1140–1149 CrossRef CAS PubMed.
- J. Arizti-Sanz, A. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije, M. E. Grunberg, T. F. Kosoko-Thoroddsen, N. L. Welch, P. P. Pillai and S. Mantena, Nat. Biomed. Eng., 2022, 1–12 Search PubMed.
- M. Patchsung, A. Homchan, K. Aphicho, S. Suraritdechachai, T. Wanitchanon, A. Pattama, K. Sappakhaw, P. Meesawat, T. Wongsatit and A. Athipanyasilp, medRxiv, 2022, preprint, DOI:10.1101/2022.03.17.22272589.
- B. Casati, J. P. Verdi, A. Hempelmann, M. Kittel, A. G. Klaebisch, B. Meister, S. Welker, S. Asthana, S. Di Giorgio and P. Boskovic, Nat. Commun., 2022, 13, 1–11 Search PubMed.
- A. Mahas, T. Marsic, M. Lopez-Portillo Masson, Q. Wang, R. Aman, C. Zheng, Z. Ali, M. Alsanea, A. Al-Qahtani and B. Ghanem, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2118260119 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li, R. V. Lalla, E. Ballesteros, M. M. Sfeir and C. Liu, Nat. Commun., 2020, 11, 1–10 CrossRef PubMed.
- S. Lu, X. Tong, Y. Han, K. Zhang, Y. Zhang, Q. Chen, J. Duan, X. Lei, M. Huang and Y. Qiu, Nat. Biomed. Eng., 2022, 1–12 Search PubMed.
- R. A. Lee, H. De Puig, P. Q. Nguyen, N. M. Angenent-Mari, N. M. Donghia, J. P. McGee, J. D. Dvorin, C. M. Klapperich, N. R. Pollock and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25722–25731 CrossRef CAS PubMed.
- H. de Puig, R. A. Lee, D. Najjar, X. Tan, L. R. Soenksen, N. M. Angenent-Mari, N. M. Donghia, N. E. Weckman, A. Ory and C. F. Ng, Sci. Adv., 2021, 7, eabh2944 CrossRef PubMed.
- J. S. Park, K. Hsieh, L. Chen, A. Kaushik, A. Y. Trick and T. Wang, Adv. Sci., 2021, 8, 2003564 CrossRef CAS PubMed.
- X. Wu, J. K. Tay, C. K. Goh, C. Chan, Y. H. Lee, S. L. Springs, K. S. Loh, T. K. Lu and H. Yu, Biomaterials, 2021, 274, 120876 CrossRef CAS PubMed.
- J. Li, J. Macdonald and F. von Stetten, Analyst, 2018, 144, 31–67 RSC.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados and A. Sotomayor-Gonzalez, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- Y. Bao, Y. Jiang, E. Xiong, T. Tian, Z. Zhang, J. Lv, Y. Li and X. Zhou, ACS Sens., 2020, 5, 1082–1091 CrossRef CAS PubMed.
- L. T. Nguyen, N. C. Macaluso, B. L. Pizzano, M. N. Cash, J. Spacek, J. Karasek, M. R. Miller, J. A. Lednicky, R. R. Dinglasan and M. Salemi, EBioMedicine, 2022, 77, 103926 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li, M. M. Sfeir and C. Liu, Biosens. Bioelectron., 2021, 184, 113218 CrossRef CAS PubMed.
- B. Tian, G. A. S. Minero, J. Fock, M. Dufva and M. F. Hansen, Nucleic Acids Res., 2020, 48, e30 CrossRef CAS PubMed.
- G. Wang, W. Tian, X. Liu, W. Ren and C. Liu, Anal. Chem., 2020, 92, 6702–6708 CrossRef CAS PubMed.
- W. Tian, X. Liu, G. Wang and C. Liu, Chem. Commun., 2020, 56, 13445–13448 RSC.
- X. Wang, X. Chen, C. Chu, Y. Deng, M. Yang, Z. Ji, F. Xu, D. Huo, Y. Luo and C. Hou, Sens. Actuators, B, 2020, 323, 128653 CrossRef CAS.
- Y. Deng, G. Cao, X. Chen, M. Yang, D. Huo and C. Hou, Talanta, 2021, 232, 122415 CrossRef CAS PubMed.
- C. Qian, R. Wang, H. Wu, F. Zhang, J. Wu and L. Wang, Anal. Chem., 2019, 91, 11362–11366 CrossRef CAS PubMed.
- K. Yin, X. Ding, Z. Li, H. Zhao, K. Cooper and C. Liu, Anal. Chem., 2020, 92, 8561–8568 CrossRef CAS PubMed.
- O. Mukama, J. Wu, Z. Li, Q. Liang, Z. Yi, X. Lu, Y. Liu, Y. Liu, M. Hussain and G. G. Makafe, Biosens. Bioelectron., 2020, 159, 112143 CrossRef CAS PubMed.
- Y. Chen, Y. Shi, Y. Chen, Z. Yang, H. Wu, Z. Zhou, J. Li, J. Ping, L. He and H. Shen, Biosens. Bioelectron., 2020, 169, 112642 CrossRef CAS PubMed.
- H. Wu, C. Qian, C. Wu, Z. Wang, D. Wang, Z. Ye, J. Ping, J. Wu and F. Ji, Biosens. Bioelectron., 2020, 157, 112153 CrossRef CAS PubMed.
- R. A. Lee, H. De Puig, P. Q. Nguyen, N. M. Angenent-Mari, N. M. Donghia, J. P. McGee, J. D. Dvorin, C. M. Klapperich, N. R. Pollock and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25722–25731 CrossRef CAS PubMed.
- R. Zhou, Y. Li, T. Dong, Y. Tang and F. Li, Chem. Commun., 2020, 56, 3536–3538 RSC.
- O. Mukama, T. Yuan, Z. He, Z. Li, J. de Dieu Habimana, M. Hussain, W. Li, Z. Yi, Q. Liang and L. Zeng, Sens. Actuators, B, 2020, 316, 128119 CrossRef CAS.
- C. Qian, H. Wu, Y. Shi, J. Wu and H. Chen, Sens. Actuators, B, 2020, 305, 127440 CrossRef CAS.
- B. Pang, J. Xu, Y. Liu, H. Peng, W. Feng, Y. Cao, J. Wu, H. Xiao, K. Pabbaraju and G. Tipples, Anal. Chem., 2020, 92, 16204–16212 CrossRef CAS PubMed.
- Z. Ali, R. Aman, A. Mahas, G. S. Rao, M. Tehseen, T. Marsic, R. Salunke, A. K. Subudhi, S. M. Hala and S. M. Hamdan, Virus Res., 2020, 288, 198129 CrossRef CAS PubMed.
- W. S. Zhang, J. Pan, F. Li, M. Zhu, M. Xu, H. Zhu, Y. Yu and G. Su, Anal. Chem., 2021, 93, 4126–4133 CrossRef CAS PubMed.
- Y. Jiang, M. Hu, A. Liu, Y. Lin, L. Liu, B. Yu, X. Zhou and D. Pang, ACS Sens., 2021, 6, 1086–1093 CrossRef CAS PubMed.
- H. Wu, Y. Chen, Y. Shi, L. Wang, M. Zhang, J. Wu and H. Chen, Biosens. Bioelectron., 2021, 178, 113001 CrossRef CAS PubMed.
- R. Wang, C. Qian, Y. Pang, M. Li, Y. Yang, H. Ma, M. Zhao, F. Qian, H. Yu and Z. Liu, Biosens. Bioelectron., 2021, 172, 112766 CrossRef CAS PubMed.
- R. Ding, J. Long, M. Yuan, X. Zheng, Y. Shen, Y. Jin, H. Yang, H. Li, S. Chen and G. Duan, Int. J. Mol. Sci., 2021, 22, 4842 CrossRef CAS PubMed.
- X. Wu, J. K. Tay, C. K. Goh, C. Chan, Y. H. Lee, S. L. Springs, K. S. Loh, T. K. Lu and H. Yu, Biomaterials, 2021, 274, 120876 CrossRef CAS PubMed.
- K. H. Ooi, M. M. Liu, J. W. D. Tay, S. Y. Teo, P. Kaewsapsak, S. Jin, C. K. Lee, J. Hou, S. Maurer-Stroh and W. Lin, Nat. Commun., 2021, 12, 1–23 CrossRef PubMed.
- M. Zhang, H. Wang, H. Wang, F. Wang and Z. Li, Anal. Chem., 2021, 93, 7942–7948 CrossRef CAS PubMed.
- Y. Zhang, M. Chen, C. Liu, J. Chen, X. Luo, Y. Xue, Q. Liang, L. Zhou, Y. Tao and M. Li, Sens. Actuators, B, 2021, 345, 130411 CrossRef CAS PubMed.
- K. Yin, X. Ding, Z. Li, M. M. Sfeir, E. Ballesteros and C. Liu, Lab Chip, 2021, 21, 2730–2737 RSC.
- F. Chen, P. Lee, A. Y. Trick, J. S. Park, L. Chen, K. Shah, H. Mostafa, K. C. Carroll, K. Hsieh and T. Wang, Biosens. Bioelectron., 2021, 190, 113390 CrossRef CAS PubMed.
- X. Wu, C. Chan, S. L. Springs, Y. H. Lee, T. K. Lu and H. Yu, Anal. Chim. Acta, 2022, 339494 CrossRef CAS PubMed.
- T. Zhang, W. Zhao, X. Chen, X. Zhang, J. Zhu, S. Li, C. Wu, Z. Tian and G. Sui, Anal. Chem., 2022, 94, 15472–15480 CrossRef CAS PubMed.
- Y. Chen, X. Xu, J. Wang, Y. Zhang, W. Zeng, Y. Liu and X. Zhang, Anal. Chem., 2022, 94, 9724–9731 CrossRef CAS PubMed.
- H. Wu, X. Cao, Y. Meng, D. Richards, J. Wu, Z. Ye and A. J. deMello, Biosens. Bioelectron., 2022, 114377 CrossRef CAS PubMed.
- M. Bao, S. Zhang, C. Ten Pas, S. J. Dollery, R. V. Bushnell, F. Yuqing, R. Liu, G. Lu, G. J. Tobin and K. Du, Lab Chip, 2022, 22, 4849–4859 RSC.
- Y. Tang, T. Song, L. Gao, S. Yin, M. Ma, Y. Tan, L. Wu, Y. Yang, Y. Wang and T. Lin, Nat. Commun., 2022, 13, 1–10 CAS.
- Z. Xu, D. Chen, T. Li, J. Yan, J. Zhu, T. He, R. Hu, Y. Li, Y. Yang and M. Liu, Nat. Commun., 2022, 13, 1–14 Search PubMed.
- M. Hu, Z. Qiu, Z. Bi, T. Tian, Y. Jiang and X. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2202034119 CrossRef CAS PubMed.
- J. Liu, H. Wang, L. Zhang, Y. Lu, X. Wang, M. Shen, N. Li, L. Feng, J. Jing and B. Cao, Small, 2022, 2200854 CrossRef CAS PubMed.
- A. Ramachandran and J. G. Santiago, Anal. Chem., 2021, 93, 7456–7464 CrossRef CAS PubMed.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. Bell, A. Boisen and A. G. Brolo, ACS Nano, 2019, 14, 28–117 CrossRef PubMed.
- D. W. Kimmel, G. LeBlanc, M. E. Meschievitz and D. E. Cliffel, Anal. Chem., 2012, 84, 685–707 CrossRef CAS PubMed.
- Z. Jin, D. Geißler, X. Qiu, K. D. Wegner and N. Hildebrandt, Angew. Chem., Int. Ed., 2015, 54, 10024–10029 CrossRef CAS PubMed.
- K. D. Wegner, Z. Jin, S. Linden, T. L. Jennings and N. Hildebrandt, ACS Nano, 2013, 7, 7411–7419 CrossRef CAS PubMed.
- K. Shi, S. Xie, R. Tian, S. Wang, Q. Lu, D. Gao, C. Lei, H. Zhu and Z. Nie, Sci. Adv., 2021, 7, eabc7802 CrossRef CAS PubMed.
- R. Hajian, S. Balderston, T. Tran, T. DeBoer, J. Etienne, M. Sandhu, N. A. Wauford, J. Chung, J. Nokes and M. Athaiya, Nat. Biomed. Eng., 2019, 3, 427–437 CrossRef CAS PubMed.
- H. Li, J. Yang, G. Wu, Z. Weng, Y. Song, Y. Zhang, J. A. Vanegas, L. Avery, Z. Gao and H. Sun, Angew. Chem., Int. Ed., 2022, 61, e202203826 CAS.
- P. Fozouni, S. Son, M. Díaz de León Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar, S. I. Stephens and M. Ott, Cell, 2021, 184, 323–333.e9 CrossRef CAS PubMed.
- T. Y. Liu, G. J. Knott, D. C. Smock, J. J. Desmarais, S. Son, A. Bhuiya, S. Jakhanwal, N. Prywes, S. Agrawal and M. Díaz de León Derby, Nat. Chem. Biol., 2021, 17, 982–988 CrossRef CAS PubMed.
- K. Guk, J. O. Keem, S. G. Hwang, H. Kim, T. Kang, E. Lim and J. Jung, Biosens. Bioelectron., 2017, 95, 67–71 CrossRef CAS PubMed.
- H. Kim, S. Lee, H. W. Seo, B. Kang, J. Moon, K. G. Lee, D. Yong, H. Kang, J. Jung and E. Lim, ACS Nano, 2020, 14, 17241–17253 CrossRef CAS PubMed.
- S. Balderston, J. J. Taulbee, E. Celaya, K. Fung, A. Jiao, K. Smith, R. Hajian, G. Gasiunas, S. Kutanovas and D. Kim, Nat. Biomed. Eng., 2021, 1–13 Search PubMed.
- Y. Dai, R. A. Somoza, L. Wang, J. F. Welter, Y. Li, A. I. Caplan and C. C. Liu, Angew. Chem., 2019, 131, 17560–17566 CrossRef.
- D. Zhang, Y. Yan, H. Que, T. Yang, X. Cheng, S. Ding, X. Zhang and W. Cheng, ACS Sens., 2020, 5, 557–562 CrossRef CAS PubMed.
- Z. Li, X. Ding, K. Yin, Z. Xu, K. Cooper and C. Liu, Biosens. Bioelectron., 2021, 192, 113498 CrossRef CAS PubMed.
- Y. Lee, J. Choi, H. Han, S. Park, S. Y. Park, C. Park, C. Baek, T. Lee and J. Min, Sens. Actuators, B, 2021, 326, 128677 CrossRef CAS.
- Y. Pang, Q. Li, C. Wang, Z. Sun and R. Xiao, Chem. Eng. J., 2022, 429, 132109 CrossRef CAS.
- J. Choi, J. Lim, M. Shin, S. Paek and J. Choi, Nano Lett., 2020, 21, 693–699 CrossRef PubMed.
- R. Zeng, W. Wang, M. Chen, Q. Wan, C. Wang, D. Knopp and D. Tang, Nano Energy, 2021, 82, 105711 CrossRef CAS.
- C. Wang, C. Han, X. Du and W. Guo, Anal. Chem., 2021, 93, 12881–12888 CrossRef CAS PubMed.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed.
- T. Tian, B. Shu, Y. Jiang, M. Ye, L. Liu, Z. Guo, Z. Han, Z. Wang and X. Zhou, ACS Nano, 2020, 15, 1167–1178 CrossRef PubMed.
- S. Son, A. Lyden, J. Shu, S. I. Stephens, P. Fozouni, G. J. Knott, D. C. Smock, T. Y. Liu, D. Boehm, C. Simoneau and D. A. Fletcher, medRxiv, 2021, preprint, DOI:10.1101/2021.08.02.21261509.
- R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer and G. A. Urban, Adv. Mater., 2019, 31, 1905311 CrossRef CAS PubMed.
- T. Zhou, R. Huang, M. Huang, J. Shen, Y. Shan and D. Xing, Adv. Sci., 2020, 7, 1903661 CrossRef CAS PubMed.
- R. Bruch, M. Johnston, A. Kling, T. Mattmüller, J. Baaske, S. Partel, S. Madlener, W. Weber, G. A. Urban and C. Dincer, Biosens. Bioelectron., 2021, 177, 112887 CrossRef CAS PubMed.
- T. Tian, B. Shu, Y. Jiang, M. Ye, L. Liu, Z. Guo, Z. Han, Z. Wang and X. Zhou, ACS Nano, 2021, 15, 1167–1178 CrossRef CAS PubMed.
- H. Shinoda, Y. Taguchi, R. Nakagawa, A. Makino, S. Okazaki, M. Nakano, Y. Muramoto, C. Takahashi, I. Takahashi and J. Ando, Commun. Biol., 2021, 4, 1–7 CrossRef PubMed.
- Y. Cui, S. Fan, Z. Yuan, M. Song, J. Hu, D. Qian, D. Zhen, J. Li and B. Zhu, Talanta, 2021, 224, 121878 CrossRef CAS PubMed.
- I. A. Iwe, W. Li, Z. Li and J. Huang, Talanta, 2021, 226, 122146 CrossRef CAS PubMed.
- Y. Zhang, L. Qian, W. Wei, Y. Wang, B. Wang, P. Lin, W. Liu, L. Xu, X. Li and D. Liu, ACS Synth. Biol., 2017, 6, 211–216 CrossRef CAS PubMed.
- H. Lee, J. Choi, E. Jeong, S. Baek, H. C. Kim, J. Chae, Y. Koh, S. W. Seo, J. Kim and S. J. Kim, Nano Lett., 2018, 18, 7642–7650 CrossRef CAS PubMed.
- J. Moon, H. Kwon, D. Yong, I. Lee, H. Kim, H. Kang, E. Lim, K. Lee, J. Jung and H. G. Park, ACS Sens., 2020, 5, 4017–4026 CrossRef CAS PubMed.
- K. N. Hass, M. Bao, Q. He, L. Liu, J. He, M. Park, P. Qin and K. Du, ACS Omega, 2020, 5, 27433–27441 CrossRef CAS PubMed.
- W. Xu, T. Jin, Y. Dai and C. C. Liu, Biosens. Bioelectron., 2020, 155, 112100 CrossRef CAS PubMed.
- Q. He, D. Yu, M. Bao, G. Korensky, J. Chen, M. Shin, J. Kim, M. Park, P. Qin and K. Du, Biosens. Bioelectron., 2020, 154, 112068 CrossRef CAS PubMed.
- X. Yang, J. Li, S. Zhang, C. Li and J. Ma, Anal. Chem., 2022, 94, 13076–13083 CrossRef CAS PubMed.
- M. Zeng, Y. Ke, Z. Zhuang, C. Qin, L. Y. Li, G. Sheng, Z. Li, H. Meng and X. Ding, Anal. Chem., 2022, 94, 10805–10812 CrossRef CAS PubMed.
- T. Luo, J. Li, Y. He, H. Liu, Z. Deng, X. Long, Q. Wan, J. Ding, Z. Gong and Y. Yang, Anal. Chem., 2022, 94, 6566–6573 CrossRef CAS PubMed.
- C. M. Green, J. Spangler, K. Susumu, D. A. Stenger, I. L. Medintz and S. A. Díaz, ACS Nano, 2022, 16, 20693–20704 CrossRef CAS PubMed.
- Z. Chen, J. Li, T. Li, T. Fan, C. Meng, C. Li, J. Kang, L. Chai, Y. Hao and Y. Tang, Natl. Sci. Rev., 2022, 9, nwac104 CrossRef PubMed.
- P. Qin, M. Park, K. J. Alfson, M. Tamhankar, R. Carrion, J. L. Patterson, A. Griffiths, Q. He, A. Yildiz and R. Mathies, ACS Sens., 2019, 4, 1048–1054 CrossRef CAS PubMed.
- Q. Chen, T. Tian, E. Xiong, P. Wang and X. Zhou, Anal. Chem., 2019, 92, 573–577 CrossRef PubMed.
- J. Li, L. Tang, T. Li, K. Li, Y. Zhang, W. Ni, M. Xiao, Y. Zhao, Z. Zhang and G. Zhang, ACS Sens., 2022, 7, 2680–2690 CrossRef CAS PubMed.
- C. Zhang, P. Zhang, H. Ren, P. Jia, J. Ji, L. Cao, P. Yang, Y. Li, J. Liu and Z. Li, Chem. Eng. J., 2022, 446, 136864 CrossRef CAS.
- H. Shinoda, T. Iida, A. Makino, M. Yoshimura, J. Ishikawa, J. Ando, K. Murai, K. Sugiyama, Y. Muramoto and M. Nakano, Commun. Biol., 2022, 5, 1–8 CrossRef PubMed.
- M. Broto, M. M. Kaminski, C. Adrianus, N. Kim, R. Greensmith, S. Dissanayake-Perera, A. J. Schubert, X. Tan, H. Kim and A. S. Dighe, Nat. Nanotechnol., 2022, 17, 1120–1126 CrossRef CAS PubMed.
- J. Yang, Y. Song, X. Deng, J. A. Vanegas, Z. You, Y. Zhang, Z. Weng, L. Avery, K. D. Dieckhaus and A. Peddi, Nat. Chem. Biol., 2022, 1–10 Search PubMed.
- S. Dube, J. Qin and R. Ramakrishnan, PLoS One, 2008, 3, e2876 CrossRef PubMed.
- C. M. Hindson, J. R. Chevillet, H. A. Briggs, E. N. Gallichotte, I. K. Ruf, B. J. Hindson, R. L. Vessella and M. Tewari, Nat. Methods, 2013, 10, 1003–1005 CrossRef CAS PubMed.
- J. S. Park, K. Hsieh, L. Chen, A. Kaushik, A. Y. Trick and T. Wang, Adv. Sci., 2021, 8, 2003564 CrossRef CAS PubMed.
- S. Teh, R. Lin, L. Hung and A. P. Lee, Lab Chip, 2008, 8, 198–220 RSC.
- J. Wang, W. Yu, Z. Wu, X. Liu and Y. Chen, Appl. Phys. Lett., 2022, 120, 204101 CrossRef CAS.
- W. Trypsteen, M. Vynck, J. De Neve, P. Bonczkowski, M. Kiselinova, E. Malatinkova, K. Vervisch, O. Thas, L. Vandekerckhove and W. De Spiegelaere, Anal. Bioanal. Chem., 2015, 407, 5827–5834 CrossRef CAS PubMed.
- X. Luo, Y. Xue, E. Ju, Y. Tao, M. Li, L. Zhou, C. Yang, J. Zhou and J. Wang, Anal. Chim. Acta, 2022, 1192, 339336 CrossRef CAS PubMed.
- J. Q. Cui, B. Cui, F. X. Liu, Y. Lin and S. Yao, Sens. Actuators, B, 2022, 132573 CrossRef CAS.
- Z. Yu, L. Xu, W. Lyu and F. Shen, Lab Chip, 2022, 22, 2954–2961 RSC.
- C. H. Y. Chung, B. Cui, R. Song, X. Liu, X. Xu and S. Yao, Micromachines, 2019, 10, 592 CrossRef PubMed.
- R. Dangla, S. C. Kayi and C. N. Baroud, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 853–858 CrossRef CAS PubMed.
- X. Xu, H. Yuan, R. Song, M. Yu, H. Y. Chung, Y. Hou, Y. Shang, H. Zhou and S. Yao, Biomicrofluidics, 2018, 12, 014103 CrossRef PubMed.
- C. Sun, L. Liu, H. N. Vasudevan, K. Chang and A. R. Abate, Anal. Chem., 2021, 93, 9974–9979 CrossRef CAS PubMed.
- X. Lin, X. Huang, K. Urmann, X. Xie and M. R. Hoffmann, ACS Sens., 2019, 4, 242–249 CrossRef CAS PubMed.
- J. E. Kreutz, J. Wang, A. M. Sheen, A. M. Thompson, J. P. Staheli, M. R. Dyen, Q. Feng and D. T. Chiu, Lab Chip, 2019, 19, 1035–1040 RSC.
- Z. Yu, W. Lyu, M. Yu, Q. Wang, H. Qu, R. F. Ismagilov, X. Han, D. Lai and F. Shen, Biosens. Bioelectron., 2020, 155, 112107 CrossRef CAS PubMed.
- M. Shehadul Islam, A. Aryasomayajula and P. R. Selvaganapathy, Micromachines, 2017, 8, 83 CrossRef.
- M. A. Lalli, J. S. Langmade, X. Chen, C. C. Fronick, C. S. Sawyer, L. C. Burcea, M. N. Wilkinson, R. S. Fulton, M. Heinz and W. J. Buchser, Clin. Chem., 2021, 67, 415–424 CrossRef PubMed.
- M. Alafeef, P. Moitra, K. Dighe and D. Pan, Nat. Protoc., 2021, 16, 3141–3162 CrossRef CAS PubMed.
- S. A. Byrnes, R. Gallagher, A. Steadman, C. Bennett, R. Rivera, C. Ortega, S. T. Motley, P. Jain, B. H. Weigl and J. T. Connelly, Anal. Chem., 2021, 93, 4160–4165 CrossRef CAS PubMed.
- B. A. Rabe and C. Cepko, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24450–24458 CrossRef CAS PubMed.
- S. Agrawal, A. Fanton, S. S. Chandrasekaran, B. Charrez, A. M. Escajeda, S. Son, R. Mcintosh, A. Bhuiya, M. Díaz de León Derby and N. A. Switz, medRxiv, 2021, preprint, DOI:10.1101/2020.12.14.20247874.
- J. Arizti-Sanz, A. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije, M. E. Grunberg, T. F. Kosoko-Thoroddsen, N. L. Welch, P. P. Pillai and S. Mantena, Nat. Biomed. Eng., 2022, 1–12 Search PubMed.
- H. Gong, I. Holcomb, A. Ooi, X. Wang, D. Majonis, M. A. Unger and R. Ramakrishnan, Bioconjugate Chem., 2016, 27, 217–225 CrossRef CAS PubMed.
- P. Zhang, L. Chen, J. Hu, A. Y. Trick, F. Chen, K. Hsieh, Y. Zhao, B. Coleman, K. Kruczynski and T. R. Pisanic II, Biosens. Bioelectron., 2022, 195, 113656 CrossRef CAS PubMed.
- I. Lee, S. Kwon, M. Sorci, P. S. Heeger and J. S. Dordick, Anal. Chem., 2021, 93, 16528–16534 CrossRef CAS PubMed.
- J. Ko, Y. Wang, J. C. Carlson, A. Marquard, J. Gungabeesoon, A. Charest, D. Weitz, M. J. Pittet and R. Weissleder, Adv. Biosyst., 2020, 4, 1900307 CrossRef CAS PubMed.
- J. Shen, X. Zhou, Y. Shan, H. Yue, R. Huang, J. Hu and D. Xing, Nat. Commun., 2020, 11, 1–10 CrossRef PubMed.
- C. Niu, C. Wang, F. Li, X. Zheng, X. Xing and C. Zhang, Biosens. Bioelectron., 2021, 183, 113196 CrossRef CAS PubMed.
- C. Li, B. Zheng, Y. Liu, J. Gao, M. Zheng, D. Pang and H. Tang, Biosens. Bioelectron., 2020, 169, 112650 CrossRef CAS PubMed.
- C. Li, B. Zheng, J. Li, J. Gao, Y. Liu, D. Pang and H. Tang, ACS Nano, 2021, 15, 8142–8154 CrossRef CAS PubMed.
- P. Wu, X. Ye, D. Wang, F. Gong, X. Wei, S. Xiang, J. Zhang, T. Kai and P. Ding, J. Hazard. Mater., 2022, 424, 127690 CrossRef CAS PubMed.
- X. Zhao, L. Zeng, Q. Mei and Y. Luo, ACS Sens., 2020, 5, 2239–2246 CrossRef CAS PubMed.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, J. Am. Chem. Soc., 2019, 142, 207–213 CrossRef PubMed.
- C. Zhang, H. Yao, Q. Ma and B. Yu, Analyst, 2021, 146, 6576–6581 RSC.
- J. Li, S. Yang, C. Zuo, L. Dai, Y. Guo and G. Xie, ACS Sens., 2020, 5, 970–977 CrossRef CAS PubMed.
- A. Mahas, Q. Wang, T. Marsic and M. M. Mahfouz, Anal. Chem., 2022, 94, 4617–4626 CrossRef CAS PubMed.
- M. Liang, Z. Li, W. Wang, J. Liu, L. Liu, G. Zhu, L. Karthik, M. Wang, K. Wang and Z. Wang, Nat. Commun., 2019, 10, 1–9 CrossRef PubMed.
- R. S. Iwasaki and R. T. Batey, Nucleic Acids Res., 2020, 48, e101 CrossRef CAS PubMed.
- J. Zhao, Z. Tan, L. Wang, C. Lei and Z. Nie, Chem. Commun., 2021, 57, 7051–7054 RSC.
- C. Zhang, H. Yao, Q. Ma and B. Yu, Analyst, 2021, 146, 6576–6581 RSC.
| This journal is © The Royal Society of Chemistry 2023 |