Open Access Article
Open Access ArticleRecovery of bacterioruberin and proteins using aqueous solutions of surface-active compounds†
Bárbara M. C. Vaz‡
a,
Mariam Kholany‡a,
Diana C. G. A. Pinto b,
Inês P. E. Macárioac,
Telma Velosoac,
Tânia Caetano
b,
Inês P. E. Macárioac,
Telma Velosoac,
Tânia Caetano c,
Joana L. Pereira
c,
Joana L. Pereira c,
João A. P. Coutinho
c,
João A. P. Coutinho a and
Sónia P. M. Ventura
a and
Sónia P. M. Ventura *a
*a
aCICECO – Aveiro Institute of Materials, Department of Chemistry, University of Aveiro, Campus Universitário de Santiago, 3810-193 Aveiro, Portugal. E-mail: spventura@ua.pt
bLAQV – REQUIMTE, Department of Chemistry, University of Aveiro, 3810-193 Aveiro, Portugal
cCESAM – Centre for Environmental and Marine Studies, Department of Biology, University of Aveiro Campus Universitário de Santiago, 3810-193 Aveiro, Portugal
First published on 24th October 2022
Abstract
Haloarchaea microorganisms are little explored marine resources that can be a promising source of valuable compounds with unique characteristics, due to their adaptation to extreme environments. In this work, the extraction of bacterioruberin and proteins from Haloferax mediterranei ATCC 33500 was investigated using aqueous solutions of ionic liquids and surfactants, which were further compared with ethanol. Despite the good performance of ethanol in the extraction of bacterioruberin, the use of aqueous solutions of surface-active compounds allowed the simultaneous release of bacterioruberin and proteins in a multi-product process, with the non-ionic surfactants being identified as the most promising. The optimum operational conditions allowed a maximum extraction yield of 0.37 ± 0.01 mgbacterioruberin gwet biomass−1 and 352 ± 9 mgprotein gwet biomass−1 with an aqueous solution of Tween® 20 (at 182.4 mM) as the extraction solvent. In addition, high purities of bacterioruberin were obtained, after performing a simple induced precipitation using ethanol as an antisolvent to recover the proteins present in the initial extract. Finally, a step for polishing the bacterioruberin was performed, to enable solvent recycling, further closing the process to maximize its circularity.
Introduction
Our continued reliance on a petroleum-based economy to satisfy humankind’s needs cannot be sustained anymore.1 The traditional model of take, make and dispose using fossil-based feedstocks has severe implications for human health, the depletion of fossil resources, and as a driver of climate change.2 In this sense, the development of sustainable technologies and practices to ensure a gradual transition to a renewable (bio-based) economy should be encouraged.1,3 By considering the concepts of circular economy and integrated biorefineries, the complete valorization of all biomass fractions could be achieved. Besides maximizing the value of raw materials, waste production could be minimized or avoided, which leads to an increase in the economic viability of downstream processes for industrial implementation.4Recently, marine resources like algae and cyanobacteria have gained significant interest as renewable feedstocks to produce several natural bioactive compounds. Their sustainable use can easily fit the priorities of the 2030 Agenda for Sustainable Development.5 They can generate biofuels and bioenergy and integrate bioactive medicinal products, food, and materials.6 In this context, halophilic microorganisms from the domain Archaea, with high commercial potential, have emerged as underexplored marine resources, easily found around the globe, mostly in hypersaline environments. Additionally, they are natural producers of several relevant high-demand products, such as proteins, poly(3-hydroxybutyrate), polyhydroxyalkanoates, and carotenoids.7
Proteins are complex molecules with a unique three-dimensional structure composed of numerous combinations of the 20 different amino acids attached to form long chains.8 There is a problem in searching for new sources of proteins for food purposes. However, due to their multiple biological roles, their biotechnological interest is vast,9,10 and thus, depending on the final application, their purification may be required. In halophilic archaea (or haloarchaea), halophilic proteins assist adaptation to extreme saline conditions by modifying their structure. The presence of acidic amino acids on the protein surface confers a more hydrophilic and flexible surface, capable of increasing the hydration of the protein. Hence, its aggregation is prevented, and the protein’s functionality is retained.11 On the other hand, carotenoids are natural colorants well known for their biotechnological applications and key properties for human health.12,13 The haloarchaea group can produce many carotenoids, including β-carotene, salinixanthin, bacterioruberin, and its precursors lycopene and phytoene.7,12 Among these, bacterioruberin represents the most abundant carotenoid produced, since all the species with sequenced genomes encode its biosynthetic pathway.14 Bacterioruberin can influence the membrane fluidity, acting as a water barrier and allowing the permeability of oxygen and other molecules needed for the cell’s survival in hypersaline environments. Consequently, the membrane structure rigidity is increased.7,15,16 Of great relevance, this C50 isoprenoid, composed of 13 conjugated double bonds and 4 hydroxyl groups, has a higher antioxidant activity than other molecules. Therefore, higher protection can be achieved against intense light, gamma irradiation, and DNA damaging agents such as radiography, UV-irradiation, and H2O2 exposure.7,15,16 These characteristics make bacterioruberin an appealing bioactive compound to be applied in the food, cosmetic and pharmaceutical industries.17 Moreover, their commercial potential goes beyond the applications previously mentioned. More recently, some of us have investigated its optical potential to develop optical sensors and retinal gene therapy formulations (ongoing work), with promising results. Despite the high potential of bacterioruberin and halophilic proteins, efficient methods of extraction and purification are still at an early stage. Most literature focuses on exploring protein structures and their adaptations to extreme environments,18–23 and optimizing bacterioruberin production and its possible applications.15,17,24–26 The studies found just quantify bacterioruberin extracted using organic solvents, without considering biomass valorization. Regarding the purification step, some works used thin-layer chromatography.17,27–32 However, this technique, despite being simple and efficient at the lab scale, is not industrially adequate. In this sense, more efforts are needed to develop efficient and sustainable approaches with scale-up potential encompassing the valorization of multiple products obtained from this biomass.
In this work, aqueous solutions of surfactants and ionic liquids (ILs) were investigated with the prospect of simultaneously recovering different products from the same biomass in a more sustainable way. As surfactants and tensioactive ILs are amphiphilic molecules,33 they become effective solvents in the extraction of both more hydrophilic compounds, e.g. proteins,10,34–37 and more hydrophobic molecules,37–41 such as bacterioruberin. Furthermore, their use has several advantages, namely milder process conditions while using water as the main solvent, which could also be attractive for cell disruption, as most haloarchaea lyse rapidly when suspended in water.7 After the extraction of multiple compounds, a purification step is usually needed to obtain each compound with the appropriate purity for the specific application. Induced protein precipitation was applied in this work. The addition of a precipitating agent lowers the solubility of proteins in water, causes their precipitation42 and allows the separation of the solvent. As well as its simplicity, it has industrial potential,43 being recognized as having a low carbon footprint and economic impact.44,45 In the end, a polishing step was also applied to bacterioruberin, allowing the solvent to be recycled.
Results and discussion
Screening of surfactants and ILs as solvents
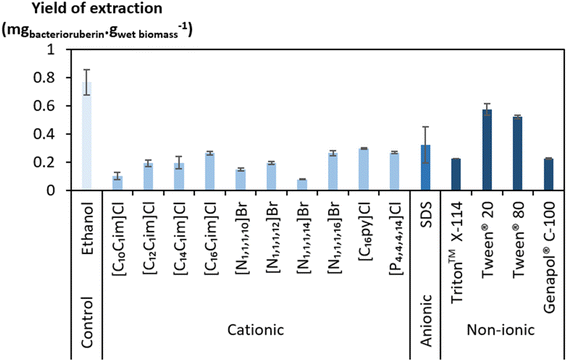
A large screening of aqueous solutions of surface-active compounds belonging to different classes (cationic, anionic, non-ionic) and other non-surfactant agents was performed at 100 mM to assess their ability to extract bacterioruberin (the most valuable compound present). The outcomes were benchmarked against ethanol. The surfactants tested were pre-selected considering previous works that identified them to be efficient in the extraction of biomolecules, namely carotenoids.37,38,46 Table S1 in the ESI† reports the ability of the surface-active compounds tested (30 in total) to extract bacterioruberin. Fig. 1 shows the yields of extraction (mgbacterioruberin gwet biomass−1) of the systems with extraction capability, in terms of the ability of the surfactants/ILs to induce the release of the target compound. Photographs of all systems can be found in Fig. S1 in the ESI.†Ethanol displayed a better bacterioruberin yield of extraction than aqueous solutions of surface-active compounds (Fig. 1). Ethanol is recognized for its excellent performance in pigment extraction and membrane solubilization.40,47 Thus, and also owing to the presence of bacterioruberin in the cell membrane, which endows it with a low water permeability,7 the performance of ethanol in bacterioruberin extraction was expected.
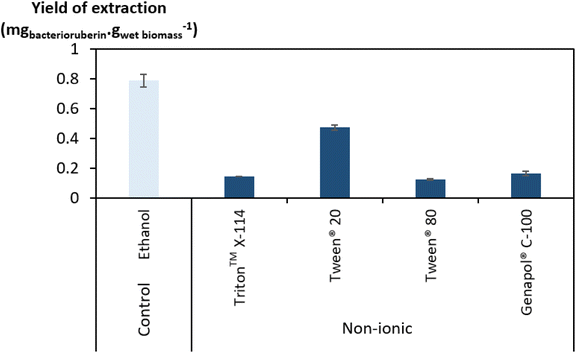
Regarding the performance of the aqueous solutions of surface-active compounds as depicted in Fig. 1 and Table S1 in the ESI,† almost none of the anionic or the non-tensioactive compounds were able to efficiently extract bacterioruberin. The non-tensioactive ILs have a low capability to disrupt the phospholipidic cell membranes due to their hydrophilic nature.38 The low performance of the anionic surfactants can be attributed to the fact that both types of tensioactive compounds and the phospholipids (components of the cell membrane) are negatively charged, creating repulsive forces.37 In contrast, almost all cationic and non-ionic compounds were able to release the compound of interest. This can be explained by their ability to establish attractive electrostatic interactions between the IL cation and the negatively charged head of the phospholipids.38,39 Bacterioruberin is almost exclusively present in its non-ionic form. This allows for more stable interactions between the pigment and the micelle hydrophobic core. However, with such conditions (with a surfactant concentration of 100 mM), a high viscosity was observed in most solutions obtained after extraction (see Table S1 in the ESI†), which is normally correlated with the release of chromosomal DNA during cell disruption.48,49 This phenomenon, whose origin is yet to be proved, imposed some experimental constraints. To overcome this issue and better assess the extraction capacity of the most efficient solvents (here, the non-ionic surfactants), a second screening at 250 mM surfactant concentration followed, with the respective results shown in Fig. 2. The UV-Vis spectra of the resulting extracts can be found in Fig. S2 in the ESI.†
Since the non-ionic surfactants tested are viscous liquids at room temperature, an increase in their concentration results in a more viscous aqueous solution. However, the viscosity of the final solutions (after bacterioruberin extraction) at 250 mM was revealed to be lower and more manageable than that of the same solutions at 100 mM. On the other hand, a very high surfactant concentration in the initial aqueous solution can also lead to viscosity issues. Therefore, the choice of an adequate concentration of surfactant seems to be a key factor in this work’s success. Fig. 2 shows that the highest performances at these higher concentrations were once again accomplished by Tween® 20 and ethanol. Nevertheless, ethanol lacks the desired capacity to simultaneously extract bacterioruberin and the proteins present in the biomass, thus failing to achieve the envisioned multi-product exploitation scenario. Aqueous solutions of surfactants provide a milder environment, thus allowing the extraction of different fractions of interest (proteins and pigments), both with applications related to the food industry. Ethanol leads to a protein extraction yield of approximately 24 mgprotein gwet biomass−1, while Tween® 20 leads to a protein extraction yield of approximately 352 mgprotein gwet biomass−1, increasing by over 14 times the amount of protein obtained. As a result, the following work used Tween® 20 as the best solvent.
Optimization of the solid–liquid extraction
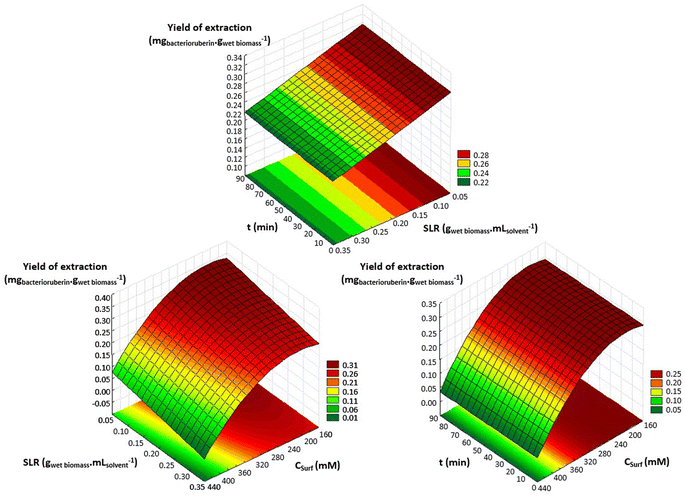
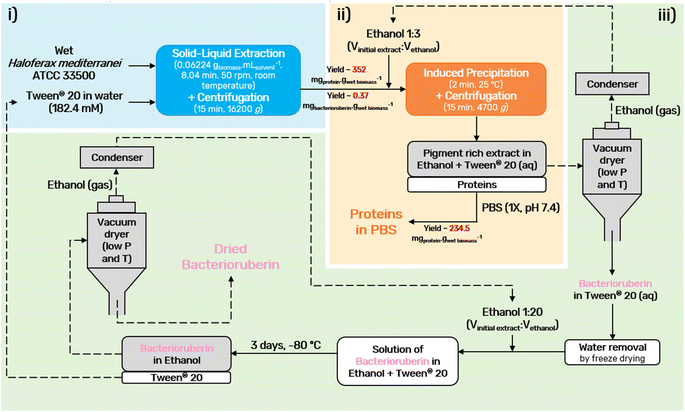
An optimization of the operational conditions was performed based on a central composite rotatable design (CCRD-23) comprising three independent variables, namely SLR (in gwet biomass mLsolvent−1, X1), time of extraction (t in min, X2) and concentration of surfactant in water (Csurf in mM, X3). Eighteen assays with four central (level 0) and axial points (−1.68 and +1.68 levels) were studied in terms of bacterioruberin yield of extraction (in mgbacterioruberin gwet biomass−1) ranging from 0.111 mgbacterioruberin gwet biomass−1 in assay 5 to 0.331 mgbacterioruberin gwet biomass−1 in assay 9 (Table S2 in the ESI†). The fitted model expressed in eqn (1), obtained using the SS residual from Analysis of Variance (ANOVA), revealed good predictability at a confidence level of 95% with R2 = 0.80912 and Fcalculated > Ftabulated.| Yield of extraction (mgbacterioruberin gwet biomass−1) = 0.147055 − 0.270072(X1) + 0.002002(X3)2 | (1) |
The response surfaces plotted in Fig. 3 show low effects on yield imposed by the extraction time. Therefore, the minimum time tested is the obvious choice to reduce the energy consumption of the process. The concentration of Tween® 20 has a great influence in the extraction process, where an optimum value was reached at 182.4 mM, the minimum level tested for this variable. These results suggest that a higher extraction yield could be achieved at concentrations of Tween® 20 below 182.4 mM, although an increase in the tested range would not be suitable due to the viscosity issues noticed (see above). SLR has a positive influence on the yield of extraction as predicted by the model, with the best response being obtained at 0.06224 gwet biomass mLsolvent−1. These results can be confirmed by the data present in Table S3† in the ESI,† the Pareto chart (Fig. S3 in the ESI†), and the predicted vs. observed values present in Fig. S4 in the ESI.† Here, it becomes evident that the solid–liquid ratio and the concentration of Tween® 20 in water were the variables that most constrained the model regarding the bacterioruberin yield of extraction. Therefore, after finding the optimal operational conditions (8.04 min, 182.4 mM, and 0.06224 gwet biomass mLsolvent−1), a model validation was developed. A bacterioruberin yield of extraction of 0.37 ± 0.01 mgbacterioruberin gwet biomass−1 was obtained experimentally (Table S4 in the ESI†), encompassing a mean relative deviation of 11.6%. Under these conditions, a protein yield of extraction of 352 ± 9 mgprotein gwet biomass−1 was achieved.
Protein induced precipitation
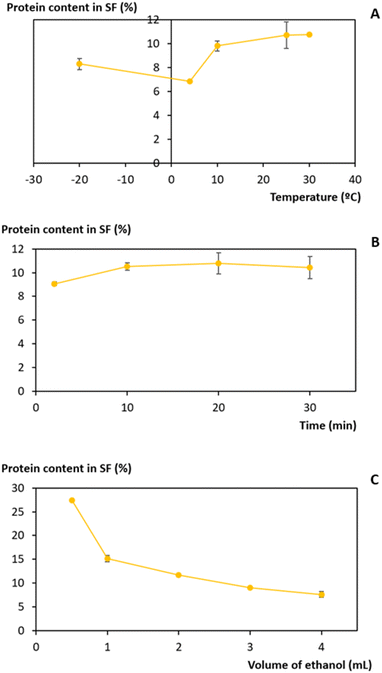
Ethanol is a common solvent used as a precipitating agent since it possesses a lower dielectric constant than water,50 which leads to an increase in the attraction forces between proteins, specifically at low temperatures.51–53 To achieve the minimum amount of protein in the supernatant fraction (SF), an evaluation of the process conditions, regarding temperature, volume of ethanol added to the initial extract, and time of the precipitation, was performed between −20 and 30 °C, 0.5 and 4 mL and 2 and 30 min, respectively (Fig. 4).The influence of temperature on the precipitation was assessed at a fixed time of 20 min and a volume of ethanol of 3 mL. Lower temperatures, particularly 4 °C, slightly reduce the protein losses of the process (Fig. 4A). However, as the difference in the protein content between 4 °C and 25 °C corresponds to only 3.9%, a temperature of 25 °C was seen as preferable, as it lowers the energetic burden of the overall process. Taking this into account, the precipitation time was evaluated at a fixed temperature of 25 °C and 3 mL of ethanol. The data obtained (Fig. 4B) show an almost constant protein content with a minimum at 2 min, and this time was chosen to proceed. Finally, the volume of ethanol was considered (Fig. 4C) under fixed conditions of 25 °C and 2 min, and it was shown that the volume of ethanol is the variable that most influences the precipitation process: protein content decreases with an increase in the ethanol volume added to the initial extract. A volume of 3 mL of ethanol was selected as the difference between 3 mL and 4 mL corresponds to only 1.5% protein content. Thus, with a set of optimum conditions of 25 °C, 2 min, and 3 mL of ethanol, a yield of 234.5 ± 0.9 mgprotein gwet biomass−1 (quantified and calculated after protein redissolution in PBS), which corresponds to 91% of the proteins present in the initial extract, was obtained.
Moreover, further studies were conducted to understand the effect of consecutive precipitations (Fig. S5 in the ESI†). After the first precipitation, and consequent recovery of the pellet fraction, a second precipitation was carried out by adding another 3 mL to the supernatant fraction and repeating the procedure. Only a very small fraction of approximately 1.0% of the total proteins present in the initial extract was obtained in the second precipitation. This corresponds to a yield of 2.7 ± 0.3 mgprotein gwet biomass−1 (quantified and calculated after protein redissolution in PBS), which is markedly lower when compared to the first, and thus the application of a second step of precipitation was disregarded. This further confirms that over 90% of the protein fraction was recovered in a single step of precipitation, as also visible by the abundance of proteins separated through SDS-PAGE electrophoresis of the precipitated proteins after dissolution in PBS (1×, pH 7.4), Fig. S6 in the ESI.† Finally, the bacterioruberin content was measured, after ethanol evaporation, to investigate bacterioruberin losses during induced precipitation, with a loss fixed at 12%.
Bacterioruberin polishing and conceptual process design
A bacterioruberin polishing step was accomplished to achieve the recycling of Tween® 20. Here, the objective was the separation of the bacterioruberin and the solvent, thus facilitating its further application. Briefly, to perform this step, the water content was first removed from the bacterioruberin extract (extract obtained after induced precipitation and ethanol evaporation) by freeze-drying. Then, the formation of two phases was achieved by adding ethanol to the sample [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (Vbacterioruberin extract
20 (Vbacterioruberin extract![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethanol)]. Afterwards, the solution obtained was stored in the freezer at −80 °C for three days. Thus, bacterioruberin migrated to the ethanol-rich (top) phase, while Tween® 20 remained in the bottom phase. At the end, the ethanol-rich phase was analyzed by 1H NMR spectroscopy (Fig. S7 in the ESI†) to evaluate if Tween® 20 was fully separated, and no surfactant was detected. The presence of bacterioruberin in the top phase of the collected sample was confirmed by UHPLC-MS analysis. It was possible to identify a mixture of bacterioruberin (m/z = 741) and monoanhydrobacterioruberin (m/z = 724) at around 12.59 min, with bacterioruberin being the major compound in the mixture (85%). However, under the conditions used (positive mode), these two compounds’ pseudomolecular ions appear as [M + H]+ and easily produce more stable fragment ions with m/z = 635 and 591, respectively (see Fig. S8 in the ESI†). Finally, the bottom phase was analyzed by UV-Vis spectroscopy, with only 2.8% of bacterioruberin detected, indicating that around 97.2% of bacterioruberin was successfully concentrated in the top phase.
Vethanol)]. Afterwards, the solution obtained was stored in the freezer at −80 °C for three days. Thus, bacterioruberin migrated to the ethanol-rich (top) phase, while Tween® 20 remained in the bottom phase. At the end, the ethanol-rich phase was analyzed by 1H NMR spectroscopy (Fig. S7 in the ESI†) to evaluate if Tween® 20 was fully separated, and no surfactant was detected. The presence of bacterioruberin in the top phase of the collected sample was confirmed by UHPLC-MS analysis. It was possible to identify a mixture of bacterioruberin (m/z = 741) and monoanhydrobacterioruberin (m/z = 724) at around 12.59 min, with bacterioruberin being the major compound in the mixture (85%). However, under the conditions used (positive mode), these two compounds’ pseudomolecular ions appear as [M + H]+ and easily produce more stable fragment ions with m/z = 635 and 591, respectively (see Fig. S8 in the ESI†). Finally, the bottom phase was analyzed by UV-Vis spectroscopy, with only 2.8% of bacterioruberin detected, indicating that around 97.2% of bacterioruberin was successfully concentrated in the top phase.
A final diagram of the process developed in this work was proposed (Fig. 5), in which all steps were considered. In the end, the colorant can be provided in ethanolic solution or in its dry form, depending on the desired end application. Drying should be performed using a vacuum dryer at low temperature and pressure. This not only prevents pigment degradation, thus allowing the recycling of the solvents, Tween® 20 and ethanol, but also allows easy scale-up.
Experimental
Materials and methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 894g for 15 min at room temperature (20–25 °C). The supernatant was discarded, and the pellet was stored in the dark at −20 °C until further use. Some natural variability was found in the data due to the use of different cultivation batches.
894g for 15 min at room temperature (20–25 °C). The supernatant was discarded, and the pellet was stored in the dark at −20 °C until further use. Some natural variability was found in the data due to the use of different cultivation batches.An additional screening of aqueous solutions of non-ionic surfactants at 250 mM was also carried out. All tests were conducted at a fixed solid–liquid ratio (SLR) of 0.1 gwet biomass mLsolvent−1, i.e. 0.1 g of wet biomass and 1 mL of the respective solvent. Finally, a centrifugation step was performed at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200g for 15 min in a VWR Microstar 17 centrifuge, at room temperature (20–25 °C). The resulting supernatant was recovered, and the biomass debris was discarded. All the assays were done in duplicate.
200g for 15 min in a VWR Microstar 17 centrifuge, at room temperature (20–25 °C). The resulting supernatant was recovered, and the biomass debris was discarded. All the assays were done in duplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dichloromethane (1
dichloromethane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used, after the solid–liquid extraction of bacterioruberin with ethanol from Haloferax mediterranei ATCC 33500, and subsequent redissolution of the resulting extract in dichloromethane.
1) was used, after the solid–liquid extraction of bacterioruberin with ethanol from Haloferax mediterranei ATCC 33500, and subsequent redissolution of the resulting extract in dichloromethane.
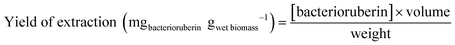 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in loading buffer (4% (w/v) SDS, 20% (w/v) glycerol, 0.5 mM Tris–HCl pH 6.8, 0.02% (w/v) bromophenol blue, 3.1% (w/v) DTT) and incubated for 5 min at 95 °C for denaturation to occur. Then, it was loaded into a polyacrylamide gel along with a molecular weight marker (Amersham™ ECL™ Rainbow™ Marker – Full range) and left to run for 2 h at 110 V. Finally, to stain the proteins, incubation with BlueSafe was carried out under mild agitation for 2 h.
1) in loading buffer (4% (w/v) SDS, 20% (w/v) glycerol, 0.5 mM Tris–HCl pH 6.8, 0.02% (w/v) bromophenol blue, 3.1% (w/v) DTT) and incubated for 5 min at 95 °C for denaturation to occur. Then, it was loaded into a polyacrylamide gel along with a molecular weight marker (Amersham™ ECL™ Rainbow™ Marker – Full range) and left to run for 2 h at 110 V. Finally, to stain the proteins, incubation with BlueSafe was carried out under mild agitation for 2 h.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (Vbacterioruberin extract
20 (Vbacterioruberin extract![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethanol) and the solution was kept for three days at −80 °C, to induce bacterioruberin release from the micellar complex present in Tween® 20. After this period, two phases were formed, a bottom viscous surfactant-rich phase and a top ethanol-rich phase, to which the pigment migrated. The bacterioruberin presence in the top phase was further evaluated by collecting a sample, removing the ethanol, and redissolving it in a controlled volume of 0.5 mL of ethanol. In addition, a 1H NMR analysis was also performed to assess Tween® 20 content in the top phase using a Bruker AC 30 spectrometer (250 MHz) at room temperature, where deuterium oxide was used as the solvent.
Vethanol) and the solution was kept for three days at −80 °C, to induce bacterioruberin release from the micellar complex present in Tween® 20. After this period, two phases were formed, a bottom viscous surfactant-rich phase and a top ethanol-rich phase, to which the pigment migrated. The bacterioruberin presence in the top phase was further evaluated by collecting a sample, removing the ethanol, and redissolving it in a controlled volume of 0.5 mL of ethanol. In addition, a 1H NMR analysis was also performed to assess Tween® 20 content in the top phase using a Bruker AC 30 spectrometer (250 MHz) at room temperature, where deuterium oxide was used as the solvent.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (70) (B).
methanol (70) (B).Conclusions
In this work, the recovery of bacterioruberin and proteins from Haloferax mediterranei was successfully achieved using tensioactive solvents. Among the aqueous solutions of surfactants and ILs studied, Tween® 20 distinguished itself as the best. This non-ionic solvent is not only able to interact with the negatively charged head of the phospholipids allowing the release of the intracellular compounds, but also increases the solubility of bacterioruberin (hydrophobic molecule) in aqueous media, by forming micelles above the CMC, thus allowing for the design of a multi-product biorefinery process.Next, through optimization of the process operational conditions, maximum extraction yields of bacterioruberin of 0.37 ± 0.01 mgbacterioruberin gwet biomass−1 and proteins of 352 ± 9 mgprotein gwet biomass−1 were achieved. Furthermore, in a single protein precipitation step, over 90% of the proteins were successfully separated from bacterioruberin. Finally, it was possible to perform a bacterioruberin polishing step and propose a conceptual design of the final process.
Author contributions
Bárbara M. C. Vaz: investigation, formal analysis, validation, writing – original draft. Mariam Kholany: conceptualization, methodology, supervision, writing – review & editing. Diana C. G. A. Pinto: resources. Inês P. E. Macário: investigation, resources. Telma Veloso: investigation, resources. Tânia Caetano: resources, writing – review & editing. Joana L. Pereira: resources, writing – review & editing. João A. P. Coutinho: resources, supervision. Sónia P. M. Ventura: conceptualization, methodology, resources, writing – review & editing, supervision, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was developed within the scope of the project CICECO-Aveiro Institute of Materials, UIDB/50011/2020, UIDP/50011/2020 & LA/P/0006/2020, financed by national funds through the FCT/MEC (PIDDAC). The authors acknowledge FTC/MCTES for the financial support to CESAM, UIDB/50017/2020 + UIDP/50017/2020 + LA/P/0094/2020, financed by national funds. The authors are also grateful to the FCT for the doctoral grants of M. Kholany (SFRH/BD/138413/2018), I. P. E. Macário (SFRH/BD/123850/2016), and T. Veloso (SFRH/BD/147346/2019). T. Caetano was funded by national funds (OE), through FCT, in the scope of the framework contract foreseen in the numbers 4, 5, and 6 of article 23, of the Decree-Law 57/2016, of August 29, changed by Law 57/2017, of July (CEECIND/01463/2017). The National NMR Network, funded within the framework of the National Program for Scientific Re-equipment, contract REDE/1517/RMN/2005 with funds from POCI 2010 (FEDER) and FCT, is also acknowledged.References
- R. Kataki, R. S. Chutia, M. Mishra, N. Bordoloi, R. Saikia and T. Bhaskar, in Recent Advances in Thermo-Chemical Conversion of Biomass, ed. A. Pandey, T. Bhaskar, M. Stöcker and R. K. Sukumaran, Elsevier, Amsterdam, 1st edn, 2015, pp. 31–74 Search PubMed.
- J. B. Zimmerman, P. T. Anastas, H. C. Erythropel and W. Leitner, Science, 2020, 367, 397–400 CrossRef CAS.
- P. Stegmann, M. Londo and M. Junginger, Resour., Conserv. Recycl., 2020, 6, 100029 Search PubMed.
- G. P. ’t Lam, M. H. Vermuë, M. H. M. Eppink, R. H. Wijffels and C. van den Berg, Trends Biotechnol., 2018, 36, 216–227 CrossRef.
- N. K. Arora and I. Mishra, Environ. Sustainability, 2019, 2, 339–342 CrossRef.
- M. I. Khan, J. H. Shin and J. D. Kim, Microb. Cell Fact., 2018, 17, 36 CrossRef.
- M. Giani, I. Garbayo, C. Vílchez and R. M. Martínez-Espinosa, Mar. Drugs, 2019, 17, 524 CrossRef CAS.
- I. Hamed, F. Özogul, Y. Özogul and J. M. Regenstein, Compr. Rev. Food Sci. Food Saf., 2015, 14, 446–465 CrossRef CAS.
- M. N. Gupta, in Natural-Based Polymers for Biomedical Applications, ed. R. L. Reis, N. M. Neves, J. F. Mano, M. E. Gomes, A. P. Marques and H. S. Azevedo, Woodhead Publishing, Cambridge, 1st edn, 2008, pp. 54–84 Search PubMed.
- J. C. F. Nunes, M. R. Almeida, J. L. Faria, C. G. Silva, M. C. Neves, M. G. Freire and A. P. M. Tavares, J. Solution Chem., 2022, 51, 243–278 CrossRef CAS.
- N. Sharma, M. S. Farooqi, K. K. Chaturvedi, S. B. Lal, M. Grover, A. Rai and P. Pandey, Database, 2014, 1–9 Search PubMed.
- M. Rodrigo-Baños, I. Garbayo, C. Vílchez, M. J. Bonete and R. M. Martínez-Espinosa, Mar. Drugs, 2015, 13, 5508–5532 CrossRef.
- C. Vílchez, E. Forján, M. Cuaresma, F. Bédmar, I. Garbayo and J. M. Vega, Mar. Drugs, 2011, 9, 319–333 CrossRef.
- S. Serrano, S. Mendo and T. Caetano, Res. Microbiol., 2022, 173, 103919 CrossRef CAS.
- M. de la Vega, A. Sayago, J. Ariza, A. G. Barneto and R. León, Biotechnol. Prog., 2016, 32, 592–600 CrossRef CAS.
- R. Calegari-Santos, R. A. Diogo, J. D. Fontana and T. M. B. Bonfim, Curr. Microbiol., 2016, 72, 641–651 CrossRef CAS PubMed.
- M. Giani, Z. Montero-Lobato, I. Garbayo, C. Vílchez, J. M. Vega and R. M. Martínez-Espinosa, Mar. Drugs, 2021, 19, 100 CrossRef CAS.
- K. L. Britton, P. J. Baker, M. Fisher, S. Ruzheinikov, D. J. Gilmour, M.-J. Bonete, J. Ferrer, C. Pire, J. Esclapez and D. W. Rice, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4846–4851 CrossRef CAS.
- M. Mevarech, F. Frolow and L. M. Gloss, Biophys. Chem., 2000, 86, 155–164 CrossRef CAS PubMed.
- G. Payá, V. Bautista, M. Camacho, M.-J. Bonete and J. Esclapez, Biochimie, 2021, 187, 33–47 CrossRef.
- C. Ebel, D. Madern and G. Zaccai, Extremophiles, 2009, 2, 278–297 Search PubMed.
- A. Kumar, A. Alam, D. Tripathi, M. Rani, H. Khatoon, S. Pandey, N. Z. Ehtesham and S. E. Hasnain, Semin. Cell Dev. Biol., 2018, 84, 147–157 CrossRef CAS PubMed.
- A. Siglioccolo, A. Paiardini, M. Piscitelli and S. Pascarella, BMC Struct. Biol., 2011, 11, 50 CrossRef CAS.
- M. Abbes, H. Baati, S. Guermazi, C. Messina, A. Santulli, N. Gharsallah and E. Ammar, BMC Complementary Altern. Med., 2013, 13, 255 CrossRef.
- L. Zalazar, P. Pagola, M. V. Miró, M. S. Churio, M. Cerletti, C. Martínez, M. Iniesta-Cuerda, A. J. Soler, A. Cesari and R. De Castro, J. Appl. Microbiol., 2019, 126, 796–810 CrossRef CAS.
- W. J. Müller, M. S. Smit, E. van Heerden, M. D. Capes and S. DasSarma, Front. Microbiol., 2018, 9, 2563 CrossRef PubMed.
- J. J. Alvares and I. J. Furtado, J. Anal. Sci. Technol., 2021, 12, 44 CrossRef CAS.
- M. Manikandan, N. Hasan and H.-F. Wu, Talanta, 2013, 107, 167–175 CrossRef CAS.
- S. Mishra, C. S. Chanotiya, K. Shanker and A. K. Tripathi, FEMS Microbiol. Lett., 2021, 368, 5 Search PubMed.
- L. Higa Herminia, P. Schilrreff, A. Martín Briski, H. Emanuel Jerez, M. Alexandre de Farias, R. Villares Portugal, E. Lilia Romero and M. Jose Morilla, Colloids Surf., B, 2020, 191, 110961 CrossRef.
- F. I. Kesbiç and N. Gültepe, Folia Microbiol., 2022, 67, 71–79 CrossRef PubMed.
- J. Biswas, F. N. Haque and A. K. Paul, J. Adv. Microbiol., 2016, 1, 1–11 CrossRef.
- S. Ghosh, A. Ray and N. Pramanik, Biophys. Chem., 2020, 265, 106429 CrossRef CAS PubMed.
- Y. Li, F. Fang, M. Sun, Q. Zhao, Y. Hu, Z. Sui, Z. Liang, L. Zhang and Y. Zhang, Talanta, 2020, 213, 120848 CrossRef CAS.
- D. Danwanichakul, O. Rattanaphan, J. Srisatjang and P. Danwanichakul, J. Appl. Polym. Sci., 2014, 131, 39900 CrossRef.
- X. Lin, Y. Wang, Q. Zeng, X. Ding and J. Chen, Analyst, 2013, 138, 6445–6453 RSC.
- M. Martins, W. C. Ooi, M. C. Neves, J. F. Pereira and J. A. Coutinho, J. Chem. Technol. Biotechnol., 2018, 93, 1864–1870 CrossRef CAS.
- M. Kholany, P. Trébulle, M. Martins, S. P. M. Ventura, J.-M. Nicaud and J. A. P. Coutinho, J. Chem. Technol. Biotechnol., 2020, 95, 1126–1134 CAS.
- M. Martins, L. M. de S. Mesquita, B. M. C. Vaz, A. C. R. V. Dias, M. A. Torres-Acosta, B. Quéguineur, J. A. P. Coutinho and S. P. M. Ventura, ACS Sustainable Chem. Eng., 2021, 9, 6599–6612 CrossRef CAS.
- M. Martins, A. P. M. Fernandes, M. A. Torres-Acosta, P. N. Collén, M. H. Abreu and S. P. M. Ventura, Sep. Purif. Technol., 2021, 254, 117589 CrossRef CAS.
- L. M. de S. Mesquita, M. Martins, É. Maricato, C. Nunes, P. S. G. N. Quinteiro, A. C. R. V. Dias, J. A. P. Coutinho, L. P. Pisani, V. V. de Rosso and S. P. M. Ventura, ACS Sustainable Chem. Eng., 2020, 8, 4085–4095 CrossRef.
- R. R. Burgess, in Methods in Enzymology, ed. R. R. Burgess and M. P. Deutscher, Elsevier, Amsterdam, 2nd edn, 2009, vol. 463, pp. 331–342 Search PubMed.
- D. J. Bell, M. Hoare and P. Dunnill, in Downstream Processing, Advances in Biochemical Engineering/Biotechnology, ed. A. Fiechter, Springer, Berlin, Heidelberg, 1st edn, 1983, pp. 1–72 Search PubMed.
- M. Martins, B. P. Soares, J. H. P. M. Santos, P. Bharmoria, M. A. Torres Acosta, A. C. R. V. Dias, J. A. P. Coutinho and S. P. M. Ventura, ACS Sustainable Chem. Eng., 2021, 9, 3942–3954 CrossRef CAS.
- M. J. Castro-Alonso, L. E. Montañez-Hernandez, M. A. Sanchez-Muñoz, M. R. M. Franco, R. Narayanasamy and N. Balagurusamy, Front. Mater., 2019, 6, 126 CrossRef.
- F. A. Vieira, R. J. R. Guilherme, M. C. Neves, H. Abreu, E. R. O. Rodrigues, M. Maraschin, J. A. P. Coutinho and S. P. M. Ventura, Sep. Purif. Technol., 2017, 172, 268–276 CrossRef CAS.
- M. Martins, F. A. Vieira, I. Correia, R. A. S. Ferreira, H. Abreu, J. A. P. Coutinho and S. P. M. Ventura, Green Chem., 2016, 18, 4287–4296 RSC.
- J. M. Newton, D. Schofield, J. Vlahopoulou and Y. Zhou, Biotechnol. Prog., 2016, 32, 1069–1076 CrossRef CAS PubMed.
- B. Balasundaram, D. Nesbeth, J. M. Ward, E. Keshavarz-Moore and D. G. Bracewell, Biotechnol. Bioeng., 2009, 104, 134–142 CrossRef CAS PubMed.
- M. Mohsen-Nia, H. Amiri and B. Jazi, J. Solution Chem., 2010, 39, 701–708 CrossRef CAS.
- C. J. van Oss, J. Protein Chem., 1989, 8, 661–668 CrossRef CAS PubMed.
- S. Taskila, M. Ahokas, J. Järvinen, J. Toivanen and J. P. Tanskanen, Scientifica, 2017, 5120947 Search PubMed.
- G. Schilcher, A. Schlagenhauf, D. Schneditz, H. Scharnagl, W. Ribitsch, R. Krause, A. R. Rosenkranz, T. Stojakovic and J. H. Horina, PLoS One, 2013, 8, e84869 CrossRef PubMed.
- T. Allers, H.-P. Ngo, M. Mevarech and R. G. Lloyd, Appl. Environ. Microbiol., 2004, 70, 943–953 CrossRef CAS PubMed.
- A. Dean and D. Voss, Design and Analysis of Experiments, Springer, New York, 1st edn, 1999 Search PubMed.
- M. I. Rodrigues and A. F. Lemma, Planejamento de Experimentos e Otimização de Processos, Casa do Pão Editora, 3rd edn, 2014 Search PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- L. M. De Souza Mesquita, S. P. M. Ventura, A. R. C. Braga, L. P. Pisani, A. C. R. V. Dias and V. V. De Rosso, Green Chem., 2019, 21, 2380–2391 RSC.
- B. M. C. Vaz, M. Martins, L. M. de Souza Mesquita, A. P. M. Fernandes, D. C. G. A. Pinto, M. G. P. M. S. Neves, M. C. Neves, J. A. P. Coutinho and S. P. M. Ventura, Chem. Eng. J., 2022, 428, 131073 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra02581g |
| ‡ These authors contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2022 |