Cell-derived extracellular vesicles for CRISPR/Cas9 delivery: engineering strategies for cargo packaging and loading
Yujie
Liang†
 a,
Zoya
Iqbal†
b,
Jianhong
Wang
a,
Limei
Xu
b,
Xiao
Xu
b,
Kan
Ouyang
b,
Hao
Zhang
de,
Jianping
Lu
*a,
Li
Duan
*b and
Jiang
Xia
a,
Zoya
Iqbal†
b,
Jianhong
Wang
a,
Limei
Xu
b,
Xiao
Xu
b,
Kan
Ouyang
b,
Hao
Zhang
de,
Jianping
Lu
*a,
Li
Duan
*b and
Jiang
Xia
 *c
*c
aDepartment of Child and Adolescent Psychiatry, Shenzhen Kangning Hospital, Shenzhen Mental Health Center, Shenzhen 518020, China. E-mail: szlujianping@126.com; Fax: +(852) 2603 5057; Tel: +(852) 3943 6165
bDepartment of Orthopedics, the First Affiliated Hospital of Shenzhen University, Shenzhen Second People's Hospital, Shenzhen 518035, China. E-mail: duanl@szu.edu.cn
cDepartment of Chemistry, the Chinese University of Hong Kong, Shatin, Hong Kong SAR, China. E-mail: jiangxia@cuhk.edu.hk
dState Key Laboratory of Bioelectronics, National Demonstration Center for Experimental Biomedical Engineering Education, Southeast University, Nanjing 210096, Jiangsu, China
eEVLiXiR Biotech Inc., Nanjing 210032, Jiangsu, China
First published on 9th June 2022
Abstract
Genome editing technology has emerged as a potential therapeutic tool for treating incurable diseases. In particular, the discovery of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas systems and the design of single-guide RNAs (sgRNAs) have revolutionized genome editing applications. Unfortunately, compared with the rapid development of gene-editing tools, the progress in the development of delivery technologies is lagging behind and thus limiting the clinical application of genome editing. To overcome these limitations, researchers have investigated various delivery systems, including viral and non-viral vectors for delivering CRISPR/Cas and sgRNA complexes. As natural endogenous nanocarriers, extracellular vesicles (EVs) present advantages of biocompatibility, low immunogenicity, stability, and high permeability, making them one of the most promising drug delivery vehicles. This review provides an overview of the fundamental mechanisms of EVs from the aspects of biogenesis, trafficking, cargo delivery, and function as nanotherapeutic agents. We also summarize the latest trends in EV-based CRISPR/Cas delivery systems and discuss the prospects for future development. In particular, we put our emphasis on the state-of-the-art engineering strategies to realize efficient cargo packaging and loading. Altogether, EVs hold promise in bridging genome editing in the laboratory and clinical applications of gene therapies by providing a safe, effective, and targeted delivery vehicle.
1. Introduction
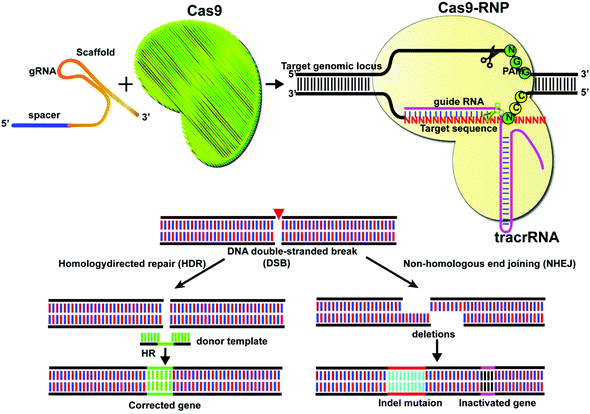
Gene editing technologies have been developed to modify the target gene sequence at a specific point, with site-directed mutation, insertion, or knockout as the main editing methods. The CRISPR/Cas system consists of a single guide RNA (sgRNA) that identifies the DNA sequence to be edited and a Cas nuclease that binds to its target sequence before the protospacer adjacent motif (PAM).1,2 The Cas9 system at its target site can act as a molecular scissor upon activation by the PAM sequence. Currently, the most widely used Cas9 is Streptococcus pyogenes Cas9 (SpCas9) derived from Streptococcus pyogenes. However, other types of Cas9, including SaCas from Staphylococcus aureus,3,4 FnCas9 from Francisella,5,6 St1Cas9 from Streptococcus thermophilus,7 CjCas9 from Campylobacter jejuni,8 and NmCas9 from Neisseria meningitidis,9 have also been reported. In CRISPR/Cas9 mediated genome editing, sgRNA guides the Cas9 proteins to identify the target site at DNA and induce double-strand breakage (DSB), which activates the pathways for cellular DNA repair for precise genome editing.10,11 The classical pathways of DSB repair include non-homologous end joining (NHEJ) and homologous-directed repair (HDR), together with alternative non-homologous end joining (alt-NHEJ) and single-strand annealing (SSA) and other methods (Fig. 1).Intracellular delivery of CRISPR/Cas systems can be achieved by direct delivery of the CRISPR/Cas9 ribonucleoprotein (RNP), which is a complex of ribonucleic acid and RNA-binding protein Cas9. This strategy is limited by the large size of the Cas9 protein (for example, SpCas9 is a large protein with 1368 amino acids). One way to tackle this problem is to engineer Cas9 variants or identify Cas proteins with smaller sizes. Recently, a small Cas9 ortholog was derived from Staphylococcus auricularis and named SauriCas9,12 with high specificity and on-target editing efficiency. Other examples include the mini CRISPR-Cas12f1 system which has only 422 amino acids.13
Instead of directly delivering the Cas9 protein, the delivery of gene plasmids that express the components of the CRISPR/Cas9 system is also viable, the key to which are the carriers. However, the clinical translation of many carriers/vectors faces two major problems: safety and the rapid clearance by the reticuloendothelial system (RES) or the mononuclear phagocyte system (MPS). Compared with nanocarriers constructed from synthetic materials, cell-derived nanovesicles are superior in both aspects. Collectively called extracellular vesicles (EVs) (including exosomes as the most well-studied type of EVs), the naturally borne nanovesicles have several advantages, including a membrane structure resembling that of the cell membranes and their ability to avoid phagocytosis, escape immune response, prolong the circulation time, and penetrate to deep tissue regions. All these properties make them advantageous nanomaterials for drug delivery, including small molecules, nucleic acid drugs, and proteins for a wide variety of diseases. The use of EVs or exosomes for drug delivery and therapy has been extensively reviewed.14–19 Recently we together with other labs reported that targeting moieties can be installed on the surface of exosomes through various engineering and modification methods to realize cell and tissue-specific delivery of the cargo. This area has also been extensively reviewed.18,20–23 EV/exosome-mediated deliveries of the CRISPR/Cas9 systems are emerging but are underdeveloped. One major hurdle in this regard is the packaging or loading of the CRISPR/Cas9 systems within EVs and exosomes, because of the large size and heterogeneity of the gene editing systems and the limited capacity within EVs and exosomes. Besides giving an overview of how cell-derived EVs can be harnessed for CRISPR/Cas9 delivery, this review focuses on cargo packaging and loading within EVs/exosomes, which is a critical hurdle toward the clinical applications of EV/exosome-mediated CRISPR/Cas9 delivery. We also discuss the quality control, regulation, and limitations of EV/exosome-mediated CRISPR/Cas9 delivery toward clinical applications.
2. Overview of EVs
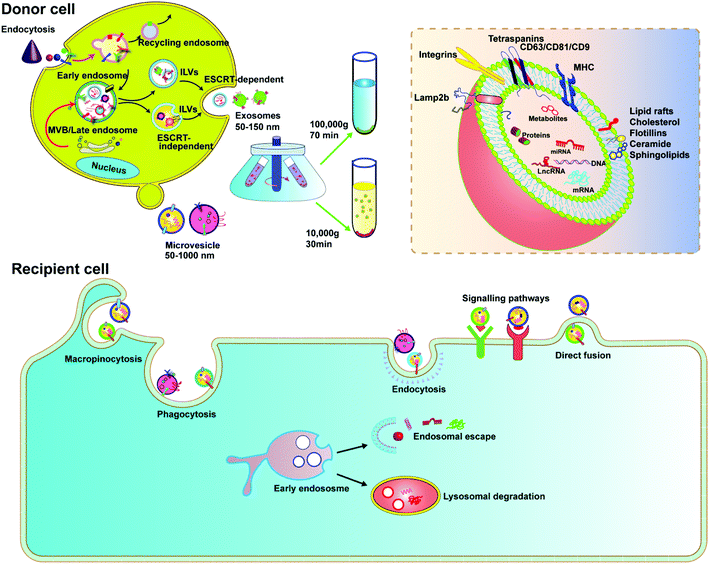
EVs are membrane-derived vesicles released by cells into the extracellular space, which play an important role in intercellular communication and regulate a range of biological processes. Over the past decade, knowledge about the characteristics and function of EVs has accumulated rapidly. Due to the heterogeneous nature of EVs in origin, size, and composition, there is currently no gold standard to define EVs. However, the general consensus is that EVs are lipid vesicles derived from endosomal compartments and are secreted to the extracellular environment after the fusion of the endosomal compartment with the plasma membrane. According to the minimal information for studies of extracellular vesicles 2018 (MISEV2018) and the updated guidelines, EVs are categorized based on the following: (a) size (small EVs are <200 nm and medium/large EVs are >200 nm), (b) density (low, middle, high), and (c) marker protein (e.g., CD63+/CD81+-EVs).24,25 In this review, we focus on two types of EVs, exosomes and microvesicles, as delivery vehicles.Exosomes are classically defined by their size (usually <150 nm in diameter) and endosome associated marker proteins. Exosomes are typically obtained by ultracentrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g after removing apoptotic bodies and microvesicles. Separation with Optiprep or sucrose gradients can achieve higher homogeneity. Biogenesis of exosomes is generally divided into three stages: (1) formation of endocytic vesicles by cell membrane depression, (2) formation of multivesicular bodies (MVBs) by inward budding of endocytic vesicle membranes, and (3) fusion of MVBs with the cell membrane to form intraluminal vesicles (ILVs) and ILVs are released as exosomes (Fig. 2). During MVB formation, ILVs can package Golgi-processed proteins, cytoplasmic proteins, and genomic cargoes from their parental cells. The endosomal sorting complex required for endosomal sorting and transport (ESCRT) proteins, especially AIP1/Alix/Vps31 and Tsg101/Vps23, regulate cargo loading. Among them, ESCRT-0, ESCRT-I and ESCRT-II are responsible for recognizing and delivering ubiquitinated cargoes into the lumen of endosomes. Besides the ESCRT-dependent pathway, an ESCRT-independent pathway also exists.26,27 The exosomal contents, including lipids, such as cholesterol, sphingomyelin, and phosphoglycerides, proteins, DNA, and RNA are released by inclusion in exosomes (Fig. 2). The surface of exosomes is enriched with multiple protein families such as tetraspanins (CD63, CD9 and CD81), heat shock proteins (Hsp70), membrane transporters (RAB proteins, flotillin and annexin) and proteins involved in MVB formation (ALIX, TSG101 and clathrin), lysosomal protein (Lamp2b), etc. These proteins have been widely used as exosomal markers. Exosomes are natural carriers for cell–cell communications, transporting materials and information from donor cells to recipient cells; this intrinsic feature makes them potential drug carriers.
000g after removing apoptotic bodies and microvesicles. Separation with Optiprep or sucrose gradients can achieve higher homogeneity. Biogenesis of exosomes is generally divided into three stages: (1) formation of endocytic vesicles by cell membrane depression, (2) formation of multivesicular bodies (MVBs) by inward budding of endocytic vesicle membranes, and (3) fusion of MVBs with the cell membrane to form intraluminal vesicles (ILVs) and ILVs are released as exosomes (Fig. 2). During MVB formation, ILVs can package Golgi-processed proteins, cytoplasmic proteins, and genomic cargoes from their parental cells. The endosomal sorting complex required for endosomal sorting and transport (ESCRT) proteins, especially AIP1/Alix/Vps31 and Tsg101/Vps23, regulate cargo loading. Among them, ESCRT-0, ESCRT-I and ESCRT-II are responsible for recognizing and delivering ubiquitinated cargoes into the lumen of endosomes. Besides the ESCRT-dependent pathway, an ESCRT-independent pathway also exists.26,27 The exosomal contents, including lipids, such as cholesterol, sphingomyelin, and phosphoglycerides, proteins, DNA, and RNA are released by inclusion in exosomes (Fig. 2). The surface of exosomes is enriched with multiple protein families such as tetraspanins (CD63, CD9 and CD81), heat shock proteins (Hsp70), membrane transporters (RAB proteins, flotillin and annexin) and proteins involved in MVB formation (ALIX, TSG101 and clathrin), lysosomal protein (Lamp2b), etc. These proteins have been widely used as exosomal markers. Exosomes are natural carriers for cell–cell communications, transporting materials and information from donor cells to recipient cells; this intrinsic feature makes them potential drug carriers.
Microvesicles are heterogeneous, membrane-bound vesicles shed from the cell surface of various cell types, normally larger than exosomes. Microvesicles can be obtained from conditioned media and biological fluids by removing apoptotic bodies and then collecting the supernatant by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, also a speed slower than that required for exosomes. The biogenesis of microvesicles involves vertical transport of molecular cargo to the plasma membrane, membrane lipid redistribution, and the use of different contractile mechanisms at the surface to permit vesicles to contract.26 Since microvesicles are formed by direct budding from the plasma membrane, their sizes are more heterogenous, ranging from 150 nm to greater than 1000 nm in diameter. In recent years, microvesicles have been recognized to play important roles in altering the extracellular environment, intercellular signalling, and promoting tumor invasion. Microvesicles encapsulate and transfer various types of cargoes, including proteins, miRNAs and DNA/mRNA fragments, lipid components, plasma membrane receptors, ribosomal RNA (rRNA), mitochondrial DNA, and their membrane surface markers including selectins, integrins, ARF6 and CD40.27 Although there is much similarity between exosomes and microvesicles, it was reported that only microvesicles, but not exosomes, could transfer plasmid DNA from donor to recipient cells.28 Microvesicles therefore can be a platform to deliver CRISPR plasmids.
000g, also a speed slower than that required for exosomes. The biogenesis of microvesicles involves vertical transport of molecular cargo to the plasma membrane, membrane lipid redistribution, and the use of different contractile mechanisms at the surface to permit vesicles to contract.26 Since microvesicles are formed by direct budding from the plasma membrane, their sizes are more heterogenous, ranging from 150 nm to greater than 1000 nm in diameter. In recent years, microvesicles have been recognized to play important roles in altering the extracellular environment, intercellular signalling, and promoting tumor invasion. Microvesicles encapsulate and transfer various types of cargoes, including proteins, miRNAs and DNA/mRNA fragments, lipid components, plasma membrane receptors, ribosomal RNA (rRNA), mitochondrial DNA, and their membrane surface markers including selectins, integrins, ARF6 and CD40.27 Although there is much similarity between exosomes and microvesicles, it was reported that only microvesicles, but not exosomes, could transfer plasmid DNA from donor to recipient cells.28 Microvesicles therefore can be a platform to deliver CRISPR plasmids.
3. EVs as drug delivery vehicles
In recent years, many synthetic nano-delivery platforms have emerged as delivery vehicles for CRISPR/cas9 tools. Since EVs are naturally released from all cells, they have high biocompatibility and lower immunogenicity.29 Encapsulating protein or RNA drugs within the lipid bilayer membrane of EVs protects them from enzymatic degradation during circulation, prolongs the half-life, and improves their stability. The particle size and negative charge of EVs avoid phagocytosis by the reticuloendothelial system and rapid renal clearance, increasing the retention at the disease site.30 Intrinsically expressed membrane-anchored ligands or targeting moieties on the surface of EVs can also achieve specific targeting.23 Unlike other synthetic nanoparticle systems, endocytosed EVs can escape the endosomal and lysosomal degradation pathway, thus facilitating direct cytoplasmic uptake of encapsulated contents.31 EVs can also cross tissue boundaries and diffuse into deep tissues, including the blood–brain and intestinal barriers. Taken together, these features make EVs unique among nano-sized drug delivery vehicles (Fig. 3).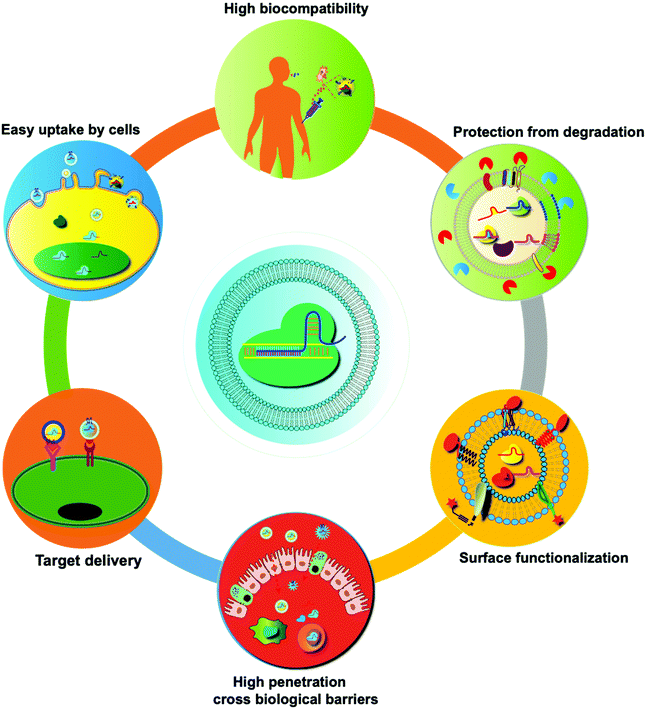 | ||
| Fig. 3 Summary of the unique features of EVs as drug delivery vehicles. Compared with synthetic nanoparticles, EVs show low toxicity and the ability to cross biological barriers, amenability of surface modification, and other advantages.32 | ||
4. Delivery of CRISPR/Cas9 tools as plasmid DNA
EVs have been used to mediate the delivery of plasmid DNA. Kanada and co-workers reported MV mediated delivery of DNA encoding the cre recombinase in vivo.28 K. M. McAndrews and co-workers reported that non-autologous exosomes could encapsulate CRISPR/Cas9 plasmid DNA by transfection reagents and deliver to pancreatic cancer cells to knockout mutant cell KrasG12D oncogenic alleles, thereby inhibiting proliferation and suppressing tumor growth in orthotopic pancreatic tumor models.33 He and co-workers reported that epithelial cell-derived MVs can act as a safe, natural and efficient delivery vehicle for CRISPR/Cas9 as compared to cancer-derived MVs and can effectively inhibit cell proliferation.34 Xu and co-workers developed an epithelial cell-derived EV-based platform for CRISPR/Cas9 plasmid delivery and genetically engineered EVs with chimeric antigen receptors (CARs) to enhance their tumor targeting ability. The result showed that CRISPR/Cas9-loaded CAR-EVs accumulated in cancer cells quickly and efficiently released CRISPR/Cas9 targeting the MYC oncogene to induce cytotoxicity in malignant B-cells.35 The combination of CAR and EVs was proved to be a novel gene therapy approach in tumor treatment.Since cancer-derived MVs have tumor cell tropism ability, Kim and co-workers developed cancer-derived exosomes as natural carriers for efficient delivery of CRISPR/Cas9 in cancer.36 These cancer-cell-derived exosomes loaded with CRISPR/Cas9 plasmids targeting poly (ADP-ribose) polymerase-1 significantly inhibited PARP-1 expression and enhanced chemosensitivity to cisplatin. Furthermore, cancer-derived exosomes can selectively accumulate in ovarian cancer xenografts more than epithelial cell-derived exosomes, probably due to their cell tropism, providing a potential vehicle for efficient in vivo target delivery. We have summarized the present studies about the Cas9 DNA loading into EVs in Table 1.
| EV source | Cargo | Function | Ref. |
|---|---|---|---|
| HEK293T-derived exosomes | CRISPR/SpCas9 sgRNA KrasG12D | To knock out the mutant KrasG12D oncogenic allele to attenuate tumor progression | 33 |
| Epithelial cell-derived microvesicles | CRISPR/Cas9-sgRNA IQGAP1 | Safe, natural delivery with synergistic cytotoxicity effect with sorafenib | 34 |
| CD19-CAR-modified epithelial cell-derived EVs | CRISPR/Cas9-sgRNA MYC | To induce mutations and toxicities in malignant B-cells | 35 |
| Cancer-derived exosomes | CRISPR/Cas9-sgRNA PARP-1 | Cell tropism, apoptotic cell death of SKOV3, enhances synergistic anti-tumor effect with cisplatin | 36 |
| HEK293FT-derived exosomes (hybrid exosomes) | CRISPR/SpCas9 sgRNA Runx2 | To deliver CRISPR-Cas9 system in MSCs | 39 |
Plasmid size and sequence determine the delivery efficiency of MVs to a certain extent. Kanada and co-workers reported a delivery system comprising engineered minicircle (MC) DNA loading MVs. They exhibited that MVs have greater efficiency in loading MC DNA and lead to prolonged and significantly higher transgene expression as compared to their parental plasmid counterparts.37 Cas9 MC DNA delivery by MVs constitutes a safe vector for effective gene editing in the clinic.38 Lin and co-workers reported an exosome–liposome hybrid system developed by simple incubation to improve the loading capacity and delivery efficiency of exosomes. Unlike native exosomes, hybrid exosome particles can efficiently encapsulate CRISPR/Cas9 expression plasmids. Furthermore, this hybrid system can be endocytosed and express the loaded gene in the mesenchymal cell for gene editing.39
5. Delivery of CRISPR/Cas9 RNPs by EVs
The plasmid-mediated approach provides the most stable Cas9 protein expression, but the gene DNA must be delivered directly into the nucleus to undergo transcription to achieve a therapeutic effect, resulting in delayed gene editing. mRNA does not need to be delivered to the nucleus and can be directly translated to proteins in the cytoplasm. However, mRNAs are easily degraded by enzymes. The delivery of Cas9 protein and sgRNA is another viable approach. Saving the transcription and translation steps, the editing process is faster and more efficient, with a lower off-target effect. The difficulty of this method lies in the loading of the CRISPR/Cas9 RNP into EVs. Currently, a variety of small molecule drugs, proteins, and nucleic acid drugs have been loaded into EVs using ultrasound, electroporation, transfection, incubation, extrusion, freeze–thaw cycling, heat shock, pH gradient method, and hypotonic dialysis. Overexpressed Cas9 protein in the donor cell can be packaged in the EVs of the donor cells. Kim and co-workers reported that EVs derived from HL-60 cells showed efficient loading of different cargoes and higher effector functions. EVs collected from donor cells were used to transfect the cells with GFP-tagged Cas9. The recipient cells showed the highest GFP fluorescence.40 This indicated that the EVs produced from dHL-60 cells could be a potential source for CRISPR/Cas9 RNP delivery. Besides, various engineering technologies have been developed to increase loading efficiency. Here we systemically describe two below.5.1 Engineering exosomal proteins for CRISPR/Cas9 RNP loading
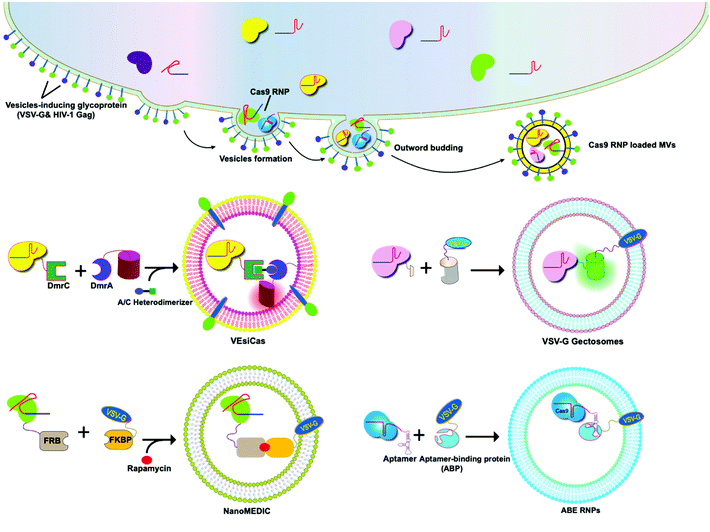
A variety of transmembrane and luminal proteins are found to be specifically expressed on EVs; some of them, called scaffold proteins, can be used to directly package exogenous proteins during exosome biogenesis. Through these scaffold proteins (listed in Fig. 4), cargoes can be loaded on the surface or in the lumen. These scaffold proteins include the immunoglobulin superfamily (IGSF, IGSF8, and PTGFRN),41 MARCKS protein family (BASP1, MARCKS, and MARKCSL1),42 exosomal sorting proteins (syntenin, Alix, TSG101 and Flotilin)43 and transmembrane proteins (PDGFR, Lamp-2b, CD63, CD9, and CD44) (Fig. 4). Fusion of Cas9 with either the intracellular or extracellular domain of the exosomal scaffolding proteins can realize the loading. Dooley and co-workers identified that PTGFRN and BASP1 scaffold proteins were sorted into EVs and enabled the luminal loading and successfully loaded Cas9 proteins by fusing them to an N-terminal fragment of BASP1 (Fig. 4).42 Nabhan and co-workers reported a virus-independent cellular process that produces a distinct type of MVs by direct plasma membrane budding (DPMB). DPMB recruits an important ESCRT component, the TSG101 protein, to the surface of the cell through interaction with a tetrapeptide motif (PS/TAP) of arrestin-domain-containing protein 1 (ARRDC1) to initiate plasma membrane budding. Subsequently, TSG101 transfers to the plasma membrane from endosomes leading to the release of ARRDC1, TSG101, and other cellular proteins containing microvesicles. Through the PPxY motifs of arrestin-domain-containing protein 1 (ARRDC1), it can interact with WW domains (∼40 amino acids each) of NEDD4 ubiquitin ligases.44 Wang and co-workers reported ARRDC1 mediated MVs. Exogenous proteins can be packaged into EVs by direct fusion to ARRDC1 or to a WW domain that can specifically interact with ARRDC1. The packaging and intracellular delivery of macromolecules, including the tumor suppressor p53 protein and the genome editing CRISPR-Cas9/guide RNA complex, have been carried out using ARRDC1 as a scaffold protein. Through its PPXY motif, ARRDC1 interacts specifically with proteins of the WW domain of the NEDD4 family, thereby allowing the recruitment of foreign proteins to EVs. In this aspect, WW-Cas9-sgRNA was combined with the ARRDC1 plasmid to co-transfect into cells, then WW-Cas9 and sgRNA were integrated inside the bilipidic membrane of EVs through molecular recognition between WW and ARRDC1 (Fig. 4). Successful delivery of EVs containing WW-Cas9 and anti-GFP sgRNA to recipient cells resulted in the disappearance of the GFP fluorescence signal, and both T7E1 assay and direct DNA sequencing confirmed GFP gene editing, suggesting that EV-mediated delivery of Cas9 enabled the knockout of the GFP gene.45 This result provided evidence that EVs secreted from the ARRDC1-Cas9 co-transfected cells represent a highly versatile platform for packaging and intracellular delivery of CRISPR-Cas9 macromolecules.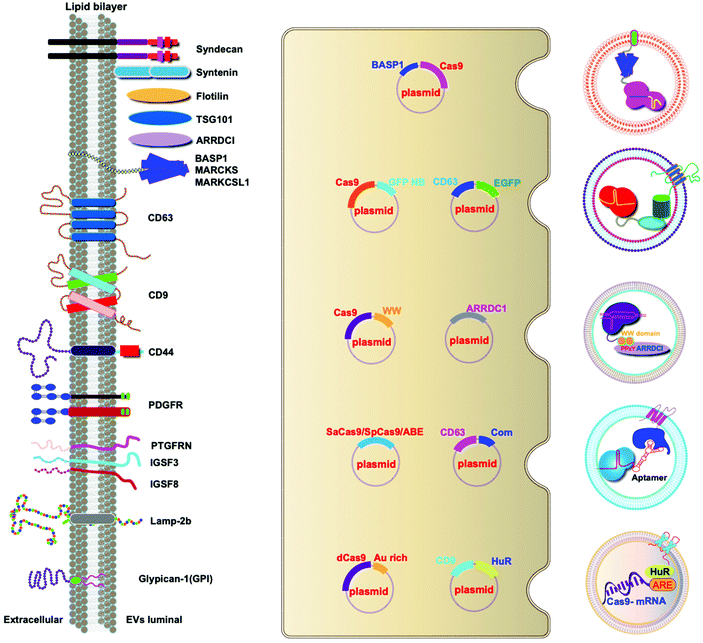 | ||
| Fig. 4 Strategies to harness exosomal scaffolding proteins for loading and delivery of CRISPR/Cas9 RNPs. | ||
Next, we select CD63 as a particular example of the scaffolding proteins. CD63 is a member of the tetraspanin family expressed on the surface of exosomal membranes. Ye and co-workers reported engineered exosomes by fusing GFP with CD63 and GFP nanobody with Cas9 protein for Cas9 protein delivery. Due to the high affinity between GFP–GFP nanobodies, Cas9 protein can be selectively captured and efficiently loaded into EVs (Fig. 4). The authors used a different tactic to visualize the function of delivered CRISPR/Cas9 components using a reporter cell line (A549stop-DsRed gave a red fluorescence signal when sgRNA-guided endonuclease deleted the stop element). The results showed that CRISPR/Cas9 surface-modified exosomes more efficiently abrogated the target gene in recipient cells.46 This exosome loading system composed of the exosomal membrane protein CD63-GFP and GFP nanobody fused to Cas9 provides a new method for the specific loading of the Cas9 protein in EVs. Li and co-workers designed surface-functionalized exosomes for loading RNA using a fusion protein, in which the exosomal membrane protein CD9 was fused to the RNA binding protein HuR.47 CD9-HuR exosomes can efficiently load functional CRISPR/dCas9 through the interaction between HuR and AU rich elements of the RNA (Fig. 4).
Yim and co-workers developed an optogenetically engineered exosome system, EXPLORs, which has high loading capability of proteins into exosomes and delivery efficiency through the optically reversible protein–protein interaction controlled by blue light with an endogenous biogenesis process.48 This approach can achieve controllable, reversible loading, and efficient delivery of Cas9 proteins. Some studies have established that the genetic engineering of EV-producing cells to express exosome sorting domains and the N-terminal fragment of syntenin can significantly improve display efficacy.43 By combining Cas9 therapeutics and a natural delivery vehicle that can overcome delivery bottlenecks, genetically engineered EVs have great potential to be a next-generation genome editing platform.
5.2 VSV-G-assisted cargo delivery
The glycoproteins (G proteins) of vesicular stomatitis virus (VSV) mediate both cell attachment and membrane fusion of the virus. Several research groups have focused on the vesicular stomatitis virus glycoprotein (VSV-G) in assisting exosome-mediated delivery. Meyer and co-workers reported that VSV-G not only loads protein cargoes onto exosomes, but also enhances the protein delivery efficiency via a pseudotyping mechanism. The gene transfection efficiency and fluorescence tracking exhibited the successful targeting and encapsulation of VSVG fusions into exosomes. The exosome size or distribution was not influenced by the pseudotyping of exosomes. The ectodomain plays no role in protein loading, but it has a role in cell tropism. These VSV-G-modified EVs can load protein cargoes and increase their delivery capability.49 It has been shown that cells expressing VSV-G can release VSV-G efficiently into cell-derived vesicles to generate extracellular vesicles called “gectosomes” or “gesicles” (Fig. 5). Montagna and co-workers reported the efficient delivery of CRISPR/SpCas9 RNPs into target cells via decoration with fusogenic glycoprotein VSV-G. Co-expression of VSV-G with SpCas9 fused Gag or MinimalGag protein in HEK 293T cells can generate fused VSV-G vesicles (VEsiCas). VEsiCas achieved ∼60% and ∼30% knockout efficiency for CXCR4 and VEGFA, respectively. VEsiCas have higher delivery efficiency and lower toxicity compared to electroporation. It can be used to edit various types of cells including iPSCs and cardiomyocytes.50 Campbell and co-workers reported the efficient delivery of CRISPR-Cas9 via the gectosome approach for exosome-based genome editing. Recently, gectosomes have been engineered to contain Cas9 as an RNP complex. CherryPicker Red is a membrane-associated protein containing a “DmrA” domain, which could bind to the DmrC domain by the addition of an A/C heterodimeric molecule resulting in the packaging of Cas9RNPs into gectosomes. The Cas9 RNP complex thus delivered could cause both mutation and copy number loss of the HIV provirus, along with the inhibition of HIV proviral activity.51Because gectosomes are genetically encoded, highly programmable, easy to prepare, and amenable to purification, this facile approach can be adapted for a wide range of research and possible therapeutic applications. Utilizing self-associating split GFP fragments,52 Zhang and co-workers developed gectosomes by co-transfecting a plasmid-encoded VSV-G C-terminal GFP11 (VSV-G-GFP11) and SaCas9-GFP1-10 in 293 cells. The binding of GFP11 to GFP1-10 could reconstitute a functional fluorescent GFP molecule so that VSV-G-GFP11 could recognize GFP1-10-tagged cargo protein. This approach based on GFP complementation greatly improves the efficiency of cargo delivery to target cells and reduces nonspecific encapsulation of cellular proteins. Using SaCas9-encapsulating gectosomes, they demonstrated DNA recombination, RNA interference, and gene editing in cultured cells and mouse liver tissue in vivo.53 Gee and co-workers developed an all-in-one EV delivery system called NanoMEDIC (nanomembrane-derived extracellular vesicles for the delivery of macromolecular cargoes) based on VSV-G. Rapamycin-inducible FK506-binding protein FKBP12 and FKBP-rapamycin-binding domain (FRB) homing mechanisms were utilized in this system. Dimerization of FRB and FKBP12 results in the selective packaging of Cas9 protein into EVs. NanoMEDIC can effectively achieve genome editing in various human cell types, such as T cells, monocytes, iPSCs, iPSC-derived cortical neurons, and myogenic cells. Above 90% exon skipping efficiencies can be achieved in skeletal muscle cells derived from Duchenne muscular dystrophy (DMD) patient iPS cells through NanoMEDIC.54 Lu and co-workers reported that the functionalization of EVs with VSV-G protein could aid in the delivery of Cas9 RNPs to the cytosol of specific cells by escaping endocytic trafficking.55 Yao and co-workers enriched Cas9 RNPs into VSV-G-modified EVs using specific interactions between RNA aptamers and aptamer-binding proteins (ABPs). The aptamer-ABP interaction loads sgRNA inside EVs, while the intrinsic affinity between sgRNA and Cas9 protein mediates the packaging of the entire RNP complex. RNPs can be functionally delivered by EVs, and generate high editing efficiencies for multiplex genome editing. The aptamer com, ABP Com, and VSV-G proteins are all necessary elements for the functional delivery of RNPs to EVs.56 These examples manifest the potential of VSV-G based EVs as inherent drug delivery systems for CRISPR/Cas9 systems.
6. Quality control, regulations, and limitations
Quality control is critical to the mass production of EV/exosomes and their clinical uses, which can be divided into three categories. Primarily, the physicochemical properties of the EV/exosome samples, such as size and morphology, can be analyzed using particle size analyzers and transmission electron microscopes. The second category is the profiling of molecular components including specific EV/exosome markers including proteins, nucleic acids, and lipids, by high throughput sequencing, mass spectrometry techniques, or lipidomic analysis.57–59 In addition, the glycomic pattern of the carbohydrate molecules on the surface of EVs/exosomes has also emerged to be an important yet underexplored aspect.60 The third category is the activity of the EV/exosome preparations. The EV activity can be defined, in a narrow term, as the uptake rate of EVs by the recipient cells within a limited time window (often 30–60 min) in vitro (this however does not reflect the EV activity in vivo). Notably, a large variation in this regard can be found between different teams or papers, depending on the biometrics, quantification methods, isolation and purification processes, etc.61 Another understudied area is the immunogenicity of EVs/exosomes. Because EVs/exosomes have not been widely applied in clinical practice, data about the immunogenicity of EVs/exosomes are mostly from the study of non-primate animals. The R&D of clinical-level EVs/exosomes shall follow the standardized procedures of biopharmaceuticals and cell therapies, as all the manipulations such as genetic engineering operations, chemical processing, or manufacturing processes may incur immunogenicity or toxicity. The field thus urgently calls for unified standards for the quality control of EVs/exosomes and well-managed regulations of EVs/exosomes in clinical trials, cosmetic uses, or any human-related applications.7. Conclusions and perspective
Taken together, the use of CDNs for CRISPR/Cas9 delivery has made remarkable progress in the past few decades. RV/exosome-based CRISPR/Cas9 delivery exhibits several advantages over traditional drug delivery systems in general. For example, EVs/exosomes have higher stability in blood, allowing the transmission over long distances. EVs/exosomes can overcome various biological barriers and often have a natural targeting ability. The current consensus is that EVs/exosomes have lower immunogenicity compared to liposomes and viruses. Also, tissue- or cell-specific delivery of CRISPR/Cas9 tools that minimize immune activation or unexpected off-target effects are required. In this regard, the intrinsic targeting properties of EVs can be harnessed, or EVs can be functionalized to confer the targeting capability. For example, we genetically engineered exosomes to target chondrocyte or mesenchymal stem cells, which showed a precise drug delivery effect in vitro and in vivo.23,62,63 Assembly of targeting proteins or chemical modifications of EVs can significantly decrease the off-target effects, leading to increased therapeutic efficacy.22 In addition, spatiotemporal control of the CRISPR/Cas9 function could also be used to reduce off-target effects. For example, one can use external stimuli, such as light, to modulate the responses of EVs. Finally, CRISPR/Cas9 tools in the mRNA format64,65 can also be packaged within EVs/exosomes as a means of delivering CRISPR/Cas9 tools, but only a few examples can be found in the literature.47,66 Taken together, notwithstanding that clinical applications of EV-mediated genome therapy still face multiple technical hurdles, a bright future awaits ahead.Author contributions
Y. L. drew the figures. Y. L., I. Z., and J. X. wrote the paper. J. X. gave final approval of the manuscript. All authors contributed to reference collection, provided feedback, and helped in shaping the article.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was funded by the Science and Technology Innovation Committee of Shenzhen (no. JCYJ20200109150700942, SGDX20201103095800003; GJHZ20200731095606019), the Guangdong Basic and Applied Basic Research Foundation (2020A1515011581, 2021A1515010985), the Guangdong International Cooperation Project (no. 2021A0505030011), the Key-Area Research and Development Program of Guangdong Province (2019B030335001), the Sanming Project of Medicine (no. SZSM201612079), the Shenzhen Fund for Guangdong Provincial High Level Clinical Key Specialties (no. SZGSP013), and the Shenzhen Key Medical Discipline Construction Fund (no. SZXK042).Notes and references
- J. D. Sander and J. K. Joung, Nat. Biotechnol., 2014, 32, 347–355 CrossRef CAS PubMed
.
- J. A. Doudna and E. Charpentier, Science, 2014, 346, 1258096 CrossRef PubMed
.
- F. A. Ran, L. Cong, W. X. Yan, D. A. Scott, J. S. Gootenberg, A. J. Kriz, B. Zetsche, O. Shalem, X. Wu, K. S. Makarova, E. V. Koonin, P. A. Sharp and F. Zhang, Nature, 2015, 520, 186–191 CrossRef CAS PubMed
.
- Y. Tan, A. H. Y. Chu, S. Bao, D. A. Hoang, F. T. Kebede, W. Xiong, M. Ji, J. Shi and Z. Zheng, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20969–20976 CrossRef CAS PubMed
.
- E. Schunder, K. Rydzewski, R. Grunow and K. Heuner, Int. J. Med. Microbiol., 2013, 303, 51–60 CrossRef CAS PubMed
.
- S. Acharya, A. Mishra, D. Paul, A. H. Ansari, M. Azhar, M. Kumar, R. Rauthan, N. Sharma, M. Aich, D. Sinha, S. Sharma, S. Jain, A. Ray, S. Jain, S. Ramalingam, S. Maiti and D. Chakraborty, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20959–20968 CrossRef CAS PubMed
.
- M. Müller, C. M. Lee, G. Gasiunas, T. H. Davis, T. J. Cradick, V. Siksnys, G. Bao, T. Cathomen and C. Mussolino, Mol. Ther., 2016, 24, 636–644 CrossRef PubMed
.
- E. Kim, T. Koo, S. W. Park, D. Kim, K. Kim, H.-Y. Cho, D. W. Song, K. J. Lee, M. H. Jung, S. Kim, J. H. Kim, J. H. Kim and J.-S. Kim, Nat. Commun., 2017, 8, 14500 CrossRef CAS PubMed
.
- C. M. Lee, T. J. Cradick and G. Bao, Mol. Ther., 2016, 24, 645–654 CrossRef CAS PubMed
.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed
.
- P. Mali, L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville and G. M. Church, Science, 2013, 339, 823–826 CrossRef CAS PubMed
.
- Z. Hu, C. Zhang, S. Wang, S. Gao, J. Wei, M. Li, L. Hou, H. Mao, Y. Wei, T. Qi, H. Liu, D. Liu, F. Lan, D. Lu, H. Wang, J. Li and Y. Wang, Nucleic Acids Res., 2021, 49, 4008–4019 CrossRef CAS PubMed
.
- Z. Wu, Y. Zhang, H. Yu, D. Pan, Y. Wang, Y. Wang, F. Li, C. Liu, H. Nan, W. Chen and Q. Ji, Nat. Chem. Biol., 2021, 17, 1132–1138 CrossRef CAS PubMed
.
- W. Liao, Y. Du, C. Zhang, F. Pan, Y. Yao, T. Zhang and Q. Peng, Acta Biomater., 2019, 86, 1 CrossRef CAS PubMed
.
- L. Cabeza, G. Perazzoli, M. Peña, A. Cepero, C. Luque, C. Melguizo and J. Prados, J. Controlled Release, 2020, 327, 296–315 CrossRef CAS PubMed
.
- J. Wang, D. Chen and E. A. Ho, J. Controlled Release, 2021, 329, 894–906 CrossRef CAS PubMed
.
- L. Xu, Y. Liang, X. Xu, J. Xia, C. Wen, P. Zhang and L. Duan, Bioengineered, 2021, 12, 7929–7940 CrossRef CAS PubMed
.
- L. Duan, L. Xu, X. Xu, Z. Qin, X. Zhou, Y. Xiao, Y. Liang and J. Xia, Nanoscale, 2021, 13, 1387–1397 RSC
.
- L. Duan, X. Xu, L. Xu, H. Chen, X. Li, M. Alahdal, Y. Xiao, Y. Liang and J. Xia, Curr. Med. Chem., 2021, 28, 6458–6483 CrossRef CAS PubMed
.
- X. Luan, K. Sansanaphongpricha, I. Myers, H. Chen, H. Yuan and D. Sun, Acta Pharmacol. Sin., 2017, 38, 754–763 CrossRef CAS PubMed
.
- H. Chen, L. Wang, X. Zeng, H. Schwarz, H. S. Nanda, X. Peng and Y. Zhou, Front. Cell Dev. Biol., 2021, 2827 Search PubMed
.
- L. Duan, K. Ouyang, J. Wang, L. Xu, X. Xu, C. Wen, Y. Xie, Y. Liang and J. Xia, ChemBioChem, 2021, 22, 3360–3368 CrossRef CAS PubMed
.
- Y. Liang, L. Duan, J. Lu and J. Xia, Theranostics, 2021, 11, 3183 CrossRef CAS PubMed
.
- K. W. Witwer, D. C. Goberdhan, L. O'Driscoll, C. Théry, J. A. Welsh, C. Blenkiron, E. I. Buzás, D. Di Vizio, U. Erdbrügger, J. M. Falcón-Pérez, Q. L. Fu, A. F. Hill, M. Lenassi, J. Lötvall, R. Nieuwland, T. Ochiya, S. Rome, S. Sahoo and L. Zheng, J. Extracell. Vesicles, 2021, 10, e12182 CAS
.
- C. Théry, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G. K. Atkin-Smith, D. C. Ayre, J. M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N. N. Bauer, A. A. Baxter, M. Bebawy, C. Beckham, A. Bedina Zavec, A. Benmoussa, A. C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F. E. Borràs, S. Bosch, C. M. Boulanger, X. Breakefield, A. M. Breglio, M. Brennan, D. R. Brigstock, A. Brisson, M. L. Broekman, J. F. Bromberg, P. Bryl-Górecka, S. Buch, A. H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E. I. Buzás, J. B. Byrd, G. Camussi, D. R. Carter, S. Caruso, L. W. Chamley, Y. T. Chang, C. Chen, S. Chen, L. Cheng, A. R. Chin, A. Clayton, S. P. Clerici, A. Cocks, E. Cocucci, R. J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F. A. Coumans, B. Coyle, R. Crescitelli, M. F. Criado, C. D'Souza-Schorey, S. Das, A. Datta Chaudhuri, P. de Candia, E. F. De Santana, O. De Wever, H. A. Del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L. C. Dieterich, V. Dolo, A. P. Dominguez Rubio, M. Dominici, M. R. Dourado, T. A. Driedonks, F. V. Duarte, H. M. Duncan, R. M. Eichenberger, K. Ekström, S. El Andaloussi, C. Elie-Caille, U. Erdbrügger, J. M. Falcón-Pérez, F. Fatima, J. E. Fish, M. Flores-Bellver, A. Försönits, A. Frelet-Barrand, F. Fricke, G. Fuhrmann, S. Gabrielsson, A. Gámez-Valero, C. Gardiner, K. Gärtner, R. Gaudin, Y. S. Gho, B. Giebel, C. Gilbert, M. Gimona, I. Giusti, D. C. Goberdhan, A. Görgens, S. M. Gorski, D. W. Greening, J. C. Gross, A. Gualerzi, G. N. Gupta, D. Gustafson, A. Handberg, R. A. Haraszti, P. Harrison, H. Hegyesi, A. Hendrix, A. F. Hill, F. H. Hochberg, K. F. Hoffmann, B. Holder, H. Holthofer, B. Hosseinkhani, G. Hu, Y. Huang, V. Huber, S. Hunt, A. G. Ibrahim, T. Ikezu, J. M. Inal, M. Isin, A. Ivanova, H. K. Jackson, S. Jacobsen, S. M. Jay, M. Jayachandran, G. Jenster, L. Jiang, S. M. Johnson, J. C. Jones, A. Jong, T. Jovanovic-Talisman, S. Jung, R. Kalluri, S. I. Kano, S. Kaur, Y. Kawamura, E. T. Keller, D. Khamari, E. Khomyakova, A. Khvorova, P. Kierulf, K. P. Kim, T. Kislinger, M. Klingeborn, D. J. Klinke 2nd, M. Kornek, M. M. Kosanović, F. Kovács Á, E. M. Krämer-Albers, S. Krasemann, M. Krause, I. V. Kurochkin, G. D. Kusuma, S. Kuypers, S. Laitinen, S. M. Langevin, L. R. Languino, J. Lannigan, C. Lässer, L. C. Laurent, G. Lavieu, E. Lázaro-Ibáñez, S. Le Lay, M. S. Lee, Y. X. F. Lee, D. S. Lemos, M. Lenassi, A. Leszczynska, I. T. Li, K. Liao, S. F. Libregts, E. Ligeti, R. Lim, S. K. Lim, A. Linē, K. Linnemannstöns, A. Llorente, C. A. Lombard, M. J. Lorenowicz, M. Lörincz Á, J. Lötvall, J. Lovett, M. C. Lowry, X. Loyer, Q. Lu, B. Lukomska, T. R. Lunavat, S. L. Maas, H. Malhi, A. Marcilla, J. Mariani, J. Mariscal, E. S. Martens-Uzunova, L. Martin-Jaular, M. C. Martinez, V. R. Martins, M. Mathieu, S. Mathivanan, M. Maugeri, L. K. McGinnis, M. J. McVey, D. G. Meckes Jr., K. L. Meehan, I. Mertens, V. R. Minciacchi, A. Möller, M. Møller Jørgensen, A. Morales-Kastresana, J. Morhayim, F. Mullier, M. Muraca, L. Musante, V. Mussack, D. C. Muth, K. H. Myburgh, T. Najrana, M. Nawaz, I. Nazarenko, P. Nejsum, C. Neri, T. Neri, R. Nieuwland, L. Nimrichter, J. P. Nolan, E. N. Nolte-'t Hoen, N. Noren Hooten, L. O'Driscoll, T. O'Grady, A. O'Loghlen, T. Ochiya, M. Olivier, A. Ortiz, L. A. Ortiz, X. Osteikoetxea, O. Østergaard, M. Ostrowski, J. Park, D. M. Pegtel, H. Peinado, F. Perut, M. W. Pfaffl, D. G. Phinney, B. C. Pieters, R. C. Pink, D. S. Pisetsky, E. Pogge von Strandmann, I. Polakovicova, I. K. Poon, B. H. Powell, I. Prada, L. Pulliam, P. Quesenberry, A. Radeghieri, R. L. Raffai, S. Raimondo, J. Rak, M. I. Ramirez, G. Raposo, M. S. Rayyan, N. Regev-Rudzki, F. L. Ricklefs, P. D. Robbins, D. D. Roberts, S. C. Rodrigues, E. Rohde, S. Rome, K. M. Rouschop, A. Rughetti, A. E. Russell, P. Saá, S. Sahoo, E. Salas-Huenuleo, C. Sánchez, J. A. Saugstad, M. J. Saul, R. M. Schiffelers, R. Schneider, T. H. Schøyen, A. Scott, E. Shahaj, S. Sharma, O. Shatnyeva, F. Shekari, G. V. Shelke, A. K. Shetty, K. Shiba, P. R. Siljander, A. M. Silva, A. Skowronek, O. L. Snyder 2nd, R. P. Soares, B. W. Sódar, C. Soekmadji, J. Sotillo, P. D. Stahl, W. Stoorvogel, S. L. Stott, E. F. Strasser, S. Swift, H. Tahara, M. Tewari, K. Timms, S. Tiwari, R. Tixeira, M. Tkach, W. S. Toh, R. Tomasini, A. C. Torrecilhas, J. P. Tosar, V. Toxavidis, L. Urbanelli, P. Vader, B. W. van Balkom, S. G. van der Grein, J. Van Deun, M. J. van Herwijnen, K. Van Keuren-Jensen, G. van Niel, M. E. van Royen, A. J. van Wijnen, M. H. Vasconcelos, I. J. Vechetti Jr., T. D. Veit, L. J. Vella, É. Velot, F. J. Verweij, B. Vestad, J. L. Viñas, T. Visnovitz, K. V. Vukman, J. Wahlgren, D. C. Watson, M. H. Wauben, A. Weaver, J. P. Webber, V. Weber, A. M. Wehman, D. J. Weiss, J. A. Welsh, S. Wendt, A. M. Wheelock, Z. Wiener, L. Witte, J. Wolfram, A. Xagorari, P. Xander, J. Xu, X. Yan, M. Yáñez-Mó, H. Yin, Y. Yuana, V. Zappulli, J. Zarubova, V. Žėkas, J. Y. Zhang, Z. Zhao, L. Zheng, A. R. Zheutlin, A. M. Zickler, P. Zimmermann, A. M. Zivkovic, D. Zocco and E. K. Zuba-Surma, J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef PubMed
.
- F. Teng and M. Fussenegger, Adv. Sci., 2021, 8, 2003505 CrossRef CAS PubMed
.
- M. Z. Ratajczak and J. Ratajczak, Leukemia, 2020, 34, 3126–3135 CrossRef PubMed
.
- M. Kanada, M. H. Bachmann, J. W. Hardy, D. O. Frimannson, L. Bronsart, A. Wang, M. D. Sylvester, T. L. Schmidt, R. L. Kaspar, M. J. Butte, A. C. Matin and C. H. Contag, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E1433–E1442 CrossRef CAS PubMed
.
- S. El Andaloussi, S. Lakhal, I. Mäger and M. J. Wood, Adv. Drug Delivery Rev., 2013, 65, 391–397 CrossRef CAS PubMed
.
- J. Ren, W. He, L. Zheng and H. Duan, Biomater. Sci., 2016, 4, 910–921 RSC
.
- K. O'Brien, S. Ughetto, S. Mahjoum, A. V. Nair and X. O. Breakefield, Cell Rep., 2022, 39, 110651 CrossRef PubMed
.
- D. Ha, N. Yang and V. Nadithe, Acta Pharm. Sin. B, 2016, 6, 287–296 CrossRef PubMed
.
- K. M. McAndrews, F. Xiao, A. Chronopoulos, V. S. LeBleu, F. G. Kugeratski and R. Kalluri, Life Sci. Alliance, 2021, 4, e202000875 CrossRef CAS PubMed
.
- C. He, D. Jaffar Ali, H. Xu, S. Kumaravel, K. Si, Y. Li, B. Sun, J. Ma and Z. Xiao, Exp. Cell Res., 2020, 392, 112040 CrossRef CAS PubMed
.
- Q. Xu, Z. Zhang, L. Zhao, Y. Qin, H. Cai, Z. Geng, X. Zhu, W. Zhang, Y. Zhang, J. Tan, J. Wang and J. Zhou, J. Controlled Release, 2020, 326, 455–467 CrossRef CAS PubMed
.
- S. M. Kim, Y. Yang, S. J. Oh, Y. Hong, M. Seo and M. Jang, J. Controlled Release, 2017, 266, 8–16 CrossRef CAS PubMed
.
- M. Kanada, B. D. Kim, J. W. Hardy, J. A. Ronald, M. H. Bachmann, M. P. Bernard, G. I. Perez, A. A. Zarea, T. J. Ge, A. Withrow, S. A. Ibrahim, V. Toomajian, S. S. Gambhir, R. Paulmurugan and C. H. Contag, Mol. Cancer Ther., 2019, 18, 2331–2342 CrossRef CAS PubMed
.
- J. J. Kelly, M. Saee-Marand, N. N. Nyström, M. M. Evans, Y. Chen, F. M. Martinez, A. M. Hamilton and J. A. Ronald, Sci. Adv., 2021, 7, eabc3791 CrossRef CAS PubMed
.
- Y. Lin, J. Wu, W. Gu, Y. Huang, Z. Tong, L. Huang and J. Tan, Adv. Sci., 2018, 5, 1700611–1700611 CrossRef PubMed
.
- J.-K. Kim, Y.-J. Youn, Y.-B. Lee, S.-H. Kim, D.-K. Song, M. Shin, H. K. Jin, J.-S. Bae, S. Shrestha and C.-W. Hong, Sci. Rep., 2021, 11, 8289–8289 CrossRef CAS PubMed
.
- A. Usardi, K. Iyer, S. M. Sigoillot, A. Dusonchet and F. Selimi, Dev. Neurobiol., 2017, 77, 75–92 CrossRef CAS PubMed
.
- K. Dooley, R. E. McConnell, K. Xu, N. D. Lewis, S. Haupt, M. R. Youniss, S. Martin, C. L. Sia, C. McCoy, R. J. Moniz, O. Burenkova, J. Sanchez-Salazar, S. C. Jang, B. Choi, R. A. Harrison, D. Houde, D. Burzyn, C. Leng, K. Kirwin, N. L. Ross, J. D. Finn, L. Gaidukov, K. D. Economides, S. Estes, J. E. Thornton, J. D. Kulman, S. Sathyanarayanan and D. E. Williams, Mol. Ther., 2021, 29, 1729–1743 CrossRef CAS PubMed
.
- D. Gupta, O. P. B. Wiklander, A. Görgens, M. Conceição, G. Corso, X. Liang, Y. Seow, S. Balusu, U. Feldin, B. Bostancioglu, R. Jawad, D. R. Mamand, Y. X. F. Lee, J. Hean, I. Mäger, T. C. Roberts, M. Gustafsson, D. K. Mohammad, H. Sork, A. Backlund, P. Lundin, A. de Fougerolles, C. I. E. Smith, M. J. A. Wood, R. E. Vandenbroucke, J. Z. Nordin and S. El-Andaloussi, Nat. Biomed. Eng., 2021, 5, 1084–1098 CrossRef CAS PubMed
.
- J. F. Nabhan, R. Hu, R. S. Oh, S. N. Cohen and Q. Lu, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4146–4151 CrossRef CAS PubMed
.
- Q. Wang, J. Yu, T. Kadungure, J. Beyene, H. Zhang and Q. Lu, Nat. Commun., 2018, 9, 960 CrossRef PubMed
.
- Y. Ye, X. Zhang, F. Xie, B. Xu, P. Xie, T. Yang, Q. Shi, C. Y. Zhang, Y. Zhang, J. Chen, X. Jiang and J. Li, Biomater. Sci., 2020, 8, 2966–2976 RSC
.
- Z. Li, X. Zhou, M. Wei, X. Gao, L. Zhao, R. Shi, W. Sun, Y. Duan, G. Yang and L. Yuan, Nano Lett., 2019, 19, 19–28 CrossRef CAS PubMed
.
- N. Yim, S.-W. Ryu, K. Choi, K. R. Lee, S. Lee, H. Choi, J. Kim, M. R. Shaker, W. Sun, J.-H. Park, D. Kim, W. D. Heo and C. Choi, Nat. Commun., 2016, 7, 12277 CrossRef CAS PubMed
.
- C. Meyer, J. Losacco, Z. Stickney, L. Li, G. Marriott and B. Lu, Int. J. Nanomed., 2017, 12, 3153–3170 CrossRef CAS PubMed
.
- C. Montagna, G. Petris, A. Casini, G. Maule, G. M. Franceschini, I. Zanella, L. Conti, F. Arnoldi, O. R. Burrone, L. Zentilin, S. Zacchigna, M. Giacca and A. Cereseto, Mol. Ther. – Nucleic Acids, 2018, 12, 453–462 CrossRef CAS PubMed
.
- L. A. Campbell, L. M. Coke, C. T. Richie, L. V. Fortuno, A. Y. Park and B. K. Harvey, Mol. Ther., 2019, 27, 151–163 CrossRef CAS PubMed
.
- S. Cabantous, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2005, 23, 102–107 CrossRef CAS PubMed
.
- X. Zhang, Q. Xu, Z. Zi, Z. Liu, C. Wan, L. Crisman, J. Shen and X. Liu, Dev. Cell, 2020, 55, 784–801 CrossRef CAS PubMed
.
- P. Gee, M. S. Y. Lung, Y. Okuzaki, N. Sasakawa, T. Iguchi, Y. Makita, H. Hozumi, Y. Miura, L. F. Yang, M. Iwasaki, X. H. Wang, M. A. Waller, N. Shirai, Y. O. Abe, Y. Fujita, K. Watanabe, A. Kagita, K. A. Iwabuchi, M. Yasuda, H. Xu, T. Noda, J. Komano, H. Sakurai, N. Inukai and A. Hotta, Nat. Commun., 2020, 11, 1334 CrossRef CAS PubMed
.
- M. Lu, H. Xing, Z. Xun, T. Yang, X. Zhao, C. Cai, D. Wang and P. Ding, Eur. J. Pharm. Sci., 2018, 121, 34–46 CrossRef CAS PubMed
.
- X. Yao, P. Lyu, K. Yoo, M. K. Yadav, R. Singh, A. Atala and B. Lu, J. Extracell. Vesicles, 2021, 10, e12076 CAS
.
- J. Li, X. Chen, J. Yi, Y. Liu, D. Li, J. Wang, D. Hou, X. Jiang, J. Zhang, J. Wang, K. Zen, F. Yang, C.-Y. Zhang and Y. Zhang, PLoS One, 2016, e0163043 CrossRef PubMed
.
- X. Xu, L. Xu, P. Zhang, K. Ouyang, Y. Xiao, J. Xiong, D. Wang, Y. Liang and L. Duan, Oxid. Med. Cell. Longevity, 2020, 8865499 CAS
.
- A. Zebrowska, A. Skowronek, A. Wojakowska, P. Widlak and M. Pietrowska, Int. J. Mol. Sci., 2019, 20, 3461 CrossRef CAS PubMed
.
- K. M. Heffner, Q. Wang, D. B. Hizal, Ö Can and M. J. Betenbaugh, Adv. Biochem. Eng. Biotechnol., 2021, 175, 37–69 CrossRef PubMed
.
- D. Gupta, A. M. Zickler and S. E. Andaloussi, Dosing extracellular vesicles, Adv. Drug Delivery Rev., 2021, 178, 113961 CrossRef CAS PubMed
.
- X. Xu, Y. Liang, X. Li, K. Ouyang, M. Wang, T. Cao, W. Li, J. Liu, J. Xiong, B. Li, J. Xia, D. Wang and L. Duan, Biomaterials, 2021, 269, 120539 CrossRef CAS PubMed
.
- Y. Liang, X. Xu, X. Li, J. Xiong, B. Li, L. Duan, D. Wang and J. Xia, ACS Appl. Mater. Interfaces, 2020, 12, 36938–36947 CrossRef CAS PubMed
.
- C. Stadelmann, S. D. Francescantonio, A. Marg, S. Müthel, S. Spuler and H. Escobar, Mol. Ther. – Nucleic Acids, 2022, 28, 47–57 CrossRef CAS PubMed
.
- M. Qiu, Z. Glass, J. Chen, M. Haas, X. Jin, X. Zhao, X. Rui, Z. Ye, Y. Li, F. Zhang and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2021, 118 CAS
, e2020401118.
- W. M. Usman, T. C. Pham, Y. Y. Kwok, L. T. Vu, V. Ma, B. Peng, Y. S. Chan, L. Wei, S. M. Chin, A. Azad, A. B.-L. He, A. Y. H. Leung, M. Yang, N. Shyh-Chang, W. C. Cho, J. Shi and M. T. N. Le, Nat. Commun., 2018, 9, 2359–2359 CrossRef PubMed
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |