Open Access Article
Open Access ArticlePeptide sequence mediated self-assembly of molybdenum blue nanowheel superstructures†
Shan
She
 ,
Weimin
Xuan
,
Weimin
Xuan
 ,
Nicola L.
Bell
,
Nicola L.
Bell
 ,
Robert
Pow
,
Robert
Pow
 ,
Eduard Garrido
Ribo
,
Eduard Garrido
Ribo
 ,
Zoe
Sinclair
,
Zoe
Sinclair
 ,
De-Liang
Long
,
De-Liang
Long
 * and
Leroy
Cronin
* and
Leroy
Cronin
 *
*
School of Chemistry, University of Glasgow, University Avenue, Glasgow, G12 8QQ, UK. E-mail: Deliang.Long@glasgow.ac.uk; Lee.Cronin@glasgow.ac.uk
First published on 14th December 2020
Abstract
The precise control over the formation of complex nanostructures, e.g. polyoxometalates (POMs), at the sub-nanoscale is challenging but critical if non-covalent architectures are to be designed. Combining biologically-evolved systems with inorganic nanostructures could lead to sequence-mediated assembly. Herein, we exploit oligopeptides as multidentate structure-directing ligands via metal-coordination and hydrogen bonded interactions to modulate the self-assembly of POM superstructures. Six oligopeptides (GH, AH, SH, G2H, G4H and G5H) are incorporated into the cavities of Molybdenum Blue (MB) POM nanowheels. It is found that the helicity of the nanowheel can be readily switched (Δ to Λ) by simply altering the N-terminal amino acid on the peptide chain rather than their overall stereochemistry. We also reveal a delicate balance between the Mo-coordination and the hydrogen bonds found within the internal cavity of the inorganic nanowheels which results in the sequence mediated formation of two unprecedented asymmetrical nanowheel frameworks: {Mo122Ce5} and {Mo126Ce4}.
Introduction
Modulating complex nanostructures in sub-nanoscale is of great importance in the fields of single-atom catalysis,1–3 metal–organic materials,4–6 molecular machines,7,8 and directed evolution.9 Precise structural modulation can result in either new or improved material properties and functions. However, strict control over nanostructure formation is still challenging. Owing to rational ligand design, template-directed self-assembly has become one of the common strategies in the construction of nanostructures with desirable properties and functions.10 In contrast to artificial systems, nature excels at elaborate control within the nanospace by exploiting the subtle interactions between side chain amino acid residues in proteins to adaptively generate certain structures capable a vast array of biologically important functions.11–13 Exploiting the evolution of peptides, we propose that a peptide sequence mediated approach to influence the self-assembly of inorganic clusters could be used as a strategy to fabricate tailor-made nanostructures with diverse frameworks and cavities. Through introduction of multivalent peptide ligands, and control over their many non-covalent interactions within the cavity formed in the framework, it could become feasible to construct hybrid superstructures with tuneable binding stoichiometry, framework shape and symmetry via self-organisation at the sub-nanoscale.As the proof of concept, polyoxometalates (POMs) were chosen to act as the representative nanoclusters for demonstration in this research. POMs, synthesized from simple metal oxide salts, are inexpensive yet elegant structures that can be readily formed through one-pot self-assembly.14–27 Among them, gigantic Molybdenum Blue (MB) wheel-shaped nanoclusters are of great importance in this area on account of their structural complexity and functional diversity. A certain number (n) of {Mo8} as main building blocks are connected together by the same number of {Mo1} and {Mo2} linkage units to form a nanowheel architecture,28,29 exhibiting very high symmetries (e.g. D7d for {Mo154}30) and large skeletons composed of many hundreds of metal and oxygen atoms.31
Introducing lanthanide ions (LnIII) to replace several or all {Mo2} units can “break” the D7d symmetry of {Mo154} through re-organisation, yielding Lanthanide-doped Molybdenum Blue (Ln-MB) clusters with lower symmetries, for example, D3 for {Mo120Ce6},32 and D2 for {Mo128Eu4}.33–36 Recently, our group have incorporated single amino acids (e.g. histidine) as directing ligands into the cavity of Ln-MBs to construct hybrid Ln-MB wheels with inherent chirality.37 Building on this progress, we envisioned that it is possible to exploit peptide sequences, acting as multivalent bio-ligands, to tailor the internal cavity, framework shape and symmetry for Ln-MB wheels via a self-assembly strategy at the sub-nanoscale.
Herein, a set of histidine-terminated oligopeptides (GH, AH, SH, G2H, G4H, G5H) were explored as structure-directing bio-ligands and incorporated into the cavity found in the Ln-MB's nanowheel to mimic confined enzyme-like ‘active-sites’ within a simple metal oxide framework, Scheme 1. The design principle is to minimize the role of metal-coordination bonds with the carboxyl terminus and increase the binding sites for multiple hydrogen bonds via addition of glycine at the N-terminus onto the oligopeptides. Through self-assembly with metal precursors, a new series of hybrid Ln-MB nanowheel frameworks were obtained; displaying diverse substitution stoichiometry with oligopeptides, distinct helical alignment of Ln atoms and unprecedented asymmetric shapes. In this way, we demonstrated the utilisation of peptides as multi-functional bio-ligands to direct the self-assembly of inorganic nanoclusters for the first time, yielding wheel-shape structures with peptide-lined cavities. This general strategy unveils a new avenue for the design and preparation of hybrid nanostructures with tailor-made internal nanocavities and framework architectures.
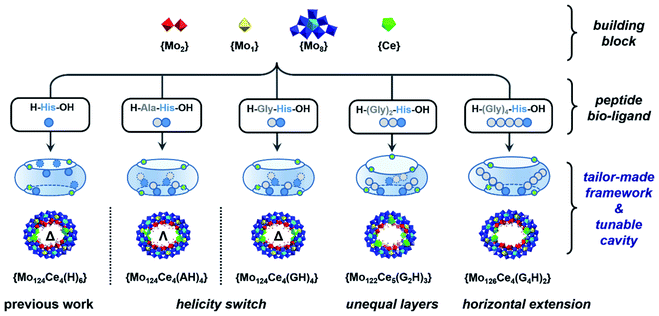 | ||
| Scheme 1 Schematic representation of peptide sequence mediated self-assembly processes for bio-hybrid MB nanowheels with tailor-made architectures. | ||
Results and discussion
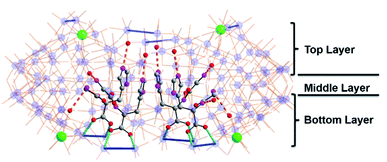
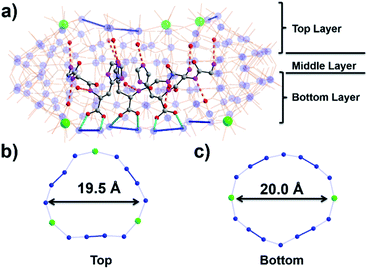
To assemble the nanowheel systems, we added the dipeptide H-Gly–L-His–OH (GH) (0.03 mmol) to a solution of sodium molybdate (1.0 mmol) and cerium(III) chloride (0.1 mmol) followed by reduction with hydrazine dihydrochloride (0.08 mmol) in an acidic environment (pH 1.1) at 90 °C for 2 hours. After crystallization at room temperature, blue crystals readily appeared. By employing previously reported methods,38,39 the structural formula was determined to be Na2(C8H14N4O3) [Δ-Mo124Ce4O376(H2O)64H12(C8H13N4O3)4]·155H2O, denoted as {Mo124Ce4(GH)4}. X-ray structural analysis reveals that {Mo124Ce4(GH)4} crystallizes in the orthorhombic system with chiral space group P21212. The crystal structural analysis (Fig. 1) plus analysis of the electron density40 (Fig. S17a†) showed that the {Mo124Ce4(GH)4} cluster-hybrid consists of an elliptical dodecamer nano-ring {Mo124Ce4}, in which four {Mo2} linkers have been replaced by CeIII and another four are coordinated by GH dipeptide ligands. Water plays an important role for metal binding to peptides.41,42 However, the MB nanowheel has an extremely large cavity and the position of the 155 water molecules inside the cavity cannot be fully resolved. Thus, it is not possible to discuss the hydration effect herein. As shown in Fig. 1, four GH dipeptide ligands are bound at the inner wall of the MB wheel, bridging the {Mo2} units and are hydrogen bonded to the terminal oxo-ligands of Mo![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O via the amide groups of the ligand. The dipeptide ligands coordinate along the elongated inner rim of the POM and arrange to maximise π-stacking interactions between the two adjacent imidazole rings, with a distance of ca. 3.6 Å, resulting in overall C2 symmetry of the whole molecule.
O via the amide groups of the ligand. The dipeptide ligands coordinate along the elongated inner rim of the POM and arrange to maximise π-stacking interactions between the two adjacent imidazole rings, with a distance of ca. 3.6 Å, resulting in overall C2 symmetry of the whole molecule.
In contrast to our previous work with single amino acids,37 the four GH dipeptides are mounted on {Mo2} units of the same layer of the MB wheel, further breaking the inversion symmetry of the MB ring, and inducing a dipole along the vertical axis of the ring. Previously coordination of amino acid ligands was shown to occur at the {Mo2} units on both layers of the MB wheel. In comparison, the current {GH} mediated framework {Mo124Ce4(GH)4}, suggests that the inherent helical chirality of the ring induces unfavourable interactions between the ammonium groups of the terminal glycine residues in the dipeptide systems resulting in a preference for homofacial binding which specifically facilitates interactions with the Ce–O moieties. Two crystallographically unique GH peptides exist in the asymmetric unit but both are anchored inside the {Mo124Ce4} wheel with a number of N–H⋯O hydrogen bonds in the range of 2.805–2.905 Å (Fig. 1 and S10†). As shown in Fig. S10,† for the first peptide the protonated N2 of the imidazole ring anchors to the {Mo2} unit on the top layer via H-bonding. And the N1 of amido group (–C(O)NH–) interacts with a Mo![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Ot of the {Mo1} unit. Finally, the protonated N4 terminal amine group links to a {Mo8} unit on the top layer. Previously we found six histidine ligands and one {Mo8} cluster can be accommodated in one {Mo124Ce4} wheel. Our work here indicates that four GH dipeptide ligands can play a similar role to direct and template {Mo124Ce4} formation, with the additional residues providing electrostatic interactions to stabilize the framework. Comparing with the use of single amino acid ligands, these results imply that oligopeptides can be used as both directing ligands and templates in the assembly of MB nanoclusters.
Ot of the {Mo1} unit. Finally, the protonated N4 terminal amine group links to a {Mo8} unit on the top layer. Previously we found six histidine ligands and one {Mo8} cluster can be accommodated in one {Mo124Ce4} wheel. Our work here indicates that four GH dipeptide ligands can play a similar role to direct and template {Mo124Ce4} formation, with the additional residues providing electrostatic interactions to stabilize the framework. Comparing with the use of single amino acid ligands, these results imply that oligopeptides can be used as both directing ligands and templates in the assembly of MB nanoclusters.
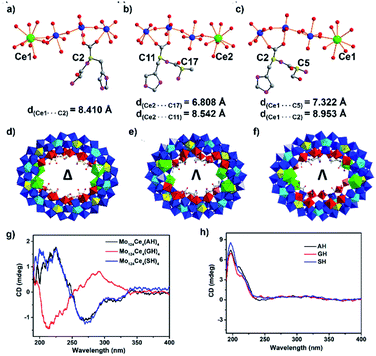
To elucidate the effects of the peptide N-terminus on MB formation, we utilised two additional dipeptide ligands H-Ala–L-His–OH (AH) and H-Ser–L-His–OH (SH) to produce {Λ-Mo124Ce4(AH)4} and {Λ-Mo124Ce4(SH)4}. The subtle changes in the side-chains in these three systems result in complete switching of the overall helicity (Δ to Λ)43 of the MB framework. Specifically, the absolute configuration of {Mo124Ce4(GH)4} is identified as Δ in the crystal structure determination, while {Mo124Ce4(AH)4} and {Mo124Ce4(SH)4} are both identified as Λ, shown in Fig. 2. The solution CD behaviours (Fig. 2g) of these three compounds, recorded in triplicate on the crystals and their corresponding mother liquors, are also in accordance with above results confirming these observations are a result of chiral induction rather than any racemisation processes.
This helical change demonstrates the role of the individual residues of the peptide ligands in directing the self-assembly process. As shown in Fig. 2a, since L-GH has one chiral centre, on the C-terminal residue, and the overall structure has Δ helicity, so the C-terminal residue can be determined to have a heterochiral directing effect. In contrast, the other peptides (L-AH and L-SH) have two chiral centres. For L-AH (Fig. 2b), the chiral carbon C* on N-terminal residue is closer to the Ce atom (d = 6.81 Å) compared with C* on C-terminal residue (d = 8.54 Å). Since replacement of a {Mo2} unit with Ce results in chiral induction, it can be surmised that the closer a residue is to this site, the more strongly it will influence helicity of the ring. Thus, the Λ chirality of the latter two rings suggests the strong homochiral directing effect of the N-terminal residue. This is also consistent with our previous observation that coordination of six L-His ligands within {Mo124Ce4} wheels yielded only {Δ-Mo124Ce4(Mo8)(His)6} while D-His yielded the corresponding Λ-isomer.37,44
To further probe the effects of the peptide residues, we investigated the effects of longer oligopeptide chains on the wheel and cavity structure. For this purpose, we utilized the tripeptide ligand H–Gly–Gly–His–OH (G2H) in the fabrication of MB wheel. Surprisingly, a new structure {Mo122Ce5(G2H)3} with formula Na2(C10H17N5O4)[Mo122Ce5O371(H2O)69H12(C10H16N5O4)3]·160H2O was obtained. This represents the unique example where five lanthanide ions have been incorporated into a MB wheel and this arises because an additional {Mo2} unit has been replaced by a CeIII ion cf. {Mo124Ce4(GH)4}. The refined crystal structure shows incorporation of only three G2H tripeptide ligands which, again, bind on the same layer of the MB wheel (Fig. 3). Two Ce atoms are located on the bottom layer and three other Ce atoms on the top layer of the wheel. On the uncoordinated top layer, the extra lanthanide ion causes the ring to contract, changing shape from an ellipsoid to a more cyclical form, with roughly 3-fold symmetry (Fig. 3b and c). Despite the inclusion of further lanthanides, the MB wheel metal oxide framework itself with Cs symmetry is asymmetrical but crystalizes in a chiral space group (P21212), solely due to the presence of enantiopure peptides.
 | ||
| Fig. 3 (a) Molecular structure of {Mo122Ce5(G2H)3}; cerium-substituted rings on (b) the top layer (uncoordinated) and (c) the bottom layer (coordinated). Colour code is identical to that in Fig. 1. | ||
Two of the three tripeptide ligands are in a pair on two adjacent {Mo2} units with their imidazole rings separated at distance ca. 3.6 Å and each H-bonding to a top-layer cerium ion, while the third is bound opposite at the ellipse co-vertex, below the fifth cerium ion (Fig. 3a). Like {Mo124Ce4(GH)4}, multiple coordination bonds and hydrogen bonds (Fig. S12†) are observed between the protonated peptide ligands and Mo![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O moieties with the peptide effectively ‘wrapping’ around the inner rim of the wheel. As such, these interactions block incorporation of a fourth peptide ligand. Thus, the asymmetrical distribution of tripeptides led to differences in {Mo2}/{Ln} incorporation between bottom layer (ellipse) and top layer (cyclic), which cause the whole hybrid to become asymmetric.
O moieties with the peptide effectively ‘wrapping’ around the inner rim of the wheel. As such, these interactions block incorporation of a fourth peptide ligand. Thus, the asymmetrical distribution of tripeptides led to differences in {Mo2}/{Ln} incorporation between bottom layer (ellipse) and top layer (cyclic), which cause the whole hybrid to become asymmetric.
The successful encapsulation of the tripeptide species as a template to form a unique MB wheel encouraged us to further explore the potential of using even longer chain peptides (pentapeptide) as a template. Based on the tripeptide results, we anticipated that increasing the peptide chain may expand the radial size of the obtained MB wheel, thus enriching the family of metal-oxo nanoclusters. By employing pentapeptide H–Gly–Gly–Gly–Gly–His–OH (G4H) we obtained another {MoxCey} type Ce-doped MB wheel {Mo126Ce4(G4H)3}, with formula Na4[Mo126Ce4O384H17(H2O)71(C14H22N7O6)3]·150H2O. In this structure the wheel size is close to that of the previously described {Mo124Ce4} compounds, with n = 12 and four {Mo2} units replaced by four CeIII ions. However, only two pentapeptide ligands are attached to two adjacent {Mo2} units by carboxylate groups inside the clusters in a parallel mode, again with the imidazole rings separated at a distance ca. 3.6 Å. The peptide N-termini extend along the wheel's inner surface in two directions. Multiple H-bonding contacts between amide NH groups and Mo![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O moieties on the cluster are observed (Fig. 4 and S13†). Due to the limited cavity size in the wheel, only two G4H ligands are accommodated inside the cluster. A third G4H ligand is found outside the wheel cavity, which is notably seen in the electron density map of solvent area (Fig. S17c†). Through carboxylate mono-coordination this G4H ligand is anchored on a {Mo2} unit, locating on the upper layer of the wheel, close to the N-terminus of one peptide inside.
O moieties on the cluster are observed (Fig. 4 and S13†). Due to the limited cavity size in the wheel, only two G4H ligands are accommodated inside the cluster. A third G4H ligand is found outside the wheel cavity, which is notably seen in the electron density map of solvent area (Fig. S17c†). Through carboxylate mono-coordination this G4H ligand is anchored on a {Mo2} unit, locating on the upper layer of the wheel, close to the N-terminus of one peptide inside.
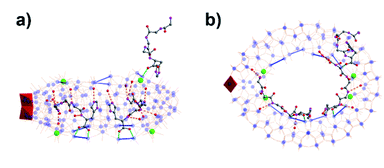 | ||
| Fig. 4 (a) Side view and (b) top view of molecular structure of {Mo126Ce4 (G4H)3}, red polyhedron = {Mo2O8}. Color code is identical to that in Fig. 1. | ||
The cluster is roughly ellipsoidal with two pairs of CeIII ions located on each end as in the {Mo124Ce4(GH)4} cluster. However, the two ends of the ellipsoid here are not identical with one end slightly more pointed giving the cluster a tear-drop shape. The tapered end shows that the two CeIII ions seem to hang on one side rather than bridging between Mo atoms; and the structure appears not closed by the expected {Mo8} building unit but by an additional {Mo2} moiety which is the case of the {Mo128Eu4} structure.34 The MB {Mo126Ce4} framework itself has a 2-fold symmetry axis passing through the two ellipsoidal ends. However, the peptide mounting points are found to be “irregular” yet unverified and thus ensures the cluster is highly asymmetrical and chiral.
Furthermore, a similar architecture was obtained by using hexapeptide (G5H) as template. Interestingly, the corresponding MB framework is almost the same as that of the pentapeptide, with formula Na3[Mo126Ce4O384H18(H2O)71(C16H25N8O7)3]·150H2O. In this structure, there are also two peptide ligands attached to two adjacent {Mo2} units inside the wheel, and another hexapeptide ligand is found outside the cavity via a mono-coordination to {Mo2} units (Fig. S14†). Here, multiple H-bonding contacts between amide NH groups and Mo![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O moieties on the cluster are observed. Since the distance between two terminal N-atoms on the two inside G5H ligands is too crowded (8.2 Å) to encapsulate another two -Gly- units (7.8 Å) inside, two hexapeptides may be the maximum feasible as peptide templates within Ln-doped MB wheels of this type. We also tried to incorporate the tetrapeptide (G3H) as a template in the synthesis of Ln-MB wheels, however we only obtained very tiny crystals not suitable for structural determination using single crystal X-ray diffraction. Despite this we were able to measure CD spectra, see Fig. S3.†
O moieties on the cluster are observed. Since the distance between two terminal N-atoms on the two inside G5H ligands is too crowded (8.2 Å) to encapsulate another two -Gly- units (7.8 Å) inside, two hexapeptides may be the maximum feasible as peptide templates within Ln-doped MB wheels of this type. We also tried to incorporate the tetrapeptide (G3H) as a template in the synthesis of Ln-MB wheels, however we only obtained very tiny crystals not suitable for structural determination using single crystal X-ray diffraction. Despite this we were able to measure CD spectra, see Fig. S3.†
To summarise our structural control over hybrid Ln-MB wheel superstructures, a comprehensive comparison between the crystal structures of {Mo124Ce4(H)6(Mo8)}, {Mo124Ce4(GH)4}, {Mo122Ce5(G2H)3}, {Mo126Ce4(G4H)3} and {Mo126Ce4(G5H)3}, was further analysed to reveal how the peptide ligands affect the assembly of the MB frameworks, see Table 1. The gradual addition of glycine residues on the N-terminus of the histidine-based oligopeptides decreased the binding stoichiometry of bio-ligands within the nanowheel from 6 to 2, on accounts of the steric hindrance and a limited cavity volume. This trend was accompanied with a decreasing number of coordination bonds and the increasing proportion of hydrogen bonds in multivalent interactions between the metal-oxide framework and peptides.
| Peptide ligand | H | GH | G2H | G4He | G5He |
|---|---|---|---|---|---|
| a The peptide inside MB cavity. b The coordination bonds are at the length of 2.0–2.3 Å. c The hydrogen bonds are at the length of 2.7–3.0 Å. d The MB framework symmetry. e Both Mo126Ce4(G4H)3 and Mo126Ce4(G5H)3 are racemic in the crystal structures, for demonstration purposes, we draw the delta one here. | |||||
| Stoichiometrya | 6 | 4 | 3 | 2 | 2 |
| Coordination bondsb | 12 | 8 | 6 | 4 | 4 |
| Hydrogen bondsc | 0 | 12 | 12 | 12 | 14 |
| Ce substitution |
|
|

|
|
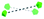
|
| Helicity | Δ | Δ | — | Δ/λ | Δ/λ |
| Symmetryd | 3× C2 | 3× C2 | C s | 1× C2 | 1× C2 |
| Shape |

|

|

|

|

|
It should be pointed out that, for histidine-coordinated MB structures in our previous work,37 all the histidines have no hydrogen bonds with the MB framework, but only with the core {Mo8} template (Table 1 and S16†), which indicates the significant importance of guest template effect. However, for peptide-based MB structures in the current research, no guest template is required because the increasing glycine residues unit (–Gly–C(O)NH–) on the oligopeptides can stabilise the nanowheel cavity by themselves through additional hydrogen bonds formed between the amide groups and the terminal oxo ligands (Mo![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Ot) on the interior surface of the MB wheels. The total number of hydrogen bonds are between 12 and 14 in all these hybrid Ln-MB structures, which may be attributed to the maximum hydrogen bonding sites formed between MB's interior terminal oxygen atoms and amide groups of peptides.
Ot) on the interior surface of the MB wheels. The total number of hydrogen bonds are between 12 and 14 in all these hybrid Ln-MB structures, which may be attributed to the maximum hydrogen bonding sites formed between MB's interior terminal oxygen atoms and amide groups of peptides.
To date, much of the previous work has focused on controlling the arrangements of four CeIII ions, within the cluster framework {Mo124Ce4}, see Fig. S16a–c.†45,46 Importantly, the encapsulation of three tripeptides into the Ln-MB wheel superstructure allows the incorporation of five lanthanide ions on the nanowheel and eliminates the helicity of the overall framework, see Fig. S16d,† which is different to other reported systems. Therefore, it can be envisaged that the {Mo122Ce5} could have more geometrically valid combinations with respect to the allowed Ce substitution sites, thus leading to a very wide variety and structural diversity of these Ln-MB superstructures.
The oligopeptides modulate the self-assembly of the various novel frameworks, not observed when using simple amino acids. For dipeptides, four ligands are mounted on {Mo2} units of the same layer of the MB wheel, further breaking the inversion symmetry of the MB ring. Because of the strong hydrogen bonding toward MB interior wall, dipeptide ligands can be used as both directing ligands and templates in the assembly of MB nanoclusters. For G2H, the extra lanthanide ion causes the ring to contract, changing shape from an ellipsoid to a half-cylinder-like form, thus bottom layer (ellipse) and top layer (cyclic) become unequal, which results in Cs symmetry for the overall framework. For G4H and G5H, the longer peptide chain forces the ellipsoidal {Mo124Ce4} to expand with an additional {Mo2} moiety, thus leading to the shape transition of the whole wheel from symmetrical pumpkin-like wheel to asymmetrical tear-like ellipsoid. Therefore, by using oligopeptides as multidentate structure-directing ligands, we can control the self-assembly of MB superstructures by balancing the critical role of coordination and hydrogen bonds.
Conclusions
In conclusion, we have successfully exploited a series of peptide bio-ligands, from dipeptide (GH, AH, SH), tripeptide (G2H), pentapeptide (G4H) to hexapeptide (G5H), as multivalent structure-directing templates to generate self-assembled bio-hybrid nanowheels with three different frameworks, {Mo124Ce4}, {Mo122Ce5} and {Mo126Ce4}. In contrast to the single amino acid substitutions, oligopeptides are significantly important in the formation of these new wheel-shaped architectures. We found that the helicity of the {Mo124Ce4} can be readily switched from Δ to Λ by simply altering the N-terminal amino acid of the peptide rather than their overall stereochemistry. This is possibly because the C-terminal residue may have a weaker heterochirality directing effect on the MB wheel, while the N-terminal residue may have a stronger homochiral directing effect, on account of its relatively close distance to the {Mo2}/Ln substitution site.More importantly, the elaborate balance between the coordination bonds and hydrogen bonds with the internal cavity further resulted in the formation of two unprecedented asymmetrical nanowheel frameworks, {Mo122Ce5} and {Mo126Ce4}. This is the first report of a {Mo122Ce5} with an odd number of Ln substitution that could bring new possibilities for structural diversification on asymmetric superstructures. Besides, the horizontal extension of a ‘tear-like’ framework, the {Mo126Ce4} cluster, may also provide a new type of host nanocavity for potential encapsulation (or recognition) of many different neutral or cationic nanospecies.
In summary, this research unveils a simple and general approach to exploit peptide lined nanocavities in the design of bio-hybrid superstructures with great potential in peptide recognition, the design of so-called ‘inorganic proteins’ or artificial membrane channels, for example.
Author contributions
The idea for the work was conceived by LC. The synthesis and chemical analysis were performed by WX, SS, RP, EGR and ZLS. The main hypothesis development and results analysis were undertaken by SS, NLB, DL and LC. Structural analysis was done by DL, NLB and SS. NLB and DL lead the Self-Assembly team with help from LC. The manuscript was written by LC, SS, NLB and DL with input from all the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the EPSRC grants (No. EP/J015156/1; EP/L023652/1; EP/I033459/1; EP/J015156/1; EP/K023004/1; EP/L023652/1), the ERC for an Advanced Grant (ERCADG, 670467 SMART-POM). S. S. acknowledges support from the European Commission for a Marie Curie Fellowship (PPOM-PTT, 798821). We thank the Diamond Light Source for time on Beamline I19 under the proposal CY22214.Notes and references
- B. Qiao, A. Wang, X. Yang, L. F. Allard, Z. Jiang, Y. Cui, J. Liu, J. Li and T. Zhang, Nat. Chem., 2011, 3, 634–641 CrossRef CAS.
- J. Jiao, R. Lin, S. Liu, W.-C. Cheong, C. Zhang, Z. Chen, Y. Pan, J. Tang, K. Wu, S.-F. Hung, H. M. Chen, L. Zheng, Q. Lu, X. Yang, B. Xu, H. Xiao, J. Li, D. Wang, Q. Peng, C. Chen and Y. Li, Nat. Chem., 2019, 11, 222–228 CrossRef CAS.
- C. Xie, Z. Niu, D. Kim, M. Li and P. Yang, Chem. Rev., 2020, 120, 1184–1249 CrossRef CAS.
- Y. Sun, C. Chen, J. Liu and P. J. Stang, Chem. Soc. Rev., 2020, 49, 30 Search PubMed.
- M. O'Keeffe and O. M. Yaghi, Chem. Rev., 2012, 112, 675–702 CrossRef.
- J.-R. Li, J. Sculley and H.-C. Zhou, Chem. Rev., 2012, 112, 869–932 CrossRef CAS.
- D. Sluysmans and J. F. Stoddart, Proc. Natl. Acad. Sci., 2018, 115, 9359–9361 CrossRef CAS.
- S. Erbas-Cakmak, S. D. P. Fielden, U. Karaca, D. A. Leigh, C. T. McTernan, D. J. Tetlow and M. R. Wilson, Science, 2017, 358, 340–343 CrossRef CAS.
- K. Chen and F. H. Arnold, Nat. Catal., 2020, 3, 203–213 CrossRef CAS.
- E. Cadot, M. N. Sokolov, V. P. Fedin, C. Simonnet-Jegat, S. Floquet and F. Secheresse, Chem. Soc. Rev., 2012, 41, 7335–7353 RSC.
- T. Shida, Y. O. Kamatari, T. Yoda, Y. Yamaguchi, M. Feig, Y. Ohhashi, Y. Sugita, K. Kuwata and M. Tanaka, Nat. Chem. Biol., 2020, 16, 9 CrossRef.
- M. D. Shoulders, K. A. Satyshur, K. T. Forest and R. T. Raines, Proc. Natl. Acad. Sci., 2010, 107, 559–564 CrossRef CAS.
- K. Mohan, G. Ueda, A. R. Kim, K. M. Jude, J. A. Fallas, Y. Guo, M. Hafer, Y. Miao, R. A. Saxton, J. Piehler, V. G. Sankaran, D. Baker and K. C. Garcia, Science, 2019, 364, eaav7532 CrossRef CAS.
- M. T. Pope and A. Müller, in Polyoxometalate Chemistry From Topology via Self-Assembly to Applications, ed. M. T. Pope and A. Müller, Springer Netherlands, Dordrecht, 2001, pp. 1–6, DOI:10.1007/0-306-47625-8_1.
- P. Yang and U. Kortz, Acc. Chem. Res., 2018, 51, 1599–1608 CrossRef CAS.
- L. Cronin and A. Muller, Chem. Soc. Rev., 2012, 41, 7333–7334 RSC.
- J. Li, Z. Chen, M. Zhou, J. Jing, W. Li, Y. Wang, L. Wu, L. Wang, Y. Wang and M. Lee, Angew. Chem., Int. Ed., 2016, 55, 2592–2595 CrossRef CAS.
- S. She, M. Li, Q. Li, Z. Huang, Y. Wei and P. Yin, ChemPlusChem, 2019, 84, 1668–1672 CrossRef CAS.
- P. Yin, Z.-M. Zhang, H. Lv, T. Li, F. Haso, L. Hu, B. Zhang, J. Bacsa, Y. Wei, Y. Gao, Y. Hou, Y.-G. Li, C. L. Hill, E.-B. Wang and T. Liu, Nat. Commun., 2015, 6, 6475 CrossRef CAS.
- H. Yu, S. Ru, G. Dai, Y. Zhai, H. Lin, S. Han and Y. Wei, Angew. Chem., Int. Ed., 2017, 56, 3867–3871 CrossRef CAS.
- M. Bonchio, Z. Syrgiannis, M. Burian, N. Marino, E. Pizzolato, K. Dirian, F. Rigodanza, G. A. Volpato, G. La Ganga, N. Demitri, S. Berardi, H. Amenitsch, D. M. Guldi, S. Caramori, C. A. Bignozzi, A. Sartorel and M. Prato, Nat. Chem., 2019, 11, 146–153 CrossRef CAS.
- J. Luo, K. Chen, P. Yin, T. Li, G. Wan, J. Zhang, S. Ye, X. Bi, Y. Pang, Y. Wei and T. Liu, Angew. Chem., Int. Ed., 2018, 57, 4067–4072 CrossRef CAS.
- J. Zhao, K. Li, K. Wan, T. Sun, N. Zheng, F. Zhu, J. Ma, J. Jiao, T. Li, J. Ni, X. Shi, H. Wang, Q. Peng, J. Ai, W. Xu and S. Liu, Angew. Chem., Int. Ed., 2019, 58, 18032–18039 CrossRef CAS.
- X. Chen, Y. Zhou, V. A. L. Roy and S.-T. Han, Adv. Mater., 2018, 30, 1703950 CrossRef.
- I. Colliard, G. Morrison, H.-C. z. Loye and M. Nyman, J. Am. Chem. Soc., 2020, 142, 9039–9047 CrossRef CAS.
- N. Gao, Z. Du, Y. Guan, K. Dong, J. Ren and X. Qu, J. Am. Chem. Soc., 2019, 141, 6915–6921 CrossRef CAS.
- S. She, Z. Huang, P. Yin, A. Bayaguud, H. Jia, Y. Huang, Y. Wei and Y. Wei, Chem.–Eur. J., 2017, 23, 14860–14865 CrossRef CAS.
- A. Müller and C. Serain, Acc. Chem. Res., 2000, 33, 2–10 CrossRef.
- A. Muller and P. Gouzerh, Chem. Soc. Rev., 2012, 41, 7431–7463 RSC.
- A. Müller, E. Krickemeyer, J. Meyer, H. Bögge, F. Peters, W. Plass, E. Diemann, S. Dillinger, F. Nonnenbruch, M. Randerath and C. Menke, Angew. Chem., Int. Ed. Engl., 1995, 34, 2122–2124 CrossRef.
- A. Muller, S. Q. N. Shah, H. Bogge and M. Schmidtmann, Nature, 1999, 397, 48–50 CrossRef CAS.
- V. Duros, J. Grizou, W. Xuan, Z. Hosni, D.-L. Long, H. N. Miras and L. Cronin, Angew. Chem., Int. Ed., 2017, 56, 10815–10820 CrossRef CAS.
- A. Müller, C. Beugholt, H. Bögge and M. Schmidtmann, Inorg. Chem., 2000, 39, 3112–3113 CrossRef.
- L. Cronin, C. Beugholt, E. Krickemeyer, M. Schmidtmann, H. Bögge, P. Kögerler, T. K. K. Luong and A. Müller, Angew. Chem., Int. Ed., 2002, 41, 2805–2808 CrossRef CAS.
- W. Xuan, A. J. Surman, H. N. Miras, D.-L. Long and L. Cronin, J. Am. Chem. Soc., 2014, 136, 14114–14120 CrossRef CAS.
- T. Yamase, E. Ishikawa, Y. Abe and Y. Yano, J. Alloys Compd., 2006, 408–412, 693–700 CrossRef CAS.
- W. Xuan, R. Pow, N. Watfa, Q. Zheng, A. J. Surman, D.-L. Long and L. Cronin, J. Am. Chem. Soc., 2019, 141, 1242–1250 CrossRef CAS.
- A. Müller, E. Krickemeyer, H. Bögge, M. Schmidtmann, C. Beugholt, S. K. Das and F. Peters, Chem.–Eur. J., 1999, 5, 1496–1502 CrossRef.
- I. D. Brown, in Industrial Chemistry Library, ed. M. O'Keeffe and A. Navrotsky, Elsevier, 1981, vol. 2, pp. 1–30 Search PubMed.
- C. B. Hubschle, G. M. Sheldrick and B. Dittrich, J. Appl. Crystallogr., 2011, 44, 1281–1284 CrossRef.
- M. Li, Z. Liu, J. Ren and X. Qu, Chem. Sci., 2020, 11, 7479–7486 RSC.
- H. Yu, J. Ren and X. Qu, ChemBioChem, 2008, 9, 879–882 CrossRef CAS.
- K. A. Jensen, Inorg. Chem., 1970, 9, 1–5 CrossRef CAS.
- In our previous work (ref. 37) the helicity of our molecules was incorrectly depicted inversely to the labels in the figures. However the text and scientific of the conclusions are unaffected (i.e.D-AA yields Λ-POM and L-AA yields Δ-POM). Throughout this MS the depictions of the POM helicity correctly match the assignation.
- H. N. Miras, C. Mathis, W. Xuan, D.-L. Long, R. Pow and L. Cronin, Proc. Natl. Acad. Sci., 2020, 117, 10699–10705 CrossRef CAS.
- W. Xuan, R. Pow, Q. Zheng, N. Watfa, D.-L. Long and L. Cronin, Angew. Chem., Int. Ed., 2019, 58, 10867–10872 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2006398–2006402 and 2041759. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0sc06098d |
| This journal is © The Royal Society of Chemistry 2021 |