Open Access Article
Open Access ArticleNew antimicrobial self-assembling short lipopeptides†
César
Vicente-García
 a and
Ignacio
Colomer
a and
Ignacio
Colomer
 *ab
*ab
aIMDEA Nanociencia, Faraday 9, Campus UAM, 28049 Madrid, Spain
bInstituto de Química Orgánica General (IQOG-CSIC), Juan de la Cierva 3, 28006, Madrid, Spain. E-mail: colomer@iqog.csic.es
First published on 16th July 2021
Abstract
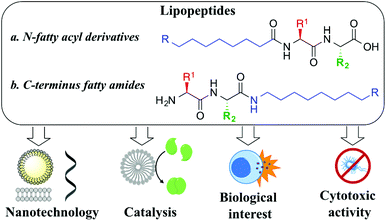
Lipopeptides are an exceptional example of amphiphilic molecules that self-assemble into functional structures with applications in the areas of nanotechnology, catalysis or medicinal chemistry. Herein, we report a library of 21 short lipopeptides, together with their supramolecular characterization and antimicrobial activity against both Gram-negative (E. coli) and Gram-positive (S. aureus) strains. This study shows that simple lipoamino acids self-assemble into micellar or vesicular structures, while incorporating dipeptides capable of stablishing hydrogen bonds results in the adoption of advanced fibrilar structures. The self-assembly effect has proven to be key to achieve antimicrobial activity.
Peptide nanotechnology has emerged as a powerful tool that allows for the rational design of functional structures whose supramolecular properties can be engineered and controlled by manipulating the peptide sequence.1 Amphiphilic peptides represent a heterogeneous category of these biomolecules that can be constituted of all amino acids, with alternating hydrophilic and lipophilic monomers, or that can incorporate appendant hydrophilic or hydrophobic chains. These amphiphiles self-assemble into miscellaneous supramolecular structures (i.e. micelles, vesicles, nanotubes, fibres, nanobelts) exhibiting versatile properties and applications.2 Lipopeptides are a particular example of self-assembling amphiphilic peptides, formed with a polar hydrophilic head and a long hydrophobic chain that can be attached either to the N-terminus position (i.e. fatty acyl moiety, Fig. 1a) or at the C-terminus site (i.e. fatty amine or alcohol, Fig. 1b). Their self-organization is highly determined by the sequence of amino acids, something that prompted rational design to achieve advanced performances, controlling their morphology and activity, a field that has rapidly grown in recent years.3
Lipopeptides have been widely used as surfactants and low-molecular weight gelators.4 Alternatively, extensive investigations have shed light on how to control the supramolecular properties by tuning the structural motifs of the amphiphilic monomers, using α- or β-amino acids,5 homochiral vs. heterochiral polymers6 or the nature and length of the hydrophobic component.7 Moreover, the influence of external stimuli on both the self-assembly and the properties displayed has been investigated, including concentration,8 solvent,9 saline10 or the pH effect.11 Their ability to respond to these stimuli and adapt over time has been used to design synthetic advanced dynamic supramolecular systems,12 nanostructured materials,13 and self-responsive delivery systems in the area of medical biotechnology14 or systems chemistry.15
Although great advances have been made in peptide engineering, identifying small (lipo)peptides with defined supramolecular behaviour,16 catalytic properties17 or biological activity18 is still a challenge. In this study, we aim to build a library of small lipopeptides to systematically explore the chemical space, covering different amino acids, complementary amino acid sequences, the number and length of the hydrophobic aliphatic chain, as well as the influence of its position (N- vs. C-terminus). Moreover, their biological activity will be investigated, in particular as antimicrobial agents.
We began our study by preparing lipoamino acids (Fig. 2a) derived from amino acids with variable side chains, bearing no substitution (glycine, 1a), and aliphatic (alanine, 1b and valine, 1c), aromatic (phenylalanine, 1d), hydrogen bond donor (serine, 1e and 1g), pH responsive (histidine, 1f and 1h) and redox active (cysteine, 1i–k) functional groups, following the Schotten–Baumann procedure, using free amino acids and decanoyl or oleoyl chlorides under basic conditions.19 Selective monofunctionalization of cysteine, 1i–j, was achieved using benzotriazole to activate decanoic acid, leading to amide 1i using a basic medium and a short reaction time (2 h), while using neutral pH and a longer time (12 h) afforded thioester 1j.20 Aiming to increase the structural complexity that would allow for additional non-covalent interactions, we prepared a set of lipodipeptides. We decided to focus on Ser and His for their potential H-bonding capabilities, acid–base properties and π–π stacking. Using lipoamino acids derived from Ser or His (1e–h), the complementary amino acid (Ser or His) was coupled at the C-terminus position, leading to four new lipodipeptides (Fig. 2a, 2a–d). The examples described, so far, display a fatty acyl moiety at the N-terminus position. Thus, we thought that evaluating complementary C-terminus derivatives will enrich the outcome of this library. A set of six new lipopeptides 3a–f (Fig. 2b) could be accessed using fatty amines. For the Ser-His sequence, semi-protected common precursor 4 (ESI†) was reacted with decylamine (3a), oleylamine (3b) or bis-oleate amine derived from serinol (3c). For His-Ser derivatives, an alternative route was needed: first incorporating the fatty amine into the C-terminus amino acid (Ser) providing serine amides 5a–c (Fig. 2b), followed by coupling of the N-terminus amino acid (His) to afford three new lipodipeptides, 3d–f.
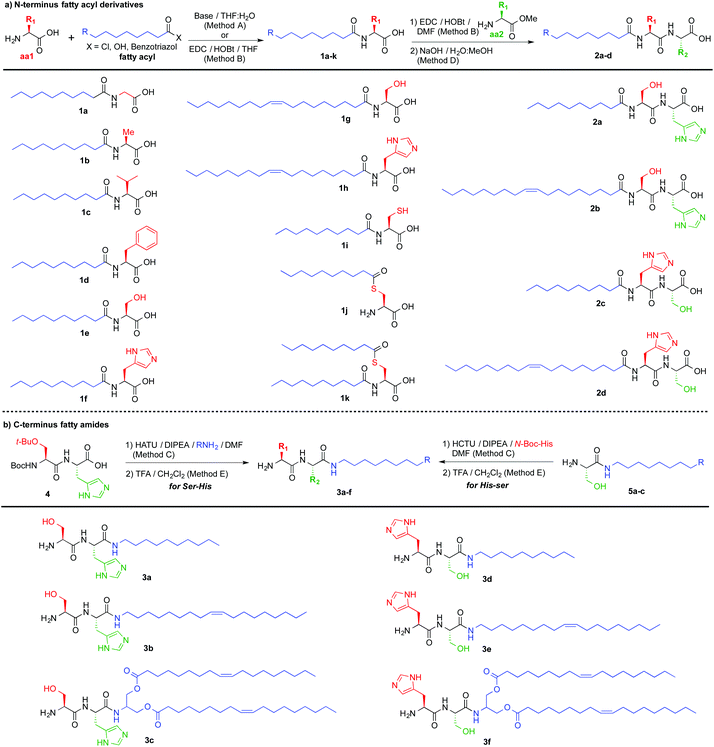 | ||
| Fig. 2 Library of lipopeptides synthesized, composed of: (a) N-terminus fatty acyl derivatives and (b) C-terminus fatty amides. | ||
With these lipopeptides in hand, we first studied their self-assembling behavior by performing systematic characterization of their supramolecular properties. For every species, we have determined the minimal concentration needed for them to aggregate, which is the Critical Aggregation Concentration (CAC). We have also investigated the particle size of the supramolecular assemblies, as well as their stability in solution. Finally, we have conducted the study of the morphology of these supramolecular aggregates using electron microscopy.
The supramolecular characterization was started by determining the CAC, using an established spectrofluorimetric method (see the ESI†). A representative example for 1f is presented in Fig. 3A. The CAC for lipoamino acids is in the range of 0.20–25.0 mM (Table 1), in agreement with the previous examples reported (see the ESI†). The lowest values of the CAC were observed for cysteine derivatives (1i–k). For lipodipeptides, the CAC data were more disperse (0.01–68.4 mM), with N-terminus acyl derivatives (2a–d) presenting much lower CAC than C-terminus fatty amides (3a–f). In general, lipoamino acids with non-polar side chains (1a–d) exhibit higher CAC values than examples bearing polar side chains capable of establishing additional non-covalent interactions. This reflects the well-known influence of the hydrogen bonding capacity and HLB to enhance the formation of supramolecular structures.21 Additionally, lipopeptides bearing longer aliphatic chains display lower CAC than their shorter counterparts, a trend extensively reported for similar lipopeptides.22
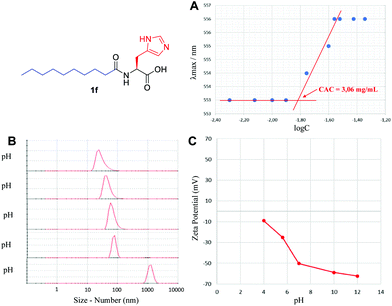 | ||
| Fig. 3 Supramolecular characterization of 1f, including (A) critical aggregation concentration, (B) particle size using DLS and (C) self-assembly stability via zeta-potential analysis. | ||
| Compound | CAC (mM) | Sizea (nm) | ZPa (mV) | |
|---|---|---|---|---|
| Number | Intensity | |||
| a Data collected at pH 11 for 1a–k and at pH 6 for 2a–d and 3a–f. | ||||
| 1a | 25.0 | 18 | 24, 142 | — |
| 1b | 4.50 | 3 | 4, 161 | −65.9 |
| 1c | 12.0 | 4 | 5, 196 | −60.0 |
| 1d | 12.5 | 68 | 74, 241 | −68.5 |
| 1e | 8.90 | 28 | 30, 164 | −85.3 |
| 1f | 9.90 | 32 | 35, 120 | −62.3 |
| 1g | 6.50 | 31 | 13, 68 | −75.3 |
| 1h | 7.60 | 37 | 39, 215 | −60.1 |
| 1i | 0.20 | 10 | 15, 95 | −67.3 |
| 1j | 3.00 | 68 | 15, 210 | — |
| 1k | 0.70 | 28 | 105 | −74.2 |
| 2a | 0.01 | 10 | 22, 255 | +42.2 |
| 2b | 0.06 | 37 | 40, 220 | +69.9 |
| 2c | 5.96 | 10 | 21, 181 | +54.0 |
| 2d | 0.01 | 190 | 8, 54, 278 | +55.9 |
| 3a | 68.4 | 68 | 70, 137 | +61.3 |
| 3b | 45.7 | 6 | 32, 236 | +66.7 |
| 3c | — | 32 | 7, 190 | +91.2 |
| 3d | 8.88 | 28 | 30, 130 | +21.6 |
| 3e | 7.17 | 5 | 24, 142 | +43.3 |
| 3f | — | 68 | 4, 161 | +78.3 |
Using Dynamic Light Scattering (DLS), we then moved to study the particle size, both by number and intensity to determine the probable polydispersity and to gain information about the different populations typically observed in this type of sample. A representative example for 1f is shown in Fig. 3B, initially screening the effect of pH, where basic pH provided monodisperse nanoparticles with an average diameter of 32 nm (see the ESI† for a detailed study); however, below pH 5, the supramolecular species starts disassembling and precipitating. Small particles in the range of 3–68 nm were generally detected for the remaining lipopeptides (Table 1, size by number), together with some larger aggregates in the range of 105–278 nm, in different ratios depending on the example (Table 1, size by intensity). For every sample, we observed exceptionally good raw correlation data (see the ESI†), with a general trend: small structures identified as micelles (3–37 nm, size by number) coexist, in some cases, with larger populations (68–278 nm, size by intensity). DLS is a powerful and non-invasive technique that works particularly well for uniform, symmetric and spherical nanoparticles and allows for studying the particle size both by number and intensity. While size by intensity constitutes a general overview of the size from all particle populations, size by number refers to smaller and more abundant particle populations and should be taken into account with good correlation functions, as is the case here.
The stability of these nanoparticles was studied by measuring the magnitude of the zeta-potential (ZP). A representative study for 1f is shown in Fig. 3C, with very stable particles above neutral pH, displaying a large absolute value of ZP (>50 mV), while they are much less stable (<30 mV) below neutral pH. Under optimal conditions of pH, a large absolute magnitude of ZP above 60 mV is obtained in most cases (Table 1). For lipoamino acids, 1a–k, a large negative ZP would agree with a deprotonated carboxylate, as a result of the basic pH used. In sharp contrast, for lipodipeptides, 2a–d and 3a–f, measured at neutral pH, a positive value of ZP is observed, pointing towards a net positive charge, suggesting that this pH is below the isoelectric point. Overall, the ZP values for this library of lipopeptides demonstrate a general and exceptionally good stability (ZP > |40| mV).
In an effort to improve this study, our attention was driven to study the infrared spectra of these lipopeptides, which exhibited the following interesting characteristics: (1) red-shifting of O–H and N–H stretching (from 3500–3000 cm−1 to 3300–2500 cm−1) and (2) band-reshaping, from smooth bell-shaped bands to speckled discrete peaks, typical of medium-to-strong hydrogen bond networks.23 Such spectral characteristics provide insight into the overall ability of this pool of lipopeptides to self-assemble into supramolecular structures, promoted not only by the hydrophobic effect of long aliphatic chains, but also by establishing a hydrogen bonding network through the backbone sequence of amino acids.
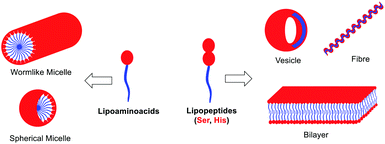
The morphology of the self-assembling lipopeptides was studied using High Resolution Transmission Electron Microscopy (HR-TEM) and Field Emission Scanning Electron Microscopy (FE-SEM). While HR-TEM shows spherical and worm-like micelles between 20 and 100 nm for lipoamino acids (Fig. 4A–C and Fig. S12 in the ESI†), multilamellar vesicles of variable sizes, micelle clusters and fibers are observed for lipodipeptides (Fig. 4D, E and Fig. S13, S14 in the ESI†). FE-SEM revealed the formation of fibers of different lengths for simple lipoamino acids, 1a–j, in most cases (Fig. 4F and G), although amorphous aggregates have also been observed (see the ESI†). Interestingly, diacylated cysteine, 1k, shows a more complex pattern (Fig. 4H), while beautiful cross-linked fibers and flower-like structures are observed for lipodipeptides (Fig. 4I and J). Moreover, when fibers are detected, it is possible to visualize that they are formed by small spherical particles with an average diameter that ranges from 20 to 50 nm. These small spherical nanoparticles match the micellar structures observed in the HR-TEM images, which might imply a hierarchical organization.
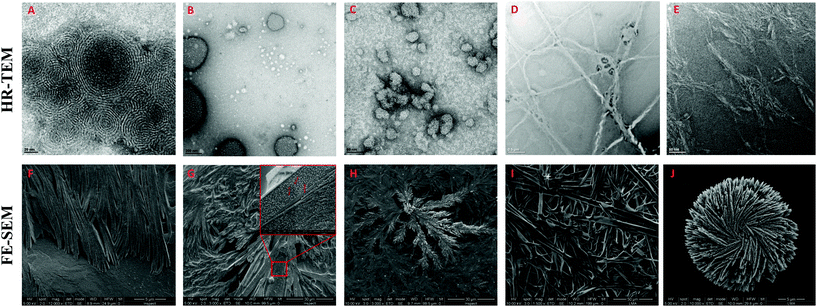 | ||
| Fig. 4 Representative HR-TEM and FE-SEM images of: 1a (A and F), 1f (B and G), 1k (C and H), 2d (D and I) and 3f (E and J). | ||
Altogether, moving from simple amino acids to dipeptides incorporating functional groups promoting non-covalent interactions allows for switching the supramolecular structure from simple micelles to more complex vesicles or fibers (Fig. 5). Moreover, the size and the presence of a double bond in the aliphatic chain play a key role in the development of a more complex pattern of self-assembly (compare 2b and 2dvs.2a and 2c). However, the sequence of amino acids can have a profound effect on the morphology (from vesicles to fibers), which cannot be anticipated, by simply reversing Ser and His residues (compare 2bvs.2d).
One of the biggest challenges concerning public health is the emergence of multidrug resistant pathogens, in particular the growth of antibiotic resistant bacteria strains.24 Understanding the resistance mechanisms25 is key to tackling this problem from different perspectives, including health education,26 optimization27 and repurposing28 of current drugs, development of new antibiotics29 or incorporating advanced genomic techniques.30 Lipopeptides have emerged as a good strategy to develop new antibacterial agents;31 most of the examples reported, so far, incorporated cationic amino acids, such as lysine or arginine.32 Mechanistic studies point towards bacterial membrane disruption;33 however, alteration of the membrane electric potential is also invoked.34
The antimicrobial activity of these new lipopeptides was evaluated in vitro against Gram-negative (E. coli CECT 516) and Gram-positive (S. aureus CECT 240) strains. The Minimal Inhibitory Concentration (MIC) was determined following the broth microdilution method (see the ESI†). While all lipoamino acids bearing a single fatty acyl chain, 1a–j, are not active at a realistic concentration (up to 1000 μg mL−1), diacylated cysteine 1k shows a notable activity and selectivity against S. aureus (Table 2). For dipeptide derivatives, we first screened non-lipidated Ser-His and His-Ser dipeptides, which proved inactive against both strains, a control experiment that demonstrates the importance of the amphiphilic nature, that allows self-assembling, key to achieving antimicrobial activity (ESI†). Considering N-terminus fatty acyl derivatives (2a–d), both N-decanoyl Ser-His (2a) and His-Ser (2c) showed modest and similar inhibitory activities against both strains (250–500 μg mL−1). However, N-oleyl derivatives (2b and 2d) were inactive. Finally, C-terminus fatty amides lead to interesting results, where all examples bearing a single hydrophobic chain (3a–b and 3d–e) are active against the two bacterial strains. From this, oleyl derivatives (3b and 3e) are exceptionally active (40 μg mL−1). Unfortunately, double hydrophobic chain derivatives, 3c and 3f, proved to be inactive.
| Compound | MICa (μg mL−1) | Compound | MICa (μg mL−1) | ||
|---|---|---|---|---|---|
| E. coli CECT 516 | S. aureus CECT 240 | E. coli CECT 516 | S. aureus CECT 240 | ||
| a Minimum concentration that inhibits proliferation after overnight incubation. | |||||
| 1a | >1000 | >1000 | 2a | 250 | 250 |
| 1b | >1000 | >1000 | 2b | >1000 | >1000 |
| 1c | >1000 | 1000 | 2c | 500 | 500 |
| 1d | >1000 | 1000 | 2d | >1000 | >1000 |
| 1e | >1000 | >1000 | 3a | 500 | 125 |
| 1f | >1000 | >1000 | 3b | 40 | 40 |
| 1g | >1000 | >1000 | 3c | >1000 | >1000 |
| 1h | >1000 | >1000 | 3d | 250 | 250 |
| 1i | >1000 | >1000 | 3e | 40 | 40 |
| 1j | >1000 | >1000 | 3f | >1000 | >1000 |
| 1k | >1000 | 40 | Gentamicin | 4.037a | 1.5637b |
From this, we can draw the following conclusions:
(1) Diacylated cysteine 1k is an outstanding example, as the only active lipoamino acid, and the only example in the library with such an impressive selectivity towards S. aureus. As far as we are aware, there is no precedent for such a simple and short lipoamino acid with this level of activity and selectivity. Shedding light on the mechanism of action of this diacylated cysteine is part of our current efforts.
(2) For dipeptides, the most active ones (2a, 2c, 3b and 3e) correlate with the smallest values of particle size (up to 10 nm size by number). However, we are aware that the Hydrophilic–Lipophilic Balance (HLB) might also play an important role.21
(3) C-terminus fatty amides 3, bearing the N-terminal ammonium group, are significantly more active than their analogous N-fatty acyl derivatives 2. Remarkably, fatty amides 3 show MIC values comparable to, or even lower than, highly cationic and structurally more complex lipopeptides previously reported.32 Previous antimicrobial lipopeptides containing both serine and histidine residues are embedded within a polycationic chain of several arginine or lysine residues.35 Alternatively, histidine gemini-lipopeptides with a highly complex pattern of substitution have been reported.36
(4) No straightforward correlation has been found between the CAC of these lipopeptides and their activity, suggesting that the antimicrobial mechanism might not be related to the detergent effect, but rather to the membrane and electrochemical disruption quenching the cell viability. This is in agreement with the higher activity displayed by polycationic peptides and their increased ability to disrupt the bacterial membrane.32,33 Moreover, elucidating whether these lipopeptides are bacteriostatic or bactericidal agents will need further investigation.
Conclusions
In conclusion, we have synthesized a library of amphiphilic short lipopeptides that combine N-fatty acyl amino acids and all possible combinations of N- and C-lipidated dipeptides from Ser and His. Their supramolecular behaviour has been systematically studied, and their Critical Aggregation Concentration (CAC), particle size, stability (zeta-potential) and morphology have been reported. In addition, their antimicrobial activity has been evaluated, showing promising results with a respectable selectivity towards Gram-positive strains for some examples. This study shows the importance of new structurally simple and short lipopeptides as a tool to develop novel, potent and selective antimicrobial agents, where the self-assembling nature is indispensable for their bioactivity.Author contributions
C.V.G. performed the experiments. C.V.G. and I.C. contributed to designing and analysing the experiments, and writing, discussing and editing the manuscript. I.C. conceived and guided the research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project received funding from the La Caixa Foundation under the Junior Leader Program (No. LCF/BQ/PI19/11690020, ID 100010434), CAM-Atracción de Talento (2019-T1/IND-12683) and Spanish MICINN (PID2019-106327GA-I00). I. C. holds a Ramón y Cajal fellowship from Spanish MICINN (RyC2019-026674-I). We also thank Prof. Isabel Rodriguez (IMDEA-Nanociencia) for providing bacterial strains and Dr Jose Miguel González (ICB-CSIC) for helping with the microscopy analysis.Notes and references
- (a) R. de la Rica and H. Matsui, Chem. Soc. Rev., 2010, 39, 3499–3509 RSC; (b) M. Zelzer and R. V. Ulijn, Chem. Soc. Rev., 2010, 39, 3351–3357 RSC; (c) A. Lampel, R. V. Ulijn and T. Tuttle, Chem. Soc. Rev., 2018, 47, 3737–3758 RSC.
- (a) D. W. P. M. Löwik and J. C. M. van Hest, Chem. Soc. Rev., 2004, 33, 234–245 RSC; (b) I. W. Hamley, Soft Matter, 2011, 7, 4122–4138 RSC; (c) S. Fleming and R. V. Ulijn, Chem. Soc. Rev., 2014, 43, 8150–8177 RSC.
- (a) I. W. Hamley, Chem. Commun., 2015, 51, 8574–8583 RSC; (b) P. Hendricks, K. Sato, L. C. Palmer and S. I. Stupp, Acc. Chem. Res., 2017, 50, 2440–2448 CrossRef PubMed; (c) N. Battista, M. Bari and T. Bisogno, Biomolecules, 2019, 9, 822 CrossRef CAS PubMed.
- (a) M. C. Morán, A. Pinazo, L. Pérez, P. Clapés, M. Angelet, M. T. García, M. P. Vinardell and M. R. Infante, Green Chem., 2004, 6, 233–240 RSC; (b) R. Bordes and K. Holmberg, Adv. Colloid Interface Sci., 2015, 222, 79–91 CrossRef CAS PubMed; (c) X. Du, J. Zhou, J. Shi and B. Xu, Chem. Rev., 2015, 115, 13165–13307 CrossRef CAS PubMed.
- D. Sivaramakrishna and M. J. Swamy, Chem. Phys. Lipids, 2016, 201, 1–10 CrossRef CAS PubMed.
- (a) A. Ohta, M. Shirai, T. Asakawa and S. Miyagishi, J. Oleo Sci., 2008, 57, 659–667 CrossRef CAS PubMed; (b) S. Lin, J. Qin, Y. Li, B. Li and Y. Yang, Langmuir, 2017, 33, 8246–8252 CrossRef CAS PubMed.
- Selected examples: (a) G. D. Smith and R. E. Barden, Chem. Phys. Lipids, 1975, 14, 1–14 CrossRef CAS; (b) A. Pinazo, L. Pérez, M. Lozano, M. Angelet, M. R. Infante, M. P. Vinardell and R. Pons, J. Phys. Chem. B, 2008, 112, 8578–8585 CrossRef CAS PubMed; (c) C. Zheng, S. Lin, Y. Chen, Y. Li, B. Li and Y. Yang, Langmuir, 2019, 35, 11406–11413 CrossRef CAS PubMed.
- C. W. Harrell, M. E. McCarroll, K. F. Morris, E. J. Billiot and I. M. Warner, Langmuir, 2003, 19, 10684–10691 CrossRef CAS.
- (a) C. Boettcher, B. Schade and J.-H. Fuhrhop, Langmuir, 2001, 17, 873–877 CrossRef CAS; (b) Y. Li, B. Li, Y. Fu, S. Lin and Y. Yang, Langmuir, 2013, 29, 9721–9726 CrossRef CAS PubMed.
- G. A. Rothbauer, E. A. Rutter, C. Reuter-Seng, S. Vera, E. J. Billiot, Y. Fang, F. H. Billiot and K. F. Morris, J. Surfactants Deterg., 2018, 21, 139–153 CrossRef CAS PubMed.
- (a) T. Kar, S. Debnath, D. Das, A. Shome and P. K. Das, Langmuir, 2009, 25, 8639–8648 CrossRef CAS PubMed; (b) M. Hatip Koc, G. Cinar Ciftci, S. Baday, V. Castelletto, I. W. Hamley and M. O. Guler, Langmuir, 2017, 33, 7947–7956 CrossRef CAS PubMed.
- Selected recent examples: (a) T. J. Moyer, J. A. Finbloom, F. Chen, D. J. Toft, V. L. Cryns and S. I. Stupp, J. Am. Chem. Soc., 2014, 136, 14746–14752 CrossRef CAS PubMed; (b) A. T. Preslar, F. Tantakitti, K. Park, S. Zhang, S. I. Stupp and T. J. Meade, ACS Nano, 2016, 10, 7376–7384 CrossRef CAS PubMed; (c) C. Gao, H. Li, Y. Li, S. Kewalramani, L. C. Palmer, V. P. Dravid, S. I. Stupp, M. Olvera De La Cruz and M. J. Bedzyk, J. Phys. Chem. B, 2017, 121, 1623–1628 CrossRef CAS PubMed.
- For a seminal outstanding contribution: J. D. Hartgerink, E. Beniash and S. I. Stupp, Science, 2001, 294, 1684 CrossRef CAS PubMed.
- K. Sato, M. P. Hendricks, L. C. Palmer and S. I. Stupp, Chem. Soc. Rev., 2018, 47, 7539–7551 RSC.
- (a) S. Bal, K. Das, S. Ahmed and D. Das, Angew. Chem., Int. Ed., 2019, 58, 244–247 CrossRef CAS PubMed; (b) S. P. Afrose, S. Bal, A. Chatterjee, K. Das and D. Das, Angew. Chem., Int. Ed., 2019, 58, 15783–15787 CrossRef CAS PubMed.
- Selected studies on short peptides’ supramolecular behaviour: (a) L. Chen, K. Morris, A. Laybourn, D. Elias, M. R. Hicks, A. Rodger, L. Serpell and D. J. Adams, Langmuir, 2010, 26, 5232–5242 CrossRef CAS PubMed; (b) L. Chen, S. Revel, K. Morris, L. C. Serpell and D. J. Adams, Langmuir, 2010, 26, 13466–13471 CrossRef CAS PubMed; (c) P. W. J. M. Frederix, R. V. Ulijn, N. T. Hunt and T. Tuttle, J. Phys. Chem. Lett., 2011, 2, 2380–2384 CrossRef CAS PubMed; (d) P. W. J. M. Frederix, G. G. Scott, Y. M. Abul-Haija, D. Kalafatovic, C. G. Pappas, N. Javid, N. T. Hunt, R. V. Ulijn and T. Tuttle, Nat. Chem., 2015, 7, 30–37 CrossRef CAS PubMed; (e) J. K. Gupta, D. J. Adams and N. G. Berry, Chem. Sci., 2016, 7, 4713–4719 RSC; (f) A. Lampel, S. A. McPhee, H.-A. Park, G. G. Scott, S. Humagain, D. R. Hekstra, B. Yoo, P. W. J. M. Frederix, T.-D. Li, R. R. Abzalimov, S. G. Greenbaum, T. Tuttle, C. Hu, C. J. Bettinger and R. V. Ulijn, Science, 2017, 356, 1064–1068 CrossRef CAS PubMed; (g) V. Castelletto, J. Seitsonen, K. M. Tewari, A. Hasan, R. M. Edkins, J. Ruokolainen, L. M. Pandey, I. W. Hamley and K. H. A. Lau, ACS Macro Lett., 2020, 9, 494–499 CrossRef CAS PubMed.
- For reviews on catalytic small peptides: (a) E. A. C. Davie, S. M. Mennen, Y. Xu and S. J. Miller, Chem. Rev., 2007, 107, 5759–5812 CrossRef PubMed; (b) A. J. Metrano and S. J. Miller, Acc. Chem. Res., 2019, 52, 199–215 CrossRef CAS PubMed. For a review partially covering catalytic lipopeptides: (c) O. Zozulia, M. A. Dolan and I. V. Korendovych, Chem. Soc. Rev., 2018, 47, 3621–3639 RSC.
- For an example of bioactive small lipopeptides: (a) A. Makovitzki, D. Avrahami and Y. Shai, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15997–16002 CrossRef CAS PubMed. For reviews: (b) R. Jerala, Expert Opin. Invest. Drugs, 2007, 16, 1159–1169 CrossRef CAS PubMed; (c) J.-J. Koh, S. Lin, R. W. Beuerman and S. Liu, Amino Acids, 2017, 49, 1653–1677 CrossRef CAS PubMed.
- Comprehensive Organic Name Reaction and Reagents, ed. Z. Wang, John Wiley and Sons, Weinheim, 2009, p. 2536 Search PubMed.
- A. R. Katritzky, S. R. Tala, N. E. Abo-Dya, K. Gyanda, B. E.-D. M. El-Gendy, Z. K. Abdel-Samii and P. J. Steel, J. Org. Chem., 2009, 74, 7165–7167 CrossRef CAS PubMed.
- For a perspective on HLB: (a) R. C. Pasquali, M. P. Taurozzi and C. Bregni, Int. J. Pharm., 2008, 356, 44–51 CrossRef CAS PubMed. For the suggested relationship between HLB and antibacterial activity: (b) K.-M. Park, S. J. Lee, H. Yu, J.-Y. Park, H.-S. Jung, K. Kim, C. J. Lee and P.-S. Chang, Food Sci. Biotechnol., 2018, 27, 401–409 CrossRef CAS PubMed; (c) A. Maneedaeng, S. Phoemboon, P. Chanthasena and N. Chudapongse, Korean J. Chem. Eng., 2018, 35, 2313–2320 CrossRef CAS.
- P. U. Sinare and J. D. Mhatre, Am. J. Chem., 2012, 2, 186–190 CrossRef.
- (a) S. Saha and G. R. Desiraju, J. Am. Chem. Soc., 2018, 140, 6361–6373 CrossRef CAS PubMed; (b) Vibrational dynamics of hydrogen bonds, ed. E. T. Nibbering, J. Dreyer, O. Kühn, J. Bredenbeck, P. Hamm and T. Elsaesser, Springer, Berlin, Heidelberg, 2007, vol. 87 Search PubMed.
- (a) H. Nikaido, Annu. Rev. Biochem., 2009, 78, 119–146 CrossRef CAS PubMed; (b) M. Frieri, K. Kumar and A. Boutin, J. Infect. Public Health, 2017, 10, 369–378 CrossRef PubMed.
- (a) C. Walsh, Nature, 2000, 406, 775–781 CrossRef CAS PubMed; (b) M. N. Alekshun and S. B. Levy, Cell, 2007, 128, 1037–1050 CrossRef CAS PubMed.
- S. Hernando-Amado, T. M. Coque, F. Baquero and J. L. Martínez, Nat. Microbiol., 2019, 4, 1432–1442 CrossRef CAS PubMed.
- M. Tyers and G. D. Wright, Nat. Rev. Microbiol., 2019, 17, 141–155 CrossRef CAS PubMed.
- M. A. Farha and E. D. Brown, Nat. Microbiol., 2019, 4, 565–577 CrossRef CAS PubMed.
- (a) D. J. Payne, M. N. Gwynn, D. J. Holmes and D. L. Pompliano, Nat. Rev. Drug Discovery, 2007, 6, 29–40 CrossRef CAS PubMed; (b) K. Lewis, Nat. Rev. Drug Discovery, 2013, 12, 371–387 CrossRef CAS PubMed; (c) U. Theuretzbacher, K. Outterson, A. Engel and A. Karlén, Nat. Rev. Microbiol., 2020, 18, 275–285 CrossRef PubMed.
- M. Boolchandani, A. W. D'Souza and G. Dantas, Nat. Rev. Genet., 2019, 20, 356–370 CAS.
- (a) A. Pinazo, M. A. Manresa, A. M. Marques, M. Bustelo, M. J. Espuny and L. Pérez, Adv. Colloid Interface Sci., 2016, 228, 17–39 CrossRef CAS PubMed; (b) F. Azmi, A. G. Elliott, Z. G. Khalil, W. M. Hussein, A. Kavanagh, J. X. Huang, M. Quezada, M. A. T. Blaskovich, R. J. Capon, M. A. Cooper, M. Skwarczynski and I. Toth, Nanomedicine, 2015, 10, 3359–3371 CrossRef CAS PubMed.
- Selected examples: (a) Y. Fang, W. Zhong, Y. Wang, T. Xun, D. Lin, W. Liu, J. Wang, L. Lv, S. Liu and J. He, Eur. J. Med. Chem., 2014, 83, 36–44 CrossRef CAS PubMed; (b) B. Mishra, T. Lushnikova and G. Wang, RSC Adv., 2015, 5, 59758–59769 RSC; (c) M. A. Paduszynska, M. Maciejewska, D. Neubauer, K. Golacki, M. Szymukowicz, M. Bauer and W. Kamysz, Pharmaceutics, 2019, 11, 506 CrossRef CAS.
- S. Nasompag, P. Dechsiri, N. Hongsing, P. Phonimdaeng, S. Daduang, S. Klaynongsruang, T. A. Camesano and R. Patramanon, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 2351–2364 CrossRef CAS PubMed.
- Y. Shai, Biopolym. – Pept. Sci. Sect., 2002, 66, 236–248 CrossRef CAS PubMed.
- (a) S. R. Nagarajan, B. Devadas, M. E. Zupec, S. K. Freeman, D. L. Brown, H.-F. Lu, P. P. Mehta, N. S. Kishore, C. A. McWherter, D. P. Getman, J. I. Gordon and J. A. Sikorski, J. Med. Chem., 1997, 40, 1422–1438 CrossRef CAS PubMed; (b) H. Yu, K. Park and P. Chang, Food Chem., 2020, 319, 126533 CrossRef CAS PubMed.
- M. Ahn, R. N. Murugan, B. Jacob, J. Hyun, C. Cheong, E. Hwang, H. Park, J. Seo, G. Srinivasrao, K. S. Lee, S. Y. Shin and J. K. Bang, Eur. J. Med. Chem., 2013, 68, 10–18 CrossRef CAS PubMed.
- (a) H.-K. Sun, A. Pang, D. C. Farr, T. Mosaiab, W. J. Britton, S. Anoopkumar-Dukie, I. D. Grice, M. J. Kiefel, N. P. West, G. D. Grant and T. A. Houston, Aust. J. Chem., 2018, 71, 716–719 CrossRef CAS; (b) T. S. Graziano, M. C. Cuzzullin, G. C. Franco, H. O. Schwartz-Filho, E. D. de Andrade, F. C. Groppo and K. Cogo-Müller, PLoS One, 2015, 10, e0128098 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, chemical and supramolecular characterization and antimicrobial assays. See DOI: 10.1039/d1ob01227d |
| This journal is © The Royal Society of Chemistry 2021 |