A helix foldamer oligopeptide improves intracellular stability and prolongs protein expression of the delivered mRNA†
Satoshi
Uchida
 *ab,
Yuto
Yamaberi
c,
Masakazu
Tanaka
c and
Makoto
Oba
*ab,
Yuto
Yamaberi
c,
Masakazu
Tanaka
c and
Makoto
Oba
 *a
*a
aMedical Chemistry, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan. E-mail: moba@koto.kpu-m.ac.jp; suchida@koto.kpu-m.ac.jp
bInnovation Center of NanoMedicine (iCONM), Kawasaki Institute of Industrial Promotion, Kawasaki, Japan
cGraduate School of Biomedical Sciences, Nagasaki University, Nagasaki, Japan
First published on 28th September 2021
Abstract
Prolonging the duration of protein expression from mRNA is a major challenge in the development of mRNA nanomedicines. mRNA complexed with helix foldamer oligopeptides consisting of arginine and α-aminoisobutyric acids showed higher intracellular stability than that complexed with oligoarginines, thereby maintaining efficient protein translation for three days.
Messenger RNA (mRNA) therapeutics have demonstrated tremendous potential in the vaccination against coronavirus disease 2019.1 The clinical approval of two vaccine formulations prompted further research and development of mRNA therapeutics in various medical fields, such as cancer vaccination, treatment of rare genetic disorders, protein supplementation and genome editing, as well as in vaccination against various infectious diseases.2–4 Prolonging the duration of protein expression from mRNA is a major challenge in the development of mRNA nanoparticles for various therapeutic applications. For example, in the treatment of rare genetic disorders using mRNA, current technologies require weekly administration of mRNA for several years.5,6 Prolonging the duration of protein expression is also required in vaccination strategies, as sustained antigen exposure enhances the immune response.7 However, the median intracellular half-life of endogenous mRNA is only 9 h,8 and exogenous mRNA is more rapidly degraded inside cells by mechanisms that discriminate exogenous from endogenous mRNA.9–11 Thus, nanoparticles that improve the intracellular stability of the delivered mRNA are in high demand.
In most studies, mRNA nanoparticles have been designed to target specific organs, protect the mRNA from enzymatic degradation before reaching the targeted cells, and facilitate endosomal escape and protein translation after cellular uptake.2–4,12–16 Only a few studies have focused on intracellular mRNA stabilization to prolong protein expression, and so far, modest stabilization has been achieved, despite the importance of this issue in the development of mRNA nanomedicines.17,18 Herein, we address this challenge using cationic oligopeptides that can be uniformly synthesized and are minimally cytotoxic due to the relatively smaller number of cationic moieties per molecule compared to that in commonly used transfection reagents, such as linear poly(ethyleneimine). We designed an oligopeptide based on oligoarginine (OligoArg), possessing guanidino groups, which strongly bind to nucleic acids, including mRNA.19,20 In addition, we introduced α-aminoisobutyric acid (Aib) to OligoArg to prepare OligoArg-Aib. Aib is the simplest form of α,α-disubstituted α-amino acid (dAA), which is a non-proteinogenic amino acid. Introduction of Aib into peptides allows the formation of helical structures21,22 and, thus, Aib is often used as a building block for functional helical foldamers.23,24 Such unique properties of Aib may influence the binding of oligopeptides to mRNA in the intracellular environment. Furthermore, helical foldamers containing dAAs are highly resistant to protease-based degradation,25 which may prolong the protective effects of peptides against mRNA degradation. The present study revealed that OligoArg-Aib drastically improved the intracellular stability of the delivered mRNA and extended the duration of protein expression.
Results and discussion
We synthesized arginine nonapeptides (OligoArg) and (Arg-Arg-Aib)5 (OligoArg-Aib), labeled with carboxyfluorescein via a glycine linker at their N terminus, as previously described.24 Carboxyfluorescein, which was introduced for monitoring oligopeptides in the cells, was reported to function as a hydrophobic group not unlike a stearyl group and increase the plasmid DNA transfection efficiency of OligoArg.26 We mixed the oligopeptides with mRNA at [guanidino groups in oligopeptides (N)]/[phosphate groups in mRNA (P)] ratios of 2 and 4. Dynamic light scattering (DLS) measurements revealed that the cumulant diameters of OligoArg/mRNA and OligoArg-Aib/mRNA were greater than 500 nm at N/P = 2 and approximately 200 nm at N/P = 4, with a relatively narrow size distribution for both oligopeptide formulations at N/P = 4 (Table 1).| OligoArg-Aib (N/P = 2) | OligoArg (N/P = 2) | OligoArg-Aib (N/P = 4) | OligoArg (N/P = 4) | |
|---|---|---|---|---|
| Size | 645 ± 13 | 1162 ± 45 | 199 ± 1 | 198 ± 1 |
| PDI | 0.45 ± 0.04 | 0.15 ± 0.07 | 0.14 ± 0.03 | 0.09 ± 0.02 |
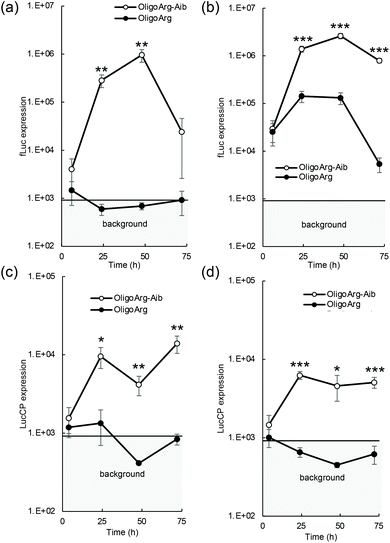
The mRNA introduction efficiency of these oligopeptides was evaluated in human hepatoma-derived HuH-7 cells using the firefly luciferase (fLuc) mRNA. Although fLuc expression levels were comparable between OligoArg and OligoArg-Aib at 4 h after mRNA addition at both N/P = 2 and 4, at 24 h or later, OligoArg-Aib exhibited a 10–1000-fold more efficient fLuc expression compared with that of OligoArg at both N/P ratios (Fig. 1a and b). Notably, cell viability was higher than 80% of that of untransfected control in every tested formulation, throughout the observation period from 24 h to 72 h after mRNA addition, demonstrating the safety of both oligopeptides (Fig. S1†). Furthermore, mRNA introduction using OligoArg and OligoArg-Aib showed negligible influence on endogenous protein expression (Fig. S2†). Notably, fLuc expression was undetected after the introduction of naked mRNA and mRNA complexed with 2.4 kDa linear poly(ethylene imine) (LPEI), which has approximately a 3-fold larger number of protonated amines, compared to that of OligoArg-Aib (Fig. S3a†), highlighting the potential of OligoArg-Aib in mRNA delivery. Although 22 kDa LPEI exhibited enhanced luciferase expression compared to OligoArg-Aib, 22 kDa LPEI induced enhanced cytotoxicity (Fig. S3b†), as was reported previously.27 Meanwhile, in a time-dependent evaluation of protein expression profile, a reporter assay using fLuc could not clearly distinguish the influence of mRNA degradation from that of protein degradation, as the fLuc protein has a relatively long intracellular half-life of around 3–4 h in mammalian cells.28 To clarify the influence of mRNA degradation, we used destabilized luciferase (Luc2CP) as the reporter. Luc2CP possesses a PEST sequence, which is rich in Pro, Glu, Ser, and Thr and decreases the intracellular half-life of the Luc protein to less than one hour, and a ubiquitination signal that further prompts intracellular degradation of luciferase.29–31 Luc2CP expression from OligoArg/mRNA was almost comparable to the background level expression throughout the observation period at both N/P = 2 and 4 (Fig. 1c and d), presumably because of the rapid intracellular degradation of the Luc2CP protein. In sharp contrast, OligoArg-Aib allowed the detection of Luc2CP at both N/P ratios, even at 72 h after mRNA addition. The results obtained with the destabilized luciferase reporter strongly suggest that OligoArg-Aib preserves the translational activity of mRNA for as long as 72 h by preventing mRNA degradation.
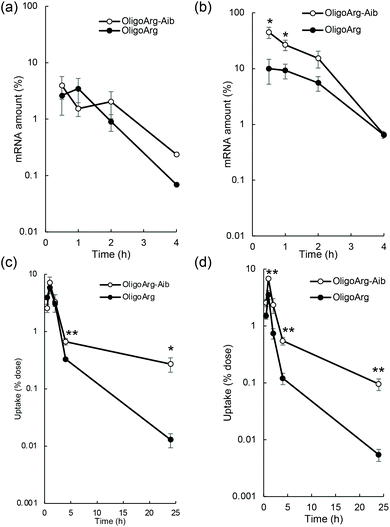
To study the mRNA degradation profile, we performed quantitative polymerase chain reaction (qPCR)-based measurement of mRNA intactness, which detects mRNA possessing an intact sequence between a primer pair. First, mRNA degradation in extracellular spaces was evaluated by quantifying fLuc mRNA degradation after incubation in 10% serum, the same serum concentration as that used in the culture medium during mRNA introduction experiments shown in Fig. 1. At N/P = 2, the detected mRNA levels were comparable between OligoArg and OligoArg-Aib (Fig. 2a). Although OligoArg-Aib showed a higher amount of detected mRNA compared to OligoArg at N/P = 4, the difference was less than five-fold (Fig. 2b), which is insufficient to completely explain the 10 to 1000-fold difference in protein expression efficiency observed between OligoArg and OligoArg-Aib (Fig. 1). Although OligoArg-Aib transfection resulted in protein expression for three days from the delivered mRNA, even with the use of destabilized luciferase (Luc2CP), the remaining amount of extracellular mRNA was less than 1% even after 4 h of incubation. Thus, extracellular mRNA stability may not be a major factor contributing to prolonged protein expression from OligoArg-Aib/mRNA.
Next, we studied the integrity of the fLuc mRNA inside the cells after mRNA addition using qPCR. In all tested formulations, the amount of fLuc mRNA peaked at one hour after mRNA addition (Fig. 2c and d), suggesting that the cellular uptake of intact mRNA was mostly completed within a few hours of mRNA addition. This is consistent with the low extracellular stability of mRNA (Fig. 2a and b). Intriguingly, OligoArg-Aib showed significantly higher amounts of mRNA inside cells at N/P = 2 and 4, compared to OligoArg both at 4 h and 24 h after mRNA addition, with more than a 10-fold difference observed between OligoArg-Aib and OligoArg at 24 h. This result may be attributed to OligoArg-Aib stabilizing mRNA in intracellular spaces, as extracellular mRNA stability was low even after complexation with OligoArg-Aib (Fig. 2a and b).
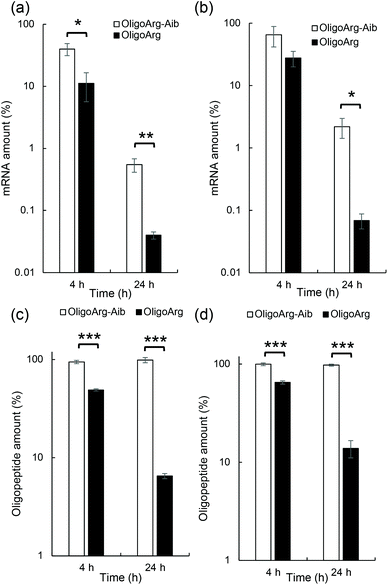
This result motivated us to directly evaluate the intracellular stability of mRNA. For this purpose, 30 min after mRNA addition, the cells were washed and supplemented with fresh culture medium to inhibit additional uptake of oligopeptide/mRNA polyplexes, and the amount of mRNA inside cells was quantified using qPCR at both 4 and 24 h after medium replacement. Fig. 3a and b show the relative mRNA amount at the indicated time points compared to that immediately after 30 min of incubation with mRNA, representing intracellular mRNA degradation profiles during the incubation period. As expected, OligoArg-Aib significantly improved mRNA intracellular stability by more than 10-fold at both N/P = 2 and 4 compared to that of OligoArg. To obtain mechanistic insight into this result, we studied the behavior of OligoArg and OligoArg-Aib in the same experimental setting by measuring fluorescein fluorescence from the oligopeptides using flow cytometry. Interestingly, the intracellular amount of OligoArg-Aib remained constant 24 h after replacing the culture medium at both N/P = 2 and 4, while the intracellular amount of OligoArg continuously decreased during the incubation period (Fig. 3c and d). Thus, the persistence of OligoArg-Aib inside the cells might aid in prolonging the protection conferred on intracellular mRNA (Fig. 3a and b).
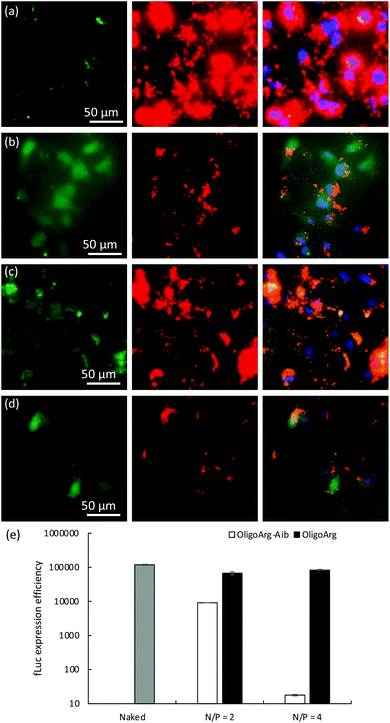
To further understand this, we first studied the intracellular behavior of mRNA and oligopeptides via fluorescence microscopic observation of transfected cells (Fig. 4a–d). Twenty-four hours after mRNA addition, OligoArg exhibited a diffusive appearance throughout the cells, while a dotted pattern of OligoArg-Aib appeared inside the cells. In the light of the results indicating enhanced intracellular stability of mRNA with the use of OligoArg-Aib (Fig. 1–3), it is possible to speculate that such different appearance of OligoArg and OligoArg-Aib might reflect more stable binding of OligoArg-Aib to mRNA or some other components inside the cells compared to that of OligoArg, although current observation is insufficient to demonstrate a direct association between OligoArg-Aib and mRNA inside cells. According to a previous study, OligoArg containing dAAs is more resistant to enzymatic degradation compared to OligoArg,25 which might also explain different appearances of these two oligopeptides. To study the association of OligoArg-Aib with mRNA, the translation efficiency of OligoArg-Aib/mRNA was studied using a cell-free translational system that mimics the intracellular environment. In rabbit reticulocyte lysate, OligoArg-Aib showed less efficient mRNA translational activity compared to naked mRNA (Fig. 4e), and this depended on the N/P ratio, while OligoArg exhibited minimal influence on the translational process. The inhibitory effect of OligoArg-Aib on translational activities was also observed in the experiment using cultured cells. Luciferase expression levels were comparable between OligoArg-Aib and OligoArg at N/P = 4, 4 h after the transfection (Fig. 1b), although OligoArg-Aib exhibited approximately 5-fold higher intracellular amount of mRNA compared to that after the addition of OligoArg/mRNA even at 4 h (Fig. 2d). Such an inhibitory effect of OligoArg-Aib on translational processes might reflect the higher affinity of OligoArg-Aib to mRNA compared to that of OligoArg. Thus, it can be suggested that stable binding of OligoArg-Aib to mRNA may protect mRNA from intracellular degradation and gradual mRNA release may prolong protein expression from mRNA, although further studies are needed to clarify this hypothesis. It should also be noted that cell-free conditions exhibited a higher extent of translation inhibition compared to that observed in cells, presumably because the cell-free experiment does not totally mimic the intracellular environment after mRNA treatment.
Conclusions
We successfully constructed an oligopeptide structure that protects mRNA from intracellular enzymatic degradation and prolongs protein expression after mRNA delivery. Notably, the protein expression profile after the introduction of mRNA encoding destabilized luciferase strongly suggests that mRNA complexed with the OligoArg-Aib helical foldamer continues to translate even after 3 days, while the intracellular half-life of exogenous mRNA was reported to be around 1–4 h.10,11 Our previous studies succeeded in modestly stabilizing mRNA inside cells by increasing the cationic charge density or increasing the amount of positively charged primary amine.17,18 In contrast, the present study drastically improved the intracellular stability of mRNA by inserting several Aib into the oligoarginine to modify the oligopeptide structure, without changing cationic moieties, providing a novel concept for future molecular designs of peptides or polymers that will aid in intracellular mRNA stabilization. Meanwhile, exact stabilization mechanisms remain to be elucidated. Nevertheless, this study demonstrates the potential of OligoArg-Aib as a building block to prepare functionalized mRNA nanoparticles. For example, size and surface charge can be controlled by PEGylation of oligopeptides or mRNA for in vivo application.32–34 OligoArg-Aib might also be useful as a building block for lipopolyplexes. Although the limited duration of protein expression is a major challenge of mRNA therapeutics for its widespread application, for example, in rare genetic disorders, OligoArg-Aib may help to overcome this challenge.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was financially supported by Grants-in-Aid for Scientific Research (A) [21H04962 to S. U.], and (B) [18K03529 to S. U.] from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) and Research and Development of Core Technologies for Gene and Cell Therapy [JP18ae0201009 to S. U.] from Japan Agency for Medical Research and Development (AMED). We thank S. Ibuki and M. Suzuki (Kyoto Prefectural University of Medicine) for technical assistance.Notes and references
- R. Verbeke, I. Lentacker, S. C. De Smedt and H. Dewitte, J. Controlled Release, 2021, 333, 511–520 CrossRef CAS PubMed.
- K. A. Hajj and K. A. Whitehead, Nat. Rev. Mater., 2017, 2, 17056 CrossRef CAS.
- Z. Zhong, S. Mc Cafferty, F. Combes, H. Huysmans, J. De Temmerman, A. Gitsels, D. Vanrompay, J. Portela Catani and N. N. Sanders, Nano Today, 2018, 23, 16–39 CrossRef CAS.
- S. Uchida, F. Perche, C. Pichon and H. Cabral, Mol. Pharm., 2020, 17, 3654–3684 CrossRef CAS PubMed.
- B. Truong, G. Allegri, X. B. Liu, K. E. Burke, X. Zhu, S. D. Cederbaum, J. Haberle, P. G. V. Martini and G. S. Lipshutz, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 21150–21159 CrossRef CAS PubMed.
- P. G. V. Martini and L. T. Guey, Hum. Gene Ther., 2019, 30, 1180–1189 CrossRef CAS PubMed.
- K. M. Cirelli, D. G. Carnathan, B. Nogal, J. T. Martin, O. L. Rodriguez, A. A. Upadhyay, C. A. Enemuo, E. H. Gebru, Y. Choe, F. Viviano, C. Nakao, M. G. Pauthner, S. Reiss, C. A. Cottrell, M. L. Smith, R. Bastidas, W. Gibson, A. N. Wolabaugh, M. B. Melo, B. Cossette, V. Kumar, N. B. Patel, T. Tokatlian, S. Menis, D. W. Kulp, D. R. Burton, B. Murrell, W. R. Schief, S. E. Bosinger, A. B. Ward, C. T. Watson, G. Silvestri, D. J. Irvine and S. Crotty, Cell, 2019, 177, 1153–1171 CrossRef CAS PubMed.
- B. Schwanhausser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, W. Chen and M. Selbach, Nature, 2011, 473, 337–342 CrossRef PubMed.
- B. R. Anderson, H. Muramatsu, B. K. Jha, R. H. Silverman, D. Weissman and K. Kariko, Nucleic Acids Res., 2011, 39, 9329–9338 CrossRef CAS PubMed.
- T. Nogimori, K. Nishiura, S. Kawashima, T. Nagai, Y. Oishi, N. Hosoda, H. Imataka, Y. Kitamura, Y. Kitade and S. I. Hoshino, Nucleic Acids Res., 2019, 47, 432–449 CrossRef CAS PubMed.
- H. Zhang, K. Rombouts, L. Raes, R. Xiong, S. C. De Smedt, K. Braeckmans and K. Remaut, Adv. Biosyst., 2020, 4, e2000057 CrossRef PubMed.
- H. Uchida, K. Itaka, S. Uchida, T. Ishii, T. Suma, K. Miyata, M. Oba, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2016, 138, 1478–1481 CrossRef CAS PubMed.
- S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova, J. J. Senn, A. Lynn, A. Bulychev, I. McFadyen, J. Chan, O. Almarsson, M. G. Stanton and K. E. Benenato, Mol. Ther., 2018, 26, 1509–1519 CrossRef CAS PubMed.
- K. Koji, N. Yoshinaga, Y. Mochida, T. Hong, T. Miyazaki, K. Kataoka, K. Osada, H. Cabral and S. Uchida, Biomaterials, 2020, 261, 120332 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed.
- H. Tanaka, T. Takahashi, M. Konishi, N. Takata, M. Gomi, D. Shirane, R. Miyama, S. Hagiwara, Y. Yamasaki, Y. Sakurai, K. Ueda, K. Higashi, K. Moribe, E. Shinsho, R. Nishida, K. Fukuzawa, E. Yonemochi, K. Okuwaki, Y. Mochizuki, Y. Nakai, K. Tange, H. Yoshioka, S. Tamagawa and H. Akita, Adv. Funct. Mater., 2020, 30, 1910575 CrossRef CAS.
- H. Uchida, K. Itaka, T. Nomoto, T. Ishii, T. Suma, M. Ikegami, K. Miyata, M. Oba, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2014, 136, 12396–12405 CrossRef CAS PubMed.
- A. Dirisala, S. Uchida, T. A. Tockary, N. Yoshinaga, J. Li, S. Osawa, L. Gorantla, S. Fukushima, K. Osada and K. Kataoka, J. Drug Targeting, 2019, 27, 670–680 CrossRef CAS PubMed.
- I. Nakase, T. Takeuchi, G. Tanaka and S. Futaki, Adv. Drug Delivery Rev., 2008, 60, 598–607 CrossRef CAS PubMed.
- T. Miyazaki, S. Uchida, H. Hatano, Y. Miyahara, A. Matsumoto and H. Cabral, Eur. Polym. J., 2020, 140, 110028 CrossRef CAS.
- M. Crisma and C. Toniolo, Pept. Sci., 2015, 104, 46–64 CrossRef CAS PubMed.
- B. A. F. Le Bailly and J. Clayden, Chem. Commun., 2016, 52, 4852–4863 RSC.
- K. Akagawa, J. Higuchi, I. Yoshikawa and K. Kudo, Eur. J. Org. Chem., 2018, 2018, 5278–5281 CrossRef CAS.
- M. Oba, Y. Ito, T. Umeno, T. Kato and M. Tanaka, ACS Biomater. Sci. Eng., 2019, 5, 5660–5668 CrossRef CAS PubMed.
- M. Oba, Y. Nagano, T. Kato and M. Tanaka, Sci. Rep., 2019, 9, 1349 CrossRef PubMed.
- M. Oba, Y. Demizu, H. Yamashita, M. Kurihara and M. Tanaka, Bioorg. Med. Chem., 2015, 23, 4911–4918 CrossRef CAS PubMed.
- S. M. Moghimi, P. Symonds, J. C. Murray, A. C. Hunter, G. Debska and A. Szewczyk, Mol. Ther., 2005, 11, 990–995 CrossRef CAS PubMed.
- G. M. Leclerc, F. R. Boockfor, W. J. Faught and L. S. Frawley, BioTechniques, 2000, 29, 590–591 CrossRef CAS PubMed.
- T. Gilon, O. Chomsky and R. G. Kulka, EMBO J., 1998, 17, 2759–2766 CrossRef CAS PubMed.
- J. B. Robertson, C. C. Stowers, E. Boczko and C. H. Johnson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17988–17993 CrossRef CAS PubMed.
- Z. Cheng, D. Garvin, A. Paguio, P. Stecha, K. Wood and F. Fan, Curr. Chem. Genomics, 2010, 4, 84–91 CrossRef CAS PubMed.
- M. Ogris, S. Brunner, S. Schüller, R. Kircheis and E. Wagner, Gene Ther., 1999, 6, 595–605 CrossRef CAS PubMed.
- S. Kurimoto, N. Yoshinaga, K. Igarashi, Y. Matsumoto, H. Cabral and S. Uchida, Molecules, 2019, 24, 1303 CrossRef CAS PubMed.
- S. Uchida and K. Kataoka, J. Biomed. Mater. Res., Part A, 2019, 107, 978–990 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr03600a |
| This journal is © The Royal Society of Chemistry 2021 |