Open Access Article
Open Access ArticlePlant-wide systems microbiology for the wastewater industry
Marta Cerruti†
a,
Bing Guo†abc,
Robert Delatolla d,
Nadieh de Jongeef,
Aleida Hommes - de Vos van Steenwijkg,
Paul Kadotah,
Christopher E. Lawsonij,
Ted Maok,
Margreet J. Oosterkampl,
Fabrizio Sabbam,
Mikkel Stokholm-Bjerregaardn,
Ian Watsono,
Dominic Frigonb and
David G. Weissbrodt†
d,
Nadieh de Jongeef,
Aleida Hommes - de Vos van Steenwijkg,
Paul Kadotah,
Christopher E. Lawsonij,
Ted Maok,
Margreet J. Oosterkampl,
Fabrizio Sabbam,
Mikkel Stokholm-Bjerregaardn,
Ian Watsono,
Dominic Frigonb and
David G. Weissbrodt† *a
*a
aDepartment of Biotechnology, Delft University of Technology, Building 58, van der Maasweg 9, 2629 HZ Delft, The Netherlands. E-mail: m.cerruti@tudelft.nl; d.g.weissbrodt@tudelft.nl; Tel: +31 15 27 81169
bDepartment of Civil Engineering, McGill University, Montréal, Canada. E-mail: dominic.frigon@mcgill.ca
cDepartment of Civil and Environmental Engineering, University of Surrey, Guildford, UK. E-mail: b.guo@surrey.ac.uk
dDepartment of Civil Engineering, University of Ottawa, Ottawa, Canada. E-mail: robert.delatolla@uOttawa.ca
eDepartment of Chemistry and Bioscience, Aalborg University, Aalborg, Denmark. E-mail: ndj@bio.aau.dk
fNiras A/S, Aalborg, Denmark
gOrvion BV, Stolwijk, The Netherlands. E-mail: adevos@orvion.nl
hMetro Vancouver, Burnaby, Canada. E-mail: paul.kadota@metrovancouver.org
iJoint BioEnergy Institute, Lawrence Berkeley National Laboratory, USA. E-mail: c.e.lawson.87@gmail.com
jDepartment of Chemical Engineering & Applied Chemistry, University of Toronto, Toronto, Canada
kMW Technologies, London, Canada. E-mail: tedmao@outlook.com
lSub-department of Environmental Technology, Wageningen University & Research, Wageningen, The Netherlands. E-mail: marjet.oosterkamp@wur.nl
mDepartment of Civil and Environmental Engineering, Northwestern University, Evanston, Illinois, USA. E-mail: fabrizio.sabba@northwestern.edu
nKrüger A/S, Aalborg, Denmark. E-mail: mxs@kruger.dk
oUSP Technologies, San Diego, California, USA. E-mail: iwatson@usptechnologies.com
First published on 11th June 2021
Abstract
The wastewater treatment sector embraces mixed-culture biotechnologies for sanitation, environmental protection, and resource recovery. Bioprocess design, monitoring and control thrive on microbial processes selected in complex microbial communities. Microbial ecology and systems microbiology help access microbiomes and characterize microorganisms, metabolisms and interactions at increased resolution and throughput. Big datasets are generated from the sequencing of informational molecules extracted from biomasses sampled across process schemes. However, they mostly remain on science benches and computing clusters, without reaching the industry in a clear engineering objective function. A bilateral bridge should actionize this information. As systems microbiologists, we miss that engineering designs and operations rely on stoichiometry and kinetics. The added-value provided by microbial ecology and systems microbiology to improve capital (CAPEX) and operating expenditures (OPEX) needs to be addressed. As engineers, we miss that microbiology can be provide powerful microbial information on top of physical–chemical measurements for quantitative process design (e.g., nutrient removal systems) with detailed scientific description of phenomena inside microbiomes. In this perspective article, we allied academia and industry to address the state of shared knowledge, successes and failures, and to establish joint investigation platforms. Our roadmap involves three milestones to (i) elaborate an essential list of microbiological information needed to implement methods at the process line; (ii) characterize microbiomes from microorganisms to metabolisms, and shape conceptual ecosystem models as primer for process ecology understanding; (iii) bridge engineering and mathematical models with an analytical toolbox for fast- vs. high-throughput analyses to discover new microbial processes and engineer assemblies. We praise for a harmonized “language of love” (incorporating common vocabulary, units, protocols) across the water and environmental biotechnology sector to team up mindsets for a sewer- and plant-wide integration of systems microbiology and engineering.
Water impactWastewater science and engineering has evolved with a continuous integration of process engineering and microbial ecology principles. The advent of high-resolution bioanalytical techniques using sequencing and mass spectrometry has enabled the unprecedented description of microbial communities across populations and metabolic networks. In this “all for digital” era, initiatives are needed to bring back the data to engineering concepts to improve process design, understanding and control or to discover new avenues for environmental biotechnology. Data generation is not the endpoint of the scientific and engineering processes. Engineers and systems microbiologists need to better integrate their investigations beyond the data, in a language of love. This will drive an efficient use of analytical and process resources at installation line to derive responsible concepts for sanitation, water protection and resource recovery. |
1 Introduction: closing the gap between engineering and microbiologically elucidating sludge
Wastewater treatment plants (WWTPs) are central to urban water cycles to clean up aqueous wastes from cities and industries, to protect public health and the environment. Following 100 years of activated sludge,1 WWTPs are reappraised as water resource recovery facilities (WRRFs) or, simply, water resource factories (WRFs, used hereafter). The environmental engineering sector transitions to tap sewage and reclaim safe water, nutrients, bulk chemicals, materials, and energy from used water streams.2–10 Compared to physical–chemical processes, open mixed-culture bioprocesses help remove or capture nutrients11–13 and produce added-value compounds and high-tech materials, at low cost.14,15Early calls of the 2000s addressed the association of environmental biotechnology and microbial ecology to manage the microbial resource in (waste)water engineering.16,17 While joint endpoints have been achieved, the disciplines are specialized, driven by computational engineering and “omics” sciences. Process engineering moved toward sewer and plant-wide mathematical modelling, and multi-scale computational fluid dynamics.18–24 Molecular biology and multi-omics dramatically expanded the microbial and metabolic resolution in microbiomes like sewers, activated sludge, granular sludge, biofilm processes, and anaerobic digesters,25–29 leading to a new era for community systems microbiology.30–33 This raises the question of how can the two fields close the gap again?.
Although microbial ecology is recognized as an important component of environmental engineering, systems microbiology is established without a clear translation into wastewater engineering. High-throughput and high-resolution bioanalytics evolved fast out of practitioners' hands. Wet-lab and dry-lab molecular developments went without conceptualizing the important analytical targets for engineering practice. Reciprocally, sewer- and plant-wide mathematical models and simulation software evolved across process scales from 1- to 2- and 3-D, primarily processing physical–chemical variables. The increasing complexity of each field rendered interactions difficult but not impossible. Both fields are now used to handle big datasets via chemometrics or bioinformatics.
The time scale from microbial ecology measurements to data processing and information delivery spans from minutes (e.g., flow cytometry) and hours (e.g., microscopy) to days (e.g., FISH, qPCR assays), weeks and months (e.g., amplicon sequencing, metagenomics, multi-omics). Data utilization for improved predictions of engineering metrics (e.g., stoichiometry, kinetics) or diagnostics of plant operations (e.g., process stability) is impeded by a lack of concrete implementation examples. Though, efficient operations would benefit from real-time feed-back/feed-forward control loops based on microbial measurements that indicate bioprocess healthiness.
This gap in mutual knowledge generates forces to pool practitioners, engineers, microbiologists, molecular biologists and bioinformaticians to develop common investigation lines. This brings challenges for interaction and integration since the respective vocabularies, questions, scales and approaches are different. The gaps need to be addressed to integrate more bioanalytics into process engineering but also more engineering into microbial ecology and systems microbiology. Closing the gaps should answer specific needs. Engineers should develop a list of essential microbiological information for process design, operation, monitoring, and control that could be matched to the state-of-the art of microbiological and molecular methods.
While capital (CAPEX) and operating (OPEX) expenditures of environmental biotechnology processes primarily rely on kinetics and stoichiometry, respectively,34 deeper insights into the microbiological blackbox can provide a systematic understanding of observed phenomena, early warnings on the unfavorable selection of microorganisms, and specific remedial solutions to manage them. Among others, this can be applied, e.g., for a better prediction and management of microorganisms and metabolisms involved in the sequential and multi-step conversions of nitrogen (e.g., nitrification, denitrification, anammox), in the emission of (unfavorable) intermediate products (e.g., NO2−, N2O), in EBPR performance and failure, in filamentous and viscous bulking phenomena, but also in discovering new metabolisms and microbial associations for innovative environmental biotechnologies.
The formulation of clear expectations from microbiological and engineering experts can tie mindsets to stimulate exchange, innovation, and impact. With this perspective article, we bridged the fields to develop a roadmap for a rational integration of (i) systems microbiology analyses to inform nutrient removal and resource recovery processes and (ii) engineering concepts to sustain microbial ecology endeavors. Our initiative benefits from a close interaction between microbiologists, molecular biologists, environmental engineers, and practitioners.
2 Bringing microbial ecology and systems microbiology in the industrial perspective
Developing a joint platform for systems microbiology and engineering raises the question on how to bridge microbial ecology and systems functions. Mixed-culture systems rely on the metabolic performance of microbial communities. Microbial populations compete and interact for resources in an ecosystem. Their growth is driven by their catabolic and anabolic conversions, so-called metabolic functions.34 Harnessing microbiomes relies on managing the metabolisms of microorganisms selected to perform the conversions of interest. Systems microbiology should not only give names and relative abundances of populations but further details on how their metabolisms are activated or repressed in the process, how a robust microbial metabolic network can be established to sustain the targeted performance, and how process failures can be prevented or overcome.Systems biology is widely applied to many engineering fields using pure cultures (e.g., metabolic engineering, industrial biotechnology, medical biotechnology and food biotechnology),35 but yet not fully exploited in the water sector. The complexity of systems and microbial communities and the lack of standardization in biomolecular methods and of bioanalytical training for environmental engineers hold back their application.
More than 30 methods are available in microbial ecology to elucidate microbial communities from populations to functions depending on investigative questions.32,34,36 Microscopy techniques are widely accepted by bringing visual information on predominant microorganisms. Systematics on morphotype observations are efficient to detect microorganisms that lead to, e.g., unfavorable deterioration of sludge properties by filamentous or viscous bulking and foaming.37–42 Microscopy, fluorescence in situ hybridization (FISH), and modern amplicon sequencing are frequently used in applied research. Implementation of higher-resolution molecular methods in engineering remains a challenge.
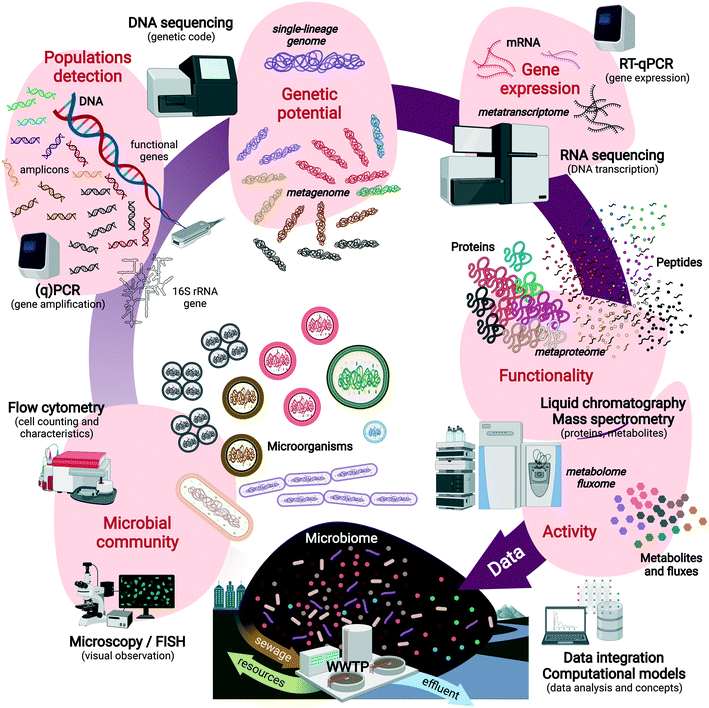
Fig. 1 displays the sequence of compositional and functional analyses that can be performed on engineered microbiomes with increasing resolution across informational (macro)molecules from DNA to RNA, proteins, metabolites and their fluxes. These can help predict the metabolic functions of single populations to microbial guilds (e.g., nitrifiers, denitrifiers, polyphosphate-accumulating organisms – PAOs), and microbial communities. Multi-omics techniques are powerful to analyze the informational content of these biomarkers, inside and outside the cells (Text box 1). Culture independence, sequencing depth, high throughput, and high resolution are strong advantages to reveal microbial community structures and capture the main metabolizers but also the low-abundance populations and their functions.
Multi-omics data help systems microbiologists capture microbial processes regulated in a microbiome at high resolution, but are not always easily linked to system performances, and therefore not considered by engineers. Besides analytical challenges related to representatively extract DNA, RNA, proteins or metabolites from the biomass matrix, the lack of joint concepts underlies the integration failure of the fields. Numerical ecology and multivariate analyses can help to this end.43 Data generation in many cases plays a leading exploratory role, rather than a supporting role for process design. Molecular datasets need to be converted into good information for factual utilization in engineering. This implies (i) the translation of data into useful concepts and models for practice (e.g., nitrification modeling, section 3.1) as well as (ii) the inception of engineering concepts into systems microbiology (e.g., controlling N2O emission, section 3.5). The inter-related effects of operational variables, microbial community compositions, and metabolic functionalities can be understood and managed on a process. Some implementation examples are discussed in section 3.
From the perspective of utilities and the private sector (e.g., environmental biotechnology companies), expectations on specific analyses and data that need to be collected to better manage the treatment processes need to be formulated. The gains of combining time series of (high-resolution) chemical and microbiological data (properly interpreted and translated) together with installation knowledge, operational patterns and process performances need to be addressed.
Text box 1. Glossary of “omics” techniques analyzing informational molecules from microorganismsAmplicon sequencing of ‘universal’ ribosomal RNA (rRNA) genes and functional marker genes is used to fingerprint microbial communities at high resolution and throughput, screen for populations and their relative abundance, and pattern their temporal and spatial dynamics.Metagenomics is used to fully sequence the pool of genomic DNA (gDNA) extracted from the microbial community in order to identify the functional genes and metabolic potential harbored by biomasses. Obtaining genomes of single populations as functional units of the microbial ecosystem is a target. Genome-centric metagenomics via binning and assembling single-lineage genomes from metagenomes44–46 (so-called metagenome-assembled genomes – MAGs) help address the functional potential of target microorganisms out of the microbiome, to predict their metabolic pathways and interactions with other populations. Metatranscriptomics involves the sequencing of the pool of messenger RNA (mRNA) to reveal gene expression patterns in a microbiome. It analyzes the expression of functional genes and indicates which genetic operons have been activated or repressed under biosystem regime shifts at fixed moments. Metaproteomics provides high-resolution characterization of the pool of proteins present in the microbial cells of a microbial community by liquid chromatography and tandem mass spectrometry. Targeted and untargeted approaches can be used to measure specific signatures or the full profiles. Proteins inform on the expressed functional properties of the organisms, on top of phylogenies. Protein activities remain to be unraveled for additional functional clues on the microorganisms. Metabolomics and fluxomics are implemented to analyze intracellular and extracellular metabolites, their dynamic concentration patterns, and fluxes generated by the conversions catalyzed by the microorganisms, for a high-resolution analysis of the metabolic state and activity of the microorganisms. However, they are not sufficient alone to understand the phylogenetic and metabolic complexity of microbial systems. For a complete picture of metabolic regulation patterns, molecular and omics techniques need plural integration, and cross-validation with ecophysiological techniques. All methods require strong and curated databases and evolved computational algorithms to process and visualize the data sets. Investigations should go beyond data generation in order to translate the “big data” into “good data” useful for scientific and engineering concepts and designs. A combination of these methods with classical microbiology, microbial ecology, ecophysiology, and analytical chemistry helps resolve microorganisms in their community and environmental contexts. |
3 Integrating microbial ecology principles and systems microbiology methods with wastewater engineering: successes and emerging topics
Biological nutrient removal (BNR) and anaerobic digestion highlight how process performances do rely on an efficient metabolic connection in microbial communities. A set of latest scientific illustrations integrating microbial ecology, systems microbiology, and wastewater engineering is provided to highlight elucidations of populations, functionalities, and metabolic regulations inside sludges. Translation at pilot and full scales is needed as a proof of success beyond the scientific concept.47,48 Scaling up the investigations will deliver the limits of operability of methods and the true analytical information they can deliver to practitioners. Experimental and analytical designs need to get adapted to the scale. Bringing methods at/on/in line should help capture and manage microbial processes in real time along with process control. Substantial developments are needed to implement advanced molecular methods into process analytical technology (PAT). PAT is used by the biotechnology industry to sustain quality by design and quality by control.49–52 For WRFs, the approaches can be applied to, e.g., control BNR processes, prevent emissions of unfavorable greenhouse gases, monitor and abate biocontaminants like antimicrobial resistance (AMR). Full-scale implementation concepts are given as well.3.1 Engineering BNR granular sludge processes using microbial ecology principles
One leading illustration of successful implementation of microbial ecology principles in engineering design relates to the development of intensified processes using granular sludge for high-rate BNR from wastewater.29 The success of BNR granular sludge processes (or commercially called “aerobic” granular sludge by contrasting to “anaerobic” granular sludge for anaerobic digestion) links to a high degree of interaction across bench, pilot, and full scales between biotechnologists, microbial ecologists, environmental engineers, and water authorities.BNR granular sludge has been early investigated by elucidating bulking activated sludge problems, and considering principles of biofilm formation under wash-out dynamics in bioreactors.53–56 Transitioning from the physical granulation phenomenon to the integration of metabolic conversions for C–N–P removal went step-wise through a sound understanding of phenomena of microbial physiology, microbial selection, and microbial niche establishment in biofilm matrices linked to anaerobic–aerobic sequencing batch reactor (SBR) engineering.57–60 Investigations moved relatively fast from bench to pilot scales, while challenges remained. Microbial ecology made a key contribution to elucidating why metabolic activities for full BNR have been lost during reactor start-ups,61,62 how PAO/GAO competition can be managed under baseline operation for robust BNR and EBPR,43,63–67 how alternating nitrification and denitrification (AND) patterns can become an important alternative to simultaneous nitrification and denitrification (SND),68 and how N2O emissions can be managed in this context.69,70 Although granulation processes and designs have been patented, granule engineering brought microbiological science on board internationally. Investigations have been performed to understand the macro/meso/micro-scale relationships. The combination of laser scanning microscopy and fluorescence in situ hybridization has been effective to unravel the dynamic microbial architectures of granule cross-sections and the phenotypic links with the predominant populations selected.71–73 High-resolution analyses of microbial population dynamics helped to capture the links between operation conditions, process performance, and variations in community compositions.43 Amplicon sequencing helped develop a detailed conceptual model of the microbial ecosystem of BNR granular sludge, as a basis for functional analyses within the microbiome.29,34,43
These thorough systems microbiology insights combined to engineering and modelling have provided keys for the development of applied methodologies to manage the microbial resource in BNR granular sludge SBRs.74 Their integration was substantially discussed with practitioners, helping practice to make steps forward a better process design, operation, and control.10,75–77 Interestingly, ecological engineering helped to bring back the granulation knowledge as a biomass densification strategy to solve filamentous bulking in activated sludges used to treat industrial wastewater.78,79 Full-scale BNR granular sludge processes are getting well established in the wastewater engineering sector,80–82 thanks to a strong interaction between engineering and microbial ecology principles. Provincial water authorities provided ground for an interactive testing of the technology.
3.2 Integrating impacts of immigrating sewer communities on nitrification in activated sludge
The natural bioaugmentation of nitrifiers in activated sludge can substantially impact process sizing and operation reliability.83 A combined survey of sewage in Canada and UK has revealed through 16S rRNA gene amplicon sequencing the presence of nitrifiers in influent wastewater when the sewers were aerobic, and how the nitrifiers immigrating from the influent populated the activated sludge.84,85According to amplicon sequencing of functional genes (amoA of ammonium oxidizers, nxrB of Nitrospira-related nitrite oxidizers), the nitrifying population structures have been highly similar in the influent and in the mixed liquor, proving that influent nitrifiers are efficiently established in the mixed liquor.84 The importance of bioaugmentation, mainly for plants operated near or beyond washout conditions (low temperature, low solid retention time – SRT),83 has been proven in lab-scale and full-scale bioreactors.84 Implementing amplicon sequencing data in activated sludge modeling has highlighted the contribution of influent nitrifying populations to activated sludge.
These investigations have provided solid foundations to implement a more aggressive design/modeling approach toward reducing costs and improving simulation accuracy.
3.3 Developing low-temperature nitrifying processes for northern climate applications
Under temperate and cold climates, the low wastewater temperatures in winter (as low as 1 °C in, e.g., Northern Canada) are detrimental to nitrifying populations. In lagoon-based passive treatment facilities, ammonium is discharged without sufficient treatment into surface water.A multi-level approach has helped unravel, anticipate and remediate the impacts of low temperatures and accelerate the design of performant alternative treatment processes, e.g., moving bed biofilm reactors (MBBRs).86,87 At the macro scale, the performance and kinetics of laboratory and pilot plants have assisted the design and optimization for an extensive nitrification at 1 °C. At the meso scale, stereomicroscopy and scanning electron microscopy (SEM) have been used to analyze the morphology of nitrifying biofilms. Optical coherence tomography (OCT) can further unravel the external architectures of biofilms. At the micro scale, FISH has allowed the relative abundance to be semi-quantified and the 3-D distribution of nitrifiers in biofilms to be visualized, combined with cell-viability tests and confocal laser scanning microscopy (CLSM). At the molecular scale, 16S rRNA gene amplicon sequencing has elucidated the bacterial community compositions, diversity, and shift under psychrophilic conditions.
The multi-scale information gained from psychrophilic nitrifying biofilms has sustained the operation of MBBR technologies for nitrification under extended periods at low temperatures.86,87
3.4 Controlling PAOs and GAOs in EBPR wastewater treatment processes
Enhanced biological phosphorus removal (EBPR) relies on the abundance and activity of PAOs. The over-proliferation of glycogen-accumulating (GAOs) competitors can hamper EBPR. Process parameters like pH, temperature, and nutrient ratios govern the PAO/GAO selection.63,88,89 Higher tropical temperatures (about 30 °C) support GAO growth. Controlling GAOs across process variations remains a challenge.Microbial population dynamics have been studied by amplicon sequencing of more than 400 samples collected over 9 years in 24 full-scale EBPR plants in Denmark to reveal connections between process design and operation and PAO/GAO competition patterns.90 Integrating a sidestream process unit configured for hydrolysis and pre-fermentation of the return sludge has stabilized a PAO-enriched microbial community by out-selecting GAOs up to temperatures as high as 30 °C.
Multi-year and high-resolution analyses of EBPR communities in multiple WRFs have identified general trends, correlated process conditions and microbial state variables, and derived key microbial parameters in the treatment process. Bringing analytical technologies into practice for routine monitoring can provide fast information to operators for fine tuning process control. The availability of curated molecular databases (e.g., 16S rRNA genes and high-quality metagenome-assembled genomes – MAGs) such as MiDAS for activated sludge and anaerobic digesters91,92 is important for microbial referencing and inter-comparison of WRF systems. Systems microbiology delivers answers to the list of key microbiological information needed for process design, operation, monitoring and control. Microbiological and molecular measurements provide sensitivity and enable early warning of process disturbances and recovery. The development of WRF dashboards may help operators and regional water authorities compare, diagnose, and troubleshoot their installations from the microbiological standpoint. It facilitates exchange of information and expertise via professional peer learning.
3.5 Tracking denitrifying PAOs in integrated BNR processes
The combined removal of COD, N and P from wastewater is increasingly important. Denitrifying PAOs (DPAOs) are capable of P-uptake under denitrifying anoxic conditions, but are prone to emissions of greenhouse nitrous oxide (N2O) when denitrification is incomplete.68,93Mechanisms of N2O formation by DPAOs have been investigated using 16S rRNA gene amplicon sequencing, genome-resolved metagenomics, and long-term kinetic analyses.94,95 An enrichment of the as-yet-uncultivated PAO “Candidatus Accumulibacter phosphatis” has reduced NO2− to N2O (70–80%) via incomplete denitrification. According to MAGs, the “Ca. Accumulibacter” enrichment was composed of two clades IA and IC, encoding genes for complete and incomplete denitrification, respectively. In another study, metabolic tests in bioreactors combined with genome-centric metagenomics have revealed that the clade IC “Ca. Accumulibacter delftensis”96 does not remove phosphorus using nitrate, but mainly using nitrite as an electron acceptor. This contrasted with common engineering correlations made on enrichment cultures of PAO I clades and nitrogen reduction conversions,89,97 stressing the need to revise clades definitions.
Insights into genomic and environmental factors underlying nitrogen-based catabolisms and N2O emissions by DPAOs have resolved the community structure and denitrification pathways of DPAO cultures, providing knowledge to design strategies to control denitrifying EBPR processes, and prevent unfavorable N2O emissions.
3.6 Solving N2O emission during partial nitritation and anammox with metatranscriptomics
PN/A is an attractive mixed-culture process that propels a complete autotrophic removal of nitrogen at low energetic and resource expenditures. The PN/A robustness relies on a close synergy of active aerobic ammonium-oxidizing (AOOs) and anammox (AMOs) organisms. Alternating redox conditions or temperature decrease are known to unfavorably lead to N2O emission.Multi-level analyses of nitrogen conversions, as well as bacterial community compositions by 16S rRNA gene amplicon sequencing, genome-centric metagenomics, and metatranscriptomics have been used to elucidate gene expression and transcriptional regulation patterns across populations in PN/A systems subjected to temperature shifts. This has helped identify misregulation of catabolic pathways in the nitrogen cycle, leading to undesired NO2− accumulation in the liquid phase and N2O emission in the off-gas (Weissbrodt et al., in prep.).98,99 Populations accounting for more than 30% of misregulated transcripts have been affiliated with AOOs, AMOs and denitrifying heterotrophic organisms (DHOs) along aerobic ammonium oxidation (up-regulated), anammox (down-regulated), and nitric oxide (NO) reduction (up-regulated) and nitrous oxide reduction (down-regulated) pathways, respectively.
Metatranscriptomics has helped identify populations and metabolisms impacted by lower temperature conditions, and propose solutions to engineer strategies for maintaining a PN/A process balance while preventing N2O emissions.
3.7 Quantifying the metabolic network fluxes of anammox bacteria with isotope tracing and metabolomics
AMOs play a central role in global nitrogen cycling and mediate energy-efficient wastewater treatment processes for autotrophic nitrogen removal. Information of their central metabolism beyond genomic predictions is needed to construct accurate metabolic models that can be used to predict their substrate utilization rates, product secretion rates, and metabolic interactions in BNR processes.Metabolic flux analysis that couples stable isotope tracing and metabolomics with computational modeling has helped predict fluxes along conversion pathways active in “Ca. Kuenenia stuttgartiensis”, fed with 13C-labeled bicarbonate.100 Intracellular metabolite samples have been measured via high-resolution liquid chromatography and mass spectrometry (LC-MS) to follow 13C incorporation into the metabolome. Metabolite labelling patterns via isotopic non-stationary metabolic flux analysis have been analyzed to compute intracellular fluxes and resolve the central carbon metabolic network. This has allowed tracking novel deviations in central metabolism beyond the initial genome-scale model, highlighting the importance of measuring fluxes experimentally.
Extending metabolic flux analysis to microbial communities enables a detailed quantification of microbiome metabolic fluxes and interactions, providing a platform for linking intracellular pathway stoichiometry and kinetics with process level stoichiometry and rates.35
3.8 Managing the microbial resource of anaerobic digestion
Optimal management of anaerobic digesters relies on collaborative strategies. Current control of digesters is largely based on empirical knowledge of operational and chemical parameters. However, the monitoring of intermediate degradation products does not provide real-time information about the state of the digester as it does not account for the microbial community.Analysis of microbial communities using 16S rRNA gene amplicon sequencing provides a wealth of information to develop novel microbial management tools. A collection of microbial community data from 50 full-scale and some lab-scale anaerobic digesters has informed reactor performance, operational and microbial stability, responses to various perturbations and indicators of malfunctions, inhibitions, substrate preferences and operational changes.101 Substrate types (manure, food waste, wastewater sludge), temperature variations, and starvation periods affect microbial community composition. Genome-centric metagenomics, metaproteomics, and substrate labelling techniques like stable isotope probing (SIP) now provide interesting grounds to elucidate metabolic functionalities distributed across the microbial communities of anaerobic digesters (and wastewater treatment processes) and their functional variations under operational regime shifts.102–108
Practical application of microbial community data has the potential to frame a holistic management of the microbial resource in anaerobic digesters.109 It paves the way for targeted studies to attain a thorough understanding of the anaerobic digestion process from the microbial standpoint.
3.9 Monitoring pathogens and antimicrobial resistance in urban water systems
Sewer systems and WWTPs have been first designed for sanitation by sewage collection and treatment to protect public health, the aquatic environment, and natural water resources. Biological contaminants like pathogens and AMR require surveillance by analytical monitoring.110–113 An important set of investigations is needed to address their fate and removal from wastewater.114 This becomes even more important for the safety of water reclamation and reuse.115–117Numerous investigations are currently performed to monitor AMR in sewage, across WRF process units, and in effluents discharged into surface waters. AMR results from the overconsumption of antibiotics via drugs and food and their emission into water catchment areas via municipal, hospital, industrial and agricultural sewage.118 Activated sludges are perceived as hotspots for the proliferation of AMR.
A battery of molecular methods is employed to track antibiotic resistant bacteria (ARB), antibiotic resistance genes (ARGs), and mobile genetic elements (MGEs) in wastewater. qPCR surveys in the Netherlands of a selected panel of ARGs and MGEs from (i) grab samples of influents and effluents of more than 60 WRFs119 and from (ii) yearly time series across process units of 3 representative WRFs120 have shown that WRFs do not amplify the AMR phenomenon. But still, on average 106 ARG copies are present per liter of effluent. Water authorities raise questions on whether this level poses an environmental and health risk. ARG levels in effluents are larger under rain events, because of higher discharge of suspended solids in effluents. The hydraulic loading of WRFs has a significant impact on microbial separation efficiency in secondary clarifiers. Besides end-of-pipe technologies,121–123 minimization of emissions at the source is an efficient mitigation method.124,125 Decentralized treatment of blackwater by anaerobic digestion has shown the positive effect of micro-aeration dosage at a higher level (150 mg O2 g−1 CODfeed per reactor cycle) to control ARG release.126 Metagenomics is becoming widely applied at a full scale to address the composition and fate of ARGs and MGEs from the intracellular and “free-floating” extracellular fraction of wastewater microorganisms.127,128
The design of legislations on AMR emissions from wastewater discharge from catchment areas still requires an accurate evaluation of exposures and risks. Deploying scalable bioanalytical methods will become an essential tool of water authorities for at-line surveillance of pathogens and AMR and for determining the efficacy of preventative measures.
4 Identifying the problem for data usage: interfacing tools to clear the black box
4.1 Bring researchers on the problems that can be solved today: diagnostics, designs, models
The integration of molecular techniques, with their high throughput and resolution, is efficient to screen microbial communities at different scales. Targeting specific biomarkers allows to address the health state of the cultures and diagnose the populations. A surveillance software platform using DNA sequence data of a microbial community can inform the dynamics of known functional groups (e.g., filaments and nitrifiers) and provide early warning and operational recommendations.129 A fast detection of microbiome status can drive a prompt response for its management versus the unavoidable fluctuating environmental and operational factors. Deep knowledge sheds light on the microbiological black box: a clearer idea of the components, network, and functionalities sustains a better informed process design, monitoring, and control. Metabolic models derived from wet-lab and dry-lab molecular analyses can predict the selection, physiology, metabolism and cellular regulation of predominant and low-abundant microbial populations and guilds, as well as the functional performance of the microbial system as a whole.89,100 Metabolic modelling provides predictive power on how an engineered microbiome might react and respond to environmental changes.35 Such prediction can reduce the response time to detect a stress event, to anticipate process failure, and to design feed-forward control strategies.Anaerobic digestion offers a leading illustration. This versatile technology has exhibited huge potential as a sustainable energy source, but plateaued in terms of system optimization and operation.130–134 An on-site/online platform for continuously monitoring the microbial population dynamics on top of metabolic conversions and process variables of full-scale systems will be informative about the microbiome responses under both stable periods and (un)planned perturbations. This could suggest operational strategies to maintain system stability or improve performance. The definition of common objectives based on overlapping interest will allow the capitalization of collaborations between engineers and microbiologists, resulting in improving the management of digesters and WRFs.
4.2 Interfacing tools to address future key challenges
An efficient interaction should help achieve a net positive energy in WRFs, to remove or recover nutrients more efficiently, to minimize emissions of greenhouse gases, and to address solutions to eliminate chemical and biological contaminants that emerged like micropollutants and antibiotic resistance.10 Process intensification and integration is needed to reduce the footprint of WRFs while meeting the emission targets and extending engineering methods to prevent their unfavorable impacts on environmental and public health.Many of the identified challenges for engineering practice (e.g., enhancing resource recovery, achieving net positive energy, minimizing greenhouse gas emission) can be addressed by using the tools for microbial ecology to unravel microbial diversity, selection and population dynamics, functional interactions and disruptions. Advanced analytical methods can fingerprint contaminants and track biological processes for their optimal conversions into harmless or even valuable products. Engineering methods are the keys to solve anthropogenic burdens on top of minimization at the source. Microbiological science and process engineering cannot substitute each other but should thrive on each other's expertise to develop technology-oriented solutions.32 Besides generation of knowledge and concepts, integrative approaches stimulate innovation and maintain the momentum for a continuous improvement.
Advanced physicochemical and systems microbiology analyses deliver a mass of data at high resolution on the system under investigation. Such data enable the consolidation of knowledge from the WRF process units underlying their global performances. As in most industrial branches, the digital hub is moving the water sector.135 Key questions to address in this context are how can data processing shift the water engineering profession and what new competences are needed to be developed136 for processing, visualizing and integrating large datasets into process understanding and control. The complex data needs to be translated into simple and specific actionable data that can be used in daily practice for operation of full-scale plants. This includes, e.g., the definition of threshold values and courses of action that need to be taken if these are exceeded.
5 Focusing on systems: engineering boundaries of investigations
5.1 From observation and description to detailed experimentation and models for new technologies
Microbial communities have transformative, limitless capabilities. They drive the Earth's biogeochemical cycles and are versatile enough to occupy every environmental niche. Even though environmental engineers have exploited their metabolic power for over a century, the majority of these transformative capabilities have yet to be unlocked and harnessed in applications. To create a next generation of WRFs, the metabolic and ecological networks underlying microbiomes have to be understood and predicted. This is currently hindered by the intrinsic complexity of microbiomes and the lack of quantitative methods and engineering tools to rationally analyze and manipulate microbiomes.Transitioning from observational and descriptive studies to detailed experimentation with simplified yet representative model systems will deliver the knowledge and tools needed to investigate and harness the complexity of microbial communities. This will enable more rigorous hypothesis generation and examination. Bridging the gap between model and full-scale systems requires close collaboration between researchers and practitioners. The cooperation will benefit both sides, by fostering scientific discovery and technology transfer and by spurring disruptive innovation in the water sector.
5.2 Fast-throughput methods and specific markers to learn and predict the systems
Full-scale facilities require fast and accurate results for a prompt reaction to operational changes. Systems are the foci. Novel molecular techniques provide an extensive set of information that can be integrated in mathematical models to predict microbial selection and activity, to better control processes. Bioanalytical methods with a fast response time should accelerate testing periods to stress and train biosystems under operational constraints.Analytical deployment for on-site and rapid infield monitoring (although not yet in real time) is an objective to minimize the buffer period before delivering the data and actuating controls. Current applications target water quality137 and coliform/pathogen monitoring (e.g., E. coli, Bacteroides, Enterococcus, Legionella)113 and are developed for surveillance of important biological parameters of activated sludges138 like foaming/bulking sludge, sensitive nitrifiers, or antibiotic resistance.112 Portable sequencing technologies like Nanopore can deliver on-site and rapid results on purpose. The mobile sequencing technology is there, while at/in-line implementation and real-time data processing are key objectives to fulfill on the next years. Innovations are required to integrate molecular methods into PAT in order to shape a fast response on the system status and to refine the full-scale operations of WRFs.
Fast analytical pipelines should enable a rapid accumulation of insightful observations that can lead to a more efficient process development and control. Similar to enzymatic assays used to evaluate the presence of E. coli in drinking water treatment plants and to flow cytometry for water quality control, on-site and real-time identification of pathogen strains can help control (waste)water-based epidemics.
Fast-throughput tools require specific markers, representative for the system. Suitable markers can be identified by digging into datasets obtained upfront from higher-throughput omics measurements. Recent techniques allow for screening communities at different scales. Markers can be identified for each level of resolution.32 The definition of such markers, useful from both scientists' and practitioners' sides, will facilitate the communication. The harmonization of parameters useful to check, monitor and control microbiome performances inside process boundaries will help develop knowledge and designs.
5.3 Development of a common vocabulary: the “language of love” between microbiologists and engineers
Microbiologists and engineers need to enhance communication to achieve joint benefits. A common language has to be established, through the standardization of protocols and definition of adequate dimensions, units and conversion factors in order to jointly approach problems and questions. Relative units have been used to describe microbial community compositions or gene expression levels, but are a limiting factor for the integration into process modelling which thrives on quantitative mass balances. Efforts have been initiated on integrating sequencing data and operational parameters to mass-balance modeling,139,140 and on near real-time surveillance through graphic interpretation of sequencing data.129 The format/readability of modeling output is particularly important, which can be a starting point of communication. The predictive promises of microbial ecology methods should embrace the scales of observations (temporal, spatial, and molecular) going beyond the homogenization of the units.Once a common vocabulary is created and common needs are identified, the molecular methods become powerful tools to complement and improve engineering practices, to explore and describe the interactions between process configurations and ecophysiology of microbial populations, and to sustain innovation in environmental biotechnologies.
The added value brought by strengthened interaction between engineering and systems microbiology can be exploited if mutual benefits are clear. Communication and mutual understanding becomes crucial.
5.4 Infiltrating the industry: education and new opportunities for cooperation
Education of operators to bioanalytical approaches and of microbiologists to engineering designs and operations is a second step toward an effective collaboration. Educating means forming experts who can think interdisciplinarily and have a comprehensive idea of the benefits of the interaction, and who work together to solve problems and implement research, adding novel knowledge and expertise. Developing joint courses and projects is required in education programs,136 such as by combining molecular biology and mathematical modelling. Interaction between systems microbiology and the wastewater industry is encouraged and pushed forward by governmental funding agencies. World-wide, national science foundations do grant several types of funding to bridge research and business, thus providing resources for training future talents in an academic-industrial context and translating invention into innovation and capital. Programs support short-term and long-term innovation projects to solve difficulties encountered in engineering practices and accelerate the integration of innovative technologies from lab-scale to existing industrial systems. Future initiatives should reach even bigger perspectives to connect other industries and agencies with built infrastructure.136 Implementing integrated approaches helps utilize knowledge to develop more circular, zero waste, zero liquid discharge, and upcycling approaches, with the assistance of life cycle assessment and techno-economic modeling.Overall, the bridge between the physics, chemistry and biology of WRF processes is the key. The black-box growth stoichiometry (relates to OPEX in process economics) and kinetics (relates to CAPEX) of environmental biotechnologies can now be efficiently complemented with modern approaches of systems microbiology.32,34 In the present life sciences era, novel wet-lab and dry-lab bioanalytical technologies deliver power to address the microbiological underpinnings at high resolution and throughput. It should now be addressed how systems microbiology can be used in practice to assist design, monitoring, control, and how the method implementation can help reduce CAPEX and OPEX. To convince practitioners of the well-founded use of microbiological information, one should answer: how does microbial ecology and systems microbiology help improve CAPEX and OPEX?
Beyond the data, novel analytical investigation methods drive innovation in the water sector, making it attractive for new digital generations beyond the urgency to safeguard and valorize water resources. At the water authority level, new bioanalytical technologies deliver keys for new legislations either based on risk assessment or prevention and precaution principles. However, safeguarding the environment and human health is not only about legislation. Critical and creative thinking should go beyond legislation to actively seek solutions to achieve water and resource protection. In the Netherlands, provincial water authorities provide an interactive ground to rapidly implement lab inventions into water innovations.
5.5 Ways of collaboration
The implementation success of innovative WRF processes thrives on academic–public–private partnerships (e.g., university–utility–company).136 The challenge consists of defining common goals and objectives to bridge the communication gap, to bring researchers and practitioners at the same table, and to acknowledge and handle selected problems, prior to clearly defining a joint workflow to address them. From a technology readiness and adoption standpoint, it is key to articulate who are customers (e.g., treatment technology developers, consulting engineers, plant managers, plant operators, regulators, etc.) and what are the unmet needs, to bring the right partners and stakeholders to invest their resources in order to advance technology readiness levels. The collaboration between systems microbiologist and engineers can be translated into various common goals, e.g., from method development to design, from knowledge collection to problem detection and solving, or from analytical to process technology innovation.Ways for collaboration include (i) problem-solving and solution driven, (ii) innovation implementation for resource/energy recovery, and (iii) adapting/redirecting ecological theories and modeling, which can be exemplified as follows. Disruptions in engineered system performance are crucial issues for engineers and are interesting topics for microbiologists. Engineers and microbiologists work together to find the root cause, understand the underlying mechanisms, and provide solutions. For instance, bulking and foaming in BNR activated sludge systems have been investigated using molecular tools and possible reasons have been identified by correlation to operational process signatures. Another aim can target the development of innovative technologies to achieve a more efficient recovery of resources and energy and to meet wastewater treatment challenges. This has been done to integrate shortcut nitrogen and biological phosphorus removal from municipal wastewater where researchers from different sectors have jointly developed process operation and modeling for a successful implementation.141 Coupled aerobic–anoxic nitrous decomposition operation (CANDO) has been developed to transform NH4+ to NO2− then to N2O and recover energy through N2O conversion to N2.142 Cooperation between utility and academia has shed light on the presence of newly discovered metabolisms such as complete ammonium oxidation (comammox) in a nitrification reactor operated at mainstream at low dissolved concentration.143 Engineered biological systems are often well controlled testing sites for microbial ecological models. Ecological niche and neutral theories139,140,144–146 have been tested using bioreactors as refined environments, providing the basis for the combination of deterministic models and probabilistic immigration models in order to manage the engineered biological systems.
6 Outlook: designing a roadmap for closer exchange
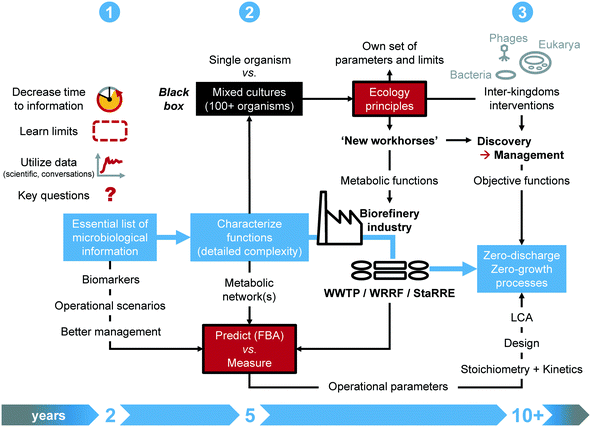
Addressing a closer collaboration between systems microbiology and wastewater engineering sounds appealing while concrete milestones have been lacking. We derived a roadmap to guide efforts, based on our interaction at the interface of science and engineering practice. The methodology (Fig. 2) targets 3 milestones on short (next 1–2 years), mid (next 5 years) and longer (next decade) terms.6.1 Milestones on short, mid and long terms
On short term, the time response of systems microbiology measurements should be substantially decreased from days/weeks/months to hours. Instead of generating a mass of data at high throughput, the identified functional biomarkers will enable fast-throughput diagnostics for monitoring, surveillance, and control. Accelerated testing designs will help push and learn the limits of the systems, and test operational scenarios. Currently, multi-year long-lasting lab-scale experiments are common. The underlying molecular measurements often remain descriptive rather than informational for engineering practice. Experimental designs should generate short-term information that is readily available for translation into engineering concepts.32,63 Targeted reflection should arise along with a sound formulation of investigations on microbiology and engineering axes prior to sampling, measuring, and generating data.The midterm milestone aims at infiltrating the industry with microbial ecology principles. Open mixed-culture environmental biotechnologies contrast to axenic pure-culture industrial biotechnologies. Although siloed over the last 50 years, the two fields harbor potential for strong connectivity. Mixed-culture approaches that thrive on microbiome investigation and engineering can deliver new workhorses for the industry. To achieve this, microbial ecology principles, parameters, and limits that govern microbiome engineering35 should be made explicit for engineering specialists. Key functional information from population and metabolic networks should be integrated into the microbial community black box. Quantitative and predictive methods, e.g., flux-balance analysis and machine learning, under development to investigate distributed metabolisms in microbiomes should become accessible to industrial engineering practice via translation into mathematical models. The definition of objective functions will drive mixed-culture biotechnology endpoints.
Long term initiatives for microbiome discovery should harness the performance of engineered microbiomes, by managing metabolic functions distributed across the microbiomes. By pushing forward the possibilities from environmental to industrial mixed-culture biotechnology, one should even identify opportunities to apply synthetic ecology with defined consortia and even directly genetically engineer microbiomes too from the perspective of recovering a diverse set of high-value molecules from wastes, since top-down microbial selection from complex environments has its limits.35,147 A closer look should be given to inter-kingdom interventions, like the use of grazing by eukaryotes or of bacteriophage therapy to shape microbial communities and biofilms, e.g., to remediate membrane biofouling. While current initiatives target more circular processes, the field of biological wastewater treatment may aim for zero-discharge, zero-emission and zero-growth processes. Along the digital informatics revolution, the massive collection of metadata from operational parameters should be bridged back to stoichiometry and kinetics fundamentals for factual integration into design. The current state of analytical methods and data processing pipelines render this objective challenging. Developments in chemometrics, online multivariate analyses, and machine learning enable the mining of data over complex time series and computation of quantitative parameters from process performances, in association with large-scale mathematical models.148–152 Finally, harmonized guidelines for life cycle assessments should become a standard to enhance the comparison of new-generation technologies.153,154
6.2 Outreach: industrial benefits and process economics
These concepts should be integrated in professional education136 for a better understanding of mutual benefits of interactive and plant-wide systems microbiology for the wastewater industry. Although primarily focusing on wastewater here, the concepts can be applied to any water system and mixed-culture biotechnology industry involving microbiomes. Water resources are important to address in the industry. Industrial manufacturing, power and food processing plants involve re-use of used water in cascade-based systems at the production process level or from the central WRF.155–159 Water quality is a key parameter of industrial processes. Advanced physical–chemical treatments are used to deliver different grades of water on purpose, to recover resources, or to foster zero liquid discharge, such as with membrane processes. Biofouling is a key biological challenge to solve and for which elucidation of microbiological processes is a key for mitigation.160–165 Beyond the WRF, unravelling microbiomes can deliver insights into human health risks associated with the recreational use of urban waters.166 Elucidating microbiomes is a further key component of the centralized/decentralized production, delivery, and biological stability of drinking water,137,167–174 and for which investigations molecular toolboxes form an important component.175,176Infiltrating systems microbiology concepts in the industry is the key to help manage beneficial and/or unwanted/harmful microbial processes that link to wastewater treatment, resource recovery, water reuse, and drinking water stability, among others. Implementing a “language of love” that fosters active communication and collaboration between microbiologists and engineers is essential. While systems microbiology jargon, methods and endpoints should be made more easily accessible in engineering practice, more engineering principles should immerse systems microbiology investigations. This should drive the translation of “big data” into informational “good data” useful for engineering concepts, plant-wide.
The deciding factor for effective integration of new discoveries in microbiological science and engineering by the (waste)water industry relates to process economics. The implementation cost is limiting practitioners and utilities from embracing novel concepts. Nutrient recovery processes are expensive and may only be implemented either in installation where the nutrient recovery (e.g., struvite) brings other benefits in operation (e.g., preventing struvite precipitation in pipes and digesters) or in progressive plants and/or if a market niche has been identified upfront to valorize/sell the recovered product. Deammonification processes are not widely implemented since the need for expensive sensor technology, process control, and qualified personnel offsets the gains on the nitrogen removal. Integration of more microbiological and bioanalytical sensing at the process line will only be implemented if leading to economic gains or to new regulatory endpoints on water quality (e.g., pathogens, antimicrobial resistance, viruses), besides widening knowledge and solving the microbiological black box.
Author contributions
Marta Cerruti, Bing Guo and David Weissbrodt outlined and wrote the manuscript, with scientific inputs and edits from the co-authors. David Weissbrodt and Marta Cerruti crafted the artworks. All the authors actively contributed to the conceptualization, development, discussion, and integration of the ideas exposed in the article from either scientific or industrial or both perspectives. The article integrates key outcomes of several international workshops organized by some of the authors on “Advances in Systems Microbiology to Inform Modeling and Operation of Nutrient Recovery and Removal Processes” (Frigon, Weissbrodt, Guo and Cerruti; IWA EcoSTP 2018), “Microbiological Methods for Waste and Water Resource Recovery” (Oosterkamp and Weissbrodt; MMWWRR 2017 in the TU Delft 175th Lustrum), and “Bridging Microbiome Science and Environmental Biotechnology” (Weissbrodt and Lawson; IWA MEWE 2016). We warmly thank Daniel Noguera from the University of Wisconsin-Madison, USA, for critical review of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This opinion article initiative was financially supported by a start-up package of the Department of Biotechnology at the TU Delft, The Netherlands (Prof. David G. Weissbrodt) and by internal fund at McGill University, Canada (Prof. Dominic Frigon). Prof. Bing Guo was supported by a FRQNT Postdoctoral Research Scholarship B3X, Canada, for research with the TU Delft. This academic-industrial initiative thrived on interactive discussions with the community of microbial ecologists and environmental engineers in three international workshops that we organized over the last five years at the 1st Symposium on Microbiological Methods for Waste and Water Resource Recovery (MMWWRR 2017, Delft, Netherlands), at the IWA Microbial Ecology in Water Engineering & Biofilms Joint Specialist Conference (MEWE 2016, Copenhagen, Denmark), and at the 4th Specialized International Conference on Ecotechnologies for Wastewater Treatment (EcoSTP 2018, London, Ontario, Canada). We thank the conference delegates for their active participation and exchange on the principles motivated by this article.References
- D. Jenkins and J. Wanner, Activated Sludge - 100 Years and Counting, IWA Publishing, London, 2014, p. 464 Search PubMed.
- M. C. M. van Loosdrecht and D. Brdjanovic, Anticipating the next century of wastewater treatment, Science, 2014, 344(6191), 1452–1453 CrossRef CAS PubMed.
- G. T. Daigger, Flexibility and adaptability: Essential elements of the WRRF of the future, Water Pract. Technol., 2017, 12(1), 156–165 CrossRef.
- J. S. Guest, S. J. Skerlos, J. L. Barnard, M. B. Beck, G. T. Daigger, H. Hilger, S. J. Jackson, K. Karvazy, L. Kelly, L. Macpherson, J. R. Mihelcic, A. Pramanik, L. Raskin, M. C. M. Van Loosdrecht, D. Yeh and N. G. Love, A new planning and design paradigm to achieve sustainable resource recovery from wastewater, Environ. Sci. Technol., 2009, 43(16), 6126–6130 CrossRef CAS PubMed.
- W. Verstraete and S. E. Vlaeminck, ZeroWasteWater: Short-cycling of wastewater resources for sustainable cities of the future, Int. J. Sustain. Dev. World Ecol., 2011, 18(3), 253–264 CrossRef.
- P. A. Marrone, D. C. Elliott, J. M. Billing, R. T. Hallen, T. R. Hart, P. Kadota, J. C. Moeller, M. A. Randel and A. J. Schmidt, Bench-Scale Evaluation of Hydrothermal Processing Technology for Conversion of Wastewater Solids to Fuels, Water Environ. Res., 2018, 90(4), 329–342 CrossRef CAS PubMed.
- H. Bürgmann, D. Frigon, W. H. Gaze, C. M. Manaia, A. Pruden, A. C. Singer, B. F. Smets and T. Zhang, Water and sanitation: An essential battlefront in the war on antimicrobial resistance, FEMS Microbiol. Ecol., 2018, 94(9), fiy101 CrossRef PubMed.
- G. Filteau, C. Whitley and I. C. Watson, Water reclamation fuels economic growth in Harlingen, Texas. Reclamation of municipal wastewater for industrial process use, Desalination, 1995, 103(1–2), 31–37 CrossRef CAS.
- A. Scott, Tapping sewage as a source of useful materials, Chem. Eng. News, 2017, vol. 94, no. 47, https://cen.acs.org/articles/95/i47/Tapping-sewage-source-useful-materials.html Search PubMed.
- D. G. Weissbrodt, StaRRE - Stations de récupération des ressources de l'eau, Aqua Gas, 2018, 1, 20–24 Search PubMed.
- M. Cerruti, B. Stevens, S. Ebrahimi, A. Alloul, S. E. Vlaeminck and D. G. Weissbrodt, Enrichment and aggregation of purple non-sulfur bacteria in a mixed-culture sequencing-batch photobioreactor for biological nutrient removal from wastewater, Front. Bioeng. Biotechnol., 2020, 8, 557234 CrossRef PubMed.
- W. Verstraete, P. Clauwaert and S. E. Vlaeminck, Used water and nutrients: Recovery perspectives in a 'panta rhei' context, Bioresour. Technol., 2016, 215, 199–208 CrossRef CAS PubMed.
- A. Alloul, M. Cerruti, D. Adamczyk, D. G. Weissbrodt and S. E. Vlaeminck, Operational Strategies to Selectively Produce Purple Bacteria for Microbial Protein in Raceway Reactors, Environ. Sci. Technol., 2021, 55(12), 8278–8286 CrossRef CAS PubMed.
- W. Verstraete, The technological side of the microbiome, npj Biofilms Microbiomes, 2015, 1, 15001 CrossRef CAS PubMed.
- L. T. Angenent, K. Karim, M. H. Al-Dahhan, B. A. Wrenn and R. Domíguez-Espinosa, Production of bioenergy and biochemicals from industrial and agricultural wastewater, Trends Biotechnol., 2004, 22(9), 477–485 CrossRef CAS PubMed.
- Key reference: B. E. Rittmann, M. Hausner, F. Loffler, N. G. Love, G. Muyzer, S. Okabe, D. B. Oerther, J. Peccia, L. Raskin and M. Wagner, A vista for microbial ecology and environmental biotechnology, Environ. Sci. Technol., 2006, 40(4), 1096–1103 CrossRef PubMed.
- Key reference: W. Verstraete, Microbial ecology and environmental biotechnology, ISME J., 2007, 1(1), 4–8 CrossRef PubMed.
- C. Vaneeckhaute, E. Remigi, F. M. G. Tack, E. Meers, E. Belia and P. A. Vanrolleghem, Optimizing the configuration of integrated nutrient and energy recovery treatment trains: A new application of global sensitivity analysis to the generic nutrient recovery model (NRM) library, Bioresour. Technol., 2018, 269, 375–383 CrossRef CAS PubMed.
- P. A. Vanrolleghem, L. Benedetti and J. Meirlaen, Modelling and real-time control of the integrated urban wastewater system, Environ. Model. Softw., 2005, 20(4 SPEC. ISS), 427–442 CrossRef.
- U. Jeppsson, M. N. Pons, I. Nopens, J. Alex, J. B. Copp, K. V. Gernaey, C. Rosen, J. P. Steyer and P. A. Vanrolleghem, Benchmark simulation model no 2: General protocol and exploratory case studies, Water Sci. Technol., 2007, 56, 67–78 CrossRef CAS PubMed.
- Key reference: P. Grau, M. de Gracia, P. A. Vanrolleghem and E. Ayesa, A new plant-wide modelling methodology for WWTPs, Water Res., 2007, 41(19), 4357–4372 CrossRef CAS PubMed.
- E. I. P. Volcke, M. C. M. van Loosdrecht and P. A. Vanrolleghem, Continuity-based model interfacing for plant-wide simulation: A general approach, Water Res., 2006, 40(15), 2817–2828 CrossRef CAS PubMed.
- I. Nopens, D. J. Batstone, J. B. Copp, U. Jeppsson, E. Volcke, J. Alex and P. A. Vanrolleghem, An ASM/ADM model interface for dynamic plant-wide simulation, Water Res., 2009, 43(7), 1913–1923 CrossRef CAS PubMed.
- H. Hauduc, L. Rieger, A. Oehmen, M. C. M. van Loosdrecht, Y. Comeau, A. Héduit, P. A. Vanrolleghem and S. Gillot, Critical review of activated sludge modeling: State of process knowledge, modeling concepts, and limitations, Biotechnol. Bioeng., 2013, 110(1), 24–46 CrossRef CAS PubMed.
- L. Wu, D. Ning, B. Zhang, Y. Li, P. Zhang, X. Shan, Q. Zhang, M. Brown, Z. Li, J. D. Van Nostrand, F. Ling, N. Xiao, Y. Zhang, J. Vierheilig, G. F. Wells, Y. Yang, Y. Deng, Q. Tu, A. Wang, T. Zhang, Z. He, J. Keller, P. H. Nielsen, P. J. J. Alvarez, C. S. Criddle, M. Wagner, J. M. Tiedje, Q. He, T. P. Curtis, D. A. Stahl, L. Alvarez-Cohen, B. E. Rittmann, X. Wen, J. Zhou, D. Acevedo, M. Agullo-Barcelo, G. L. Andersen, J. C. de Araujo, K. Boehnke, P. Bond, C. B. Bott, P. Bovio, R. K. Brewster, F. Bux, A. Cabezas, L. Cabrol, S. Chen, C. Etchebehere, A. Ford, D. Frigon, J. S. GÃ3mez, J. S. Griffin, A. Z. Gu, M. Habagil, L. Hale, S. D. Hardeman, M. Harmon, H. Horn, Z. Hu, S. Jauffur, D. R. Johnson, A. Keucken, S. Kumari, C. D. Leal, L. A. Lebrun, J. Lee, M. Lee, Z. M. P. Lee, M. Li, X. Li, Y. Liu, R. G. Luthy, L. C. Mendonça-Hagler, F. G. R. de Menezes, A. J. Meyers, A. Mohebbi, A. Oehmen, A. Palmer, P. Parameswaran, J. Park, D. Patsch, V. Reginatto, F. L. de los Reyes III, A. Noyola, S. Rossetti, J. Sidhu, W. T. Sloan, K. Smith, O. V. de Sousa, K. Stephens, R. Tian, N. B. Tooker, D. De los Cobos Vasconcelos, S. Wakelin, B. Wang, J. E. Weaver, S. West, P. Wilmes, S. G. Woo, J. H. Wu, L. Wu, C. Xi, M. Xu, T. Yan, M. Yang, M. Young, H. Yue, Q. Zhang, W. Zhang, Y. Zhang and H. Zhou, Global Water Microbiome, C., Global diversity and biogeography of bacterial communities in wastewater treatment plants, Nat. Microbiol., 2019, 4(7), 1183–1195 CrossRef CAS PubMed.
- D. Frigon, E. Arnaiz, D. B. Oerther and L. Raskin, Who eats what? Classifying microbial populations based on diurnal profiles of rRNA levels, Water Sci. Technol., 2002, 46(1–2), 1–9 CAS.
- Key reference: E. Rodríguez, P. García-Encina, A. M. Stams, F. Maphosa and D. Sousa, Meta-omics approaches to understand and improve wastewater treatment systems, Rev. Environ. Sci. Bio/Technol., 2015, 14(3), 385–406 CrossRef.
- H. Aqeel, D. G. Weissbrodt, M. Cerruti, G. M. Wolfaardt, B.-M. Wilén and S. N. Liss, Drivers of bioaggregation from flocs to biofilms and granular sludge, Environ. Sci.: Water Res. Technol., 2019, 5, 2072–2089 RSC.
- Key reference: M. K. H. Winkler, C. Meunier, O. Henriet, J. Mahillon, M. E. Suarez-Ojeda, G. Del Moro, M. De Sanctis, C. Di Iaconi and D. G. Weissbrodt, An integrative review of granular sludge for the biological removal of nutrients and of recalcitrant organic matter from wastewater, Chem. Eng. J., 2018, 336, 489–502 CrossRef CAS.
- P. H. Nielsen, A. M. Saunders, A. A. Hansen, P. Larsen and J. L. Nielsen, Microbial communities involved in enhanced biological phosphorus removal from wastewater - a model system in environmental biotechnology, Curr. Opin. Biotechnol., 2012, 23(3), 452–459 CrossRef CAS PubMed.
- K. Zengler and B. O. Palsson, A road map for the development of community systems (CoSy) biology, Nat. Rev. Microbiol., 2012, 10(5), 366–372 CrossRef CAS PubMed.
- D. G. Weissbrodt, G. F. Wells, M. Laureni, S. Agrawal, R. Goel, G. Russo, Y. Men, D. Johnson, M. Christensson, S. Lackner, A. Joss, J. L. Nielsen, H. Bürgmann and E. Morgenroth, Systems Microbiology and Engineering of Aerobic-Anaerobic Ammonium Oxidation, ChemRxiv, 2020, DOI:10.26434/chemrxiv.12243077.v1.
- M. Buckley, Systems Microbiology: Beyong Microbial Genomics, Washington, D.C., USA, 2004 Search PubMed.
- D. G. Weissbrodt, M. Laureni, M. C. M. van Loosdrecht and Y. Comeau, Basic Microbiology and Metabolism, in Biological Wastewater Treatment: Principles, Modelling and Design, ed. G. H. Chen, M. C. M. van Loosdrecht, G. A. Ekama and D. Brdjanovic, IWA Publishing, London, 2nd edn, 2020 Search PubMed.
- Key reference: C. E. Lawson, W. R. Harcombe, R. Hatzenpichler, S. R. Lindemann, F. E. Löffler, M. A. O'Malley, H. García Martín, B. F. Pfleger, L. Raskin, O. S. Venturelli, D. G. Weissbrodt, D. R. Noguera and K. D. McMahon, Common principles and best practices for engineering microbiomes, Nat. Rev. Microbiol., 2019, 17, 725–741 CrossRef CAS PubMed.
- P. H. Nielsen and K. D. McMahon, Microbiology and microbial ecology of the activated sludge process, in Activated Sludge – 100 Years and Counting, ed. D. Jenkins and J. Wanner, IWA Publishing, London, UK, 2014, pp. 53–76 Search PubMed.
- Key reference: D. H. Eikelboom, Filamentous organisms observed in activated sludge, Water Res., 1975, 9(4), 365–388 CrossRef.
- D. H. Eikelboom, Process Control of Activated Sludge Plants by Microscopic Investigation, IWA Publishing, London, UK, 2000 Search PubMed.
- S. V. Ganapati and P. M. Amin, Microbiology of the viscous scum formed by certain Indian sewages and sewage effluents, Biochem. J., 1972, 128(1), 36P CrossRef CAS PubMed.
- E. N. Banadda, I. Y. Smets, R. Jenne and J. F. Van Impe, Predicting the onset of filamentous bulking in biological wastewater treatment systems by exploiting image analysis information, Bioprocess Biosyst. Eng., 2005, 27(5), 339–348 CrossRef CAS PubMed.
- Key reference: D. Frigon, R. Michael Guthrie, G. Timothy Bachman, J. Royer, B. Bailey and L. Raskin, Long-term analysis of a full-scale activated sludge wastewater treatment system exhibiting seasonal biological foaming, Water Res., 2006, 40(5), 990–1008 CrossRef CAS PubMed.
- M. K. H. Winkler, E. Kröber, W. W. Mohn, F. Koch and D. Frigon, Comparison of microbial populations and foaming dynamics in conventional versus membrane enhanced biological phosphorous removal systems, Water Environ. J., 2016, 30(1–2), 102–112 CrossRef CAS.
- D. G. Weissbrodt, N. Shani and C. Holliger, Linking bacterial population dynamics and nutrient removal in the granular sludge biofilm ecosystem engineered for wastewater treatment, FEMS Microbiol. Ecol., 2014, 88(3), 579–595 CrossRef CAS PubMed.
- M. Albertsen, P. Hugenholtz, A. Skarshewski, K. L. Nielsen, G. W. Tyson and P. H. Nielsen, Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes, Nat. Biotechnol., 2013, 31(6), 533–538 CrossRef CAS PubMed.
- D. H. Parks, C. Rinke, M. Chuvochina, P.-A. Chaumeil, B. J. Woodcroft, P. N. Evans, P. Hugenholtz and G. W. Tyson, Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life, Nat. Microbiol., 2017, 2(11), 1533–1542 CrossRef CAS PubMed.
- Key reference: C. M. Singleton, F. Petriglieri, J. M. Kristensen, R. H. Kirkegaard, T. Y. Michaelsen, M. H. Andersen, Z. Kondrotaite, S. M. Karst, M. S. Dueholm, P. H. Nielsen and M. Albertsen, Connecting structure to function with the recovery of over 1000 high-quality metagenome-assembled genomes from activated sludge using long-read sequencing, Nat. Commun., 2021, 12(1), 2009 CrossRef CAS PubMed.
- Key reference: M. Stokholm-Bjerregaard, S. J. McIlroy, M. Nierychlo, S. M. Karst, M. Albertsen and P. H. Nielsen, A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems, Front. Microbiol., 2017, 8, 718 CrossRef PubMed.
- J. D. Muñoz Sierra, M. J. Oosterkamp, W. Wang, H. Spanjers and J. B. van Lier, Comparative performance of upflow anaerobic sludge blanket reactor and anaerobic membrane bioreactor treating phenolic wastewater: Overcoming high salinity, Chem. Eng. J., 2019, 366, 480–490 CrossRef.
- S. M. Mercier, B. Diepenbroek, R. H. Wijffels and M. Streefland, Multivariate PAT solutions for biopharmaceutical cultivation: Current progress and limitations, Trends Biotechnol., 2014, 32(6), 329–336 CrossRef CAS PubMed.
- H. Narayanan, M. F. Luna, M. von Stosch, M. N. Cruz Bournazou, G. Polotti, M. Morbidelli, A. Butté and M. Sokolov, Bioprocessing in the Digital Age: The Role of Process Models, Biotechnol. J., 2020, 15, 1900172 CrossRef CAS PubMed.
- A. S. Rathore, QbD/PAT for bioprocessing: Moving from theory to implementation, Curr. Opin. Chem. Eng., 2014, 6, 1–8 CrossRef.
- W. Sommeregger, B. Sissolak, K. Kandra, M. von Stosch, M. Mayer and G. Striedner, Quality by control: Towards model predictive control of mammalian cell culture bioprocesses, Biotechnol. J., 2017, 12, 1600546 CrossRef PubMed.
- A. M. P. Martins, J. J. Heijnen and M. C. M. van Loosdrecht, Bulking sludge in biological nutrient removal systems, Biotechnol. Bioeng., 2004, 86(2), 125–135 CrossRef CAS PubMed.
- J. J. Beun, A. Hendriks, M. C. M. van Loosdrecht, E. Morgenroth, P. A. Wilderer and J. J. Heijnen, Aerobic granulation in a sequencing batch reactor, Water Res., 1999, 33(10), 2283–2290 CrossRef CAS.
- E. Morgenroth, T. Sherden, M. C. M. van Loosdrecht, J. J. Heijnen and P. A. Wilderer, Aerobic granular sludge in a sequencing batch reactor, Water Res., 1997, 31(12), 3191–3194 CrossRef CAS.
- L. Tijhuis, E. Rekswinkel, M. C. M. Van Loosdrecht and J. J. Heijnen, Dynamics of population and biofilm structure in the biofilm airlift suspension reactor for carbon and nitrogen removal, Water Sci. Technol., 1994, 29(10–11), 377–384 CrossRef CAS.
- M. de Kreuk, J. J. Heijnen and M. C. M. van Loosdrecht, Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge, Biotechnol. Bioeng., 2005, 90(6), 761–769 CrossRef CAS PubMed.
- J. J. Beun, J. J. Heijnen and M. C. M. van Loosdrecht, N-removal in a granular sludge sequencing batch airlift reactor, Biotechnol. Bioeng., 2001, 75(1), 82–92 CrossRef CAS PubMed.
- A. Mosquera-Corral, M. K. de Kreuk, J. J. Heijnen and M. C. M. van Loosdrecht, Effects of oxygen concentration on N-removal in an aerobic granular sludge reactor, Water Res., 2005, 39(12), 2676–2686 CrossRef CAS PubMed.
- L. M. M. de Bruin, M. K. de Kreuk, H. F. R. van der Roest, C. Uijterlinde and M. C. M. van Loosdrecht, Aerobic granular sludge technology: An alternative to activated sludge?, Water Sci. Technol., 2004, 49(11–12), 1–7 CAS.
- D. G. Weissbrodt, S. Lochmatter, S. Ebrahimi, P. Rossi, J. Maillard and C. Holliger, Bacterial selection during the formation of early-stage aerobic granules in wastewater treatment systems operated under wash-out dynamics, Front. Microbiol., 2012, 3, 332 Search PubMed.
- S. Lochmatter and C. Holliger, Optimization of operation conditions for the startup of aerobic granular sludge reactors biologically removing carbon, nitrogen, and phosphorous, Water Res., 2014, 59, 58–70 CrossRef CAS PubMed.
- D. G. Weissbrodt, G. S. Schneiter, J. M. Fürbringer and C. Holliger, Identification of trigger factors selecting for polyphosphate- and glycogen-accumulating organisms in aerobic granular sludge sequencing batch reactors, Water Res., 2013, 47(19), 7006–7018 CrossRef CAS PubMed.
- M. K. H. Winkler, J. P. Bassin, R. Kleerebezem, L. M. M. de Bruin, T. P. H. van den Brand and M. C. M. van Loosdrecht, Selective sludge removal in a segregated aerobic granular biomass system as a strategy to control PAO-GAO competition at high temperatures, Water Res., 2011, 45(11), 3291–3299 CrossRef CAS PubMed.
- D. G. Weissbrodt, C. Holliger and E. Morgenroth, Modeling hydraulic transport and anaerobic uptake by PAOs and GAOs during wastewater feeding in EBPR granular sludge reactors, Biotechnol. Bioeng., 2017, 114(8), 1688–1702 CrossRef CAS PubMed.
- O. Henriet, C. Meunier, P. Henry and J. Mahillon, Improving phosphorus removal in aerobic granular sludge processes through selective microbial management, Bioresour. Technol., 2016, 211, 298–306 CrossRef CAS PubMed.
- F. De Vleeschauwer, M. Caluwé, T. Dobbeleers, H. Stes, L. Dockx, F. Kiekens, J. D'Aes, C. Copot and J. Dries, Performance and stability of a dynamically controlled EBPR anaerobic/aerobic granular sludge reactor, Bioresour. Technol., 2019, 280, 151–157 CrossRef CAS PubMed.
- S. Lochmatter, G. Gonzalez-Gil and C. Holliger, Optimized aeration strategies for nitrogen and phosphorus removal with aerobic granular sludge, Water Res., 2013, 47(16), 6187–6197 CrossRef CAS PubMed.
- S. Lochmatter, J. Maillard and C. Holliger, Nitrogen removal over nitrite by aeration control in aerobic granular sludge sequencing batch reactors, Int. J. Environ. Res. Public Health, 2014, 11(7), 6955 CrossRef PubMed.
- T. Dobbeleers, M. Caluwé, D. Daens, L. Geuens and J. Dries, Evaluation of two start-up strategies to obtain nitrogen removal via nitrite and examination of the nitrous oxide emissions for different nitritation levels during the treatment of slaughterhouse wastewater, J. Chem. Technol. Biotechnol., 2018, 93(2), 569–576 CrossRef CAS.
- D. G. Weissbrodt, T. R. Neu, U. Kuhlicke, Y. Rappaz and C. Holliger, Assessment of bacterial and structural dynamics in aerobic granular biofilms, Front. Microbiol., 2013, 4, 175 Search PubMed.
- J. J. Barr, A. E. Cook and P. L. Bond, Granule formation mechanisms within an aerobic wastewater system for phosphorus removal, Appl. Environ. Microbiol., 2010, 76(22), 7588–7597 CrossRef CAS PubMed.
- J. B. Xavier, M. K. de Kreuk, C. Picioreanu and M. C. M. van Loosdrecht, Multi-scale individual-based model of microbial and byconversion dynamics in aerobic granular sludge, Environ. Sci. Technol., 2007, 41(18), 6410–6417 CrossRef CAS PubMed.
- D. G. Weissbrodt, Bacterial Resource Management for Nutrient Removal in Aerobic Granular Sludge Wastewater Treatment Systems, Ph.D. thesis, no. 5641, Ecole Polytechnique Fédérale de Lausanne, Switzerland, 2012, DOI:10.5075/epfl-thesis-5641.
- D. G. Weissbrodt, N. Derlon, E. Morgenroth and C. Holliger, A consolidated approach of flocculent and granular sludge systems under the perspective of bacterial resource management, Proceedings of the Water Environment Federation, 2014, 2014(19), 5008–5009 CrossRef.
- D. G. Weissbrodt and C. Holliger, Intensification du traitement biologique des eaux usées : Technologie à boues granulaires et Gestion des ressources bactériennes, Bulletin de l'ARPEA - Journal Romand de l'Environnement, 2013, 256, 12–22 Search PubMed.
- P. Regmi, C. DeBarbadillo and D. G. Weissbrodt, Biofilm Reactor Technology and Design, in Design of Water Resource Recovery Facilities - MOP 8, ed. T. L. Krause, et al., Water Environment Federation, American Society of Civil Engineers and Environmental and Water Resources Institute, McGraw-Hill Education, Alexandria VA, Reston VA, New York, USA, 6th edn, 2017 Search PubMed.
- Key reference: O. Henriet, C. Meunier, P. Henry and J. Mahillon, Filamentous bulking caused by Thiothrix species is efficiently controlled in full-scale wastewater treatment plants by implementing a sludge densification strategy, Sci. Rep., 2017, 7(1), 1430 CrossRef PubMed.
- M. Caluwé, T. Dobbeleers, J. D'Aes, S. Miele, V. Akkermans, D. Daens, L. Geuens, F. Kiekens, R. Blust and J. Dries, Formation of aerobic granular sludge during the treatment of petrochemical wastewater, Bioresour. Technol., 2017, 238, 559–567 CrossRef PubMed.
- A. Giesen, R. Niermans and M. C. M. van Loosdrecht, Aerobic granular biomass: the new standard for domestic and industrial wastewater treatment?, Water 21, 2012, vol. 4, pp. 28–30 Search PubMed.
- A. Giesen, M. van Loosdrecht, S. Robertson and B. de Bruin, Aerobic Granular Biomass Technology: further innovation, system development and design optimisation, Proceedings of the Water Environment Federation, 2015, 2015(16), 1897–1917 CrossRef.
- M. Pronk, M. K. de Kreuk, B. de Bruin, P. Kamminga, R. Kleerebezem and M. C. M. van Loosdrecht, Full scale performance of the aerobic granular sludge process for sewage treatment, Water Res., 2015, 84, 207–217 CrossRef CAS PubMed.
- S. Jauffur, S. Isazadeh and D. Frigon, Should activated sludge models consider influent seeding of nitrifiers? Field characterization of nitrifying bacteria, Water Sci. Technol., 2014, 70(9), 1526–1532 CrossRef CAS PubMed.
- M. Jauffur, Phylogenetic Characterization of the Nitrifying Populations in Municipal Wastewaters and in Biological Treatment Systems to Improve Modeling Practices, McGill University Libraries, 2016 Search PubMed.
- Key reference: B. Guo, C. Liu, C. Gibson and D. Frigon, Wastewater microbial community structure and functional traits change over short timescales, Sci. Total Environ., 2019, 662, 779–785 CrossRef CAS PubMed.
- R. Delatolla, B. Young, K. Kennedy, E. Laflamme and A. Stintzi, in MBBR technology as upgrade unit for low temperature nitrification: Microscopic and molecular analysis of biofilm morphology and bacterial communities at 1°C, 88th Annual Water Environment Federation Technical Exhibition and Conference, WEFTEC, 2015, p. 2015 Search PubMed.
- B. Young, R. Delatolla, K. Kennedy, E. Laflamme and A. Stintzi, Low temperature MBBR nitrification: Microbiome analysis, Water Res., 2017, 111, 224–233 CrossRef CAS PubMed.
- C. M. Lopez-Vazquez, A. Oehmen, C. M. Hooijmans, D. Brdjanovic, H. J. Gijzen, Z. Yuan and M. C. M. van Loosdrecht, Modeling the PAO-GAO competition: Effects of carbon source, pH and temperature, Water Res., 2009, 43(2), 450–462 CrossRef CAS PubMed.
- A. Oehmen, G. Carvalho, C. M. Lopez-Vazquez, M. C. M. van Loosdrecht and M. A. M. Reis, Incorporating microbial ecology into the metabolic modelling of polyphosphate accumulating organisms and glycogen accumulating organisms, Water Res., 2010, 44(17), 4992–5004 CrossRef CAS PubMed.
- M. Stokholm-Bjerregaard, Control of GAOs in wastewater treatment plants with enhanced biological phosphorus removal, PhD thesis, Aalborg University, Aalborg, Denmark, 2016 Search PubMed.
- Key reference: M. Nierychlo, K. S. Andersen, Y. Xu, N. Green, C. Jiang, M. Albertsen, M. S. Dueholm and P. H. Nielsen, MiDAS 3: An ecosystem-specific reference database, taxonomy and knowledge platform for activated sludge and anaerobic digesters reveals species-level microbiome composition of activated sludge, Water Res., 2020, 182, 115955 CrossRef CAS PubMed.
- S. J. McIlroy, A. M. Saunders, M. Albertsen, M. Nierychlo, B. McIlroy, A. A. Hansen, S. M. Karst, J. L. Nielsen and P. H. Nielsen, MiDAS: the field guide to the microbes of activated sludge, Database, 2015, 2015, bav062 CrossRef PubMed.
- F. Sabba, A. Terada, G. Wells, B. F. Smets and R. Nerenberg, Nitrous oxide emissions from biofilm processes for wastewater treatment, Appl. Microbiol. Biotechnol., 2018, 102(22), 9815–9829 CrossRef CAS PubMed.
- H. Gao, X. Zhao, L. Zhou, F. Sabba and G. F. Wells, Differential kinetics of nitrogen oxides reduction leads to elevated nitrous oxide production by a nitrite fed granular denitrifying EBPR bioreactor, Environ. Sci.: Water Res. Technol., 2020, 6(4), 1028–1043 RSC.
- H. Gao, M. Liu, J. S. Griffin, L. Xu, D. Xiang, Y. D. Scherson, W. T. Liu and G. F. Wells, Complete Nutrient Removal Coupled to Nitrous Oxide Production as a Bioenergy Source by Denitrifying Polyphosphate-Accumulating Organisms, Environ. Sci. Technol., 2017, 51(8), 4531–4540 CrossRef CAS PubMed.
- F. J. Rubio-Rincon, D. G. Weissbrodt, C. M. Lopez-Vazquez, L. Welles, B. Abbas, M. Albertsen, P. H. Nielsen, M. C. M. van Loosdrecht and D. Brdjanovic, “Candidatus Accumulibacter delftensis”: A clade IC novel polyphosphate-accumulating organism without denitrifying activity on nitrate, Water Res., 2019, 161, 136–151 CrossRef CAS PubMed.
- A. Oehmen, G. Carvalho, F. Freitas and M. A. M. Reis, Assessing the abundance and activity of denitrifying polyphosphate accumulating organisms through molecular and chemical techniques, Water Sci. Technol., 2010, 61(8), 2061–2068 CrossRef CAS PubMed.
- D. G. Weissbrodt, G. Russo, R. Goel, M. Laureni, A. M. Minder and E. Morgenroth, in Systems microbiology of aerobic-anaerobic ammonium oxidizing biofilm processes: temperature impacts and nitrous oxide emission, 6th International Joint Specialist Conference on Microbial Ecology in Water Engineering & Biofilms, Technical University of Denmark, Copenhagen, Denmark, ed. Smets, et al., International Water Association: Technical University of Denmark, Copenhagen, Denmark, 2016 Search PubMed.
- R. Niederdorfer, D. Hausherr, A. Palomo, J. Wei, P. Magyar, B. F. Smets, A. Joss and H. Bürgmann, Temperature modulates stress response in mainstream anammox reactors, Commun. Biol., 2021, 4(1), 23 CrossRef CAS PubMed.
- C. E. Lawson, G. H. L. Nuijten, R. M. de Graaf, T. B. Jacobson, M. Pabst, D. M. Stevenson, M. S. M. Jetten, D. R. Noguera, K. D. McMahon, D. Amador-Noguez and S. Lücker, Autotrophic and mixotrophic metabolism of an anammox bacterium revealed by in vivo 13C and 2H metabolic network mapping, ISME J., 2021, 15, 673–687 CrossRef CAS PubMed.
- Key reference: N. de Jonge, V. Moset, H. B. Møller and J. L. Nielsen, Microbial population dynamics in continuous anaerobic digester systems during start up, stable conditions and recovery after starvation, Bioresour. Technol., 2017, 232, 313–320 CrossRef CAS PubMed.
- A. Hanreich, R. Heyer, D. Benndorf, E. Rapp, M. Pioch, U. Reichl and M. Klocke, Metaproteome analysis to determine the metabolically active part of a thermophilic microbial community producing biogas from agricultural biomass, Can. J. Microbiol., 2012, 58(7), 917–922 CrossRef CAS PubMed.
- S. Narayanasamy, E. E. L. Muller, A. R. Sheik and P. Wilmes, Integrated omics for the identification of key functionalities in biological wastewater treatment microbial communities, Microb. Biotechnol., 2015, 8(3), 363–368 CrossRef PubMed.
- S. Püttker, F. Kohrs, D. Benndorf, R. Heyer, E. Rapp and U. Reichl, Metaproteomics of activated sludge from a wastewater treatment plant - A pilot study, Proteomics, 2015, 15(20), 3596–3601 CrossRef PubMed.
- C. Salerno, G. Berardi, G. Laera and A. Pollice, Functional Response of MBR Microbial Consortia to Substrate Stress as Revealed by Metaproteomics, Microb. Ecol., 2019, 78(4), 873–884 CrossRef CAS PubMed.
- E. Gunnigle, J. L. Nielsen, M. Fuszard, C. H. Botting, J. Sheahan, V. O'Flaherty and F. Abram, Functional responses and adaptation of mesophilic microbial communities to psychrophilic anaerobic digestion, FEMS Microbiol. Ecol., 2015, 91(12), 1–15 Search PubMed.
- F. A. Herbst, V. Lünsmann, H. Kjeldal, N. Jehmlich, A. Tholey, M. von Bergen, J. L. Nielsen, R. L. Hettich, J. Seifert and P. H. Nielsen, Enhancing metaproteomics-The value of models and defined environmental microbial systems, Proteomics, 2016, 16(5), 783–798 CrossRef CAS PubMed.
- F. Mosbæk, H. Kjeldal, D. G. Mulat, M. Albertsen, A. J. Ward, A. Feilberg and J. L. Nielsen, Identification of syntrophic acetate-oxidizing bacteria in anaerobic digesters by combined protein-based stable isotope probing and metagenomics, ISME J., 2016, 10(10), 2405–2418 CrossRef PubMed.
- N. de Jonge, Å. Davidsson, J. la Cour Jansen and J. L. Nielsen, Characterisation of microbial communities for improved management of anaerobic digestion of food waste, Waste Manage., 2020, 117, 124–135 CrossRef CAS PubMed.
- A. Pruden, D. G. Joakim Larsson, A. Amézquita, P. Collignon, K. K. Brandt, D. W. Graham, J. M. Lazorchak, S. Suzuki, P. Silley, J. R. Snape, E. Topp, T. Zhang and Y. G. Zhu, Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment, Environ. Health Perspect., 2013, 121(8), 878–885 CrossRef PubMed.
- H. Bürgmann, D. Frigon, W. H. Gaze, C. M. Manaia, A. Pruden, A. C. Singer, B. F. Smets and T. Zhang, Water and sanitation: an essential battlefront in the war on antimicrobial resistance, FEMS Microbiol. Ecol., 2018, 94(9), fiy101 CrossRef PubMed.
- T. U. Berendonk, C. M. Manaia, C. Merlin, D. Fatta-Kassinos, E. Cytryn, F. Walsh, H. Burgmann, H. Sorum, M. Norstrom, M. N. Pons, N. Kreuzinger, P. Huovinen, S. Stefani, T. Schwartz, V. Kisand, F. Baquero and J. L. Martinez, Tackling antibiotic resistance: the environmental framework, Nat. Rev. Microbiol., 2015, 13(5), 310–317 CrossRef CAS PubMed.
- H. Sales-Ortells, G. Agostini and G. Medema, Quantification of waterborne pathogens and associated health risks in urban water, Environ. Sci. Technol., 2015, 49(11), 6943–6952 CrossRef CAS PubMed.
- N. Czekalski, T. Berthold, S. Caucci, A. Egli and H. Bürgmann, Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland, Front. Microbiol., 2012, 3, 106 Search PubMed.
- P. Y. Hong, T. R. Julian, M. L. Pype, S. C. Jiang, K. L. Nelson, D. Graham, A. Pruden and C. M. Manaia, Reusing treated wastewater: Consideration of the safety aspects associated with antibiotic-resistant bacteria and antibiotic resistance genes, Water, 2018, 10(3), 244 CrossRef.
- N. Fahrenfeld, Y. Ma, M. O'Brien and A. Pruden, Reclaimed water as a reservoir of antibiotic resistance genes: Distribution system and irrigation implications, Front. Microbiol., 2013, 4, 130 Search PubMed.
- M. M. L. Dingemans, P. W. M. H. Smeets, G. Medema, J. Frijns, K. J. Raat, A. P. van Wezel and R. P. Bartholomeus, Responsible water reuse needs an interdisciplinary approach to balance risks and benefits, Water, 2020, 12(5), 1264 CrossRef.
- Key reference: P. J. Vikesland, A. Pruden, P. J. J. Alvarez, D. Aga, H. Bürgmann, X. D. Li, C. M. Manaia, I. Nambi, K. Wigginton, T. Zhang and Y. G. Zhu, Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance, Environ. Sci. Technol., 2017, 51(22), 13061–13069 CrossRef CAS.
- Key reference: R. Pallares-Vega, H. Blaak, R. van der Plaats, A. M. de Roda Husman, L. Hernandez Leal, M. C. M. van Loosdrecht, D. G. Weissbrodt and H. Schmitt, Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: A cross-sectional study, Water Res., 2019, 161, 319–328 CrossRef CAS PubMed.
- R. Pallares-Vega, L. Hernandez Leal, B. N. Fletcher, E. Vias-Torres, M. C. M. van Loosdrecht, D. G. Weissbrodt and H. Schmitt, Annual dynamics of antimicrobials and resistance determinants in flocculent and aerobic granular sludge treatment systems, Water Res., 2021, 190, 116752 CrossRef CAS.
- D. Calderón-Franco, S. Apoorva, G. Medema, M. C. M. van Loosdrecht and D. G. Weissbrodt, Upgrading residues from wastewater and drinking water treatment plants as low-cost adsorbents to remove extracellular DNA and microorganisms carrying antibiotic resistance genes from treated effluents, Sci. Total Environ., 2021, 778, 146364 CrossRef.
- N. Czekalski, S. Imminger, E. Salhi, M. Veljkovic, K. Kleffel, D. Drissner, F. Hammes, H. Bürgmann and U. Von Gunten, Inactivation of Antibiotic Resistant Bacteria and Resistance Genes by Ozone: From Laboratory Experiments to Full-Scale Wastewater Treatment, Environ. Sci. Technol., 2016, 50(21), 11862–11871 CrossRef CAS PubMed.
- B. Bicudo, D. van Halem, S. A. Trikannad, G. Ferrero and G. Medema, Low voltage iron electrocoagulation as a tertiary treatment of municipal wastewater: removal of enteric pathogen indicators and antibiotic-resistant bacteria, Water Res., 2021, 188, 116500 CrossRef CAS PubMed.
- G. K. Paulus, L. M. Hornstra, N. Alygizakis, J. Slobodnik, N. Thomaidis and G. Medema, The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes, Int. J. Hyg. Environ. Health, 2019, 222(4), 635–644 CrossRef CAS PubMed.
- L. Kovalova, H. Siegrist, H. Singer, A. Wittmer and C. S. McArdell, Hospital wastewater treatment by membrane bioreactor: Performance and efficiency for organic micropollutant elimination, Environ. Sci. Technol., 2012, 46(3), 1536–1545 CrossRef CAS PubMed.
- B. Guo, N. Yu, D. G. Weissbrodt and Y. Liu, Effects of micro-aeration on microbial niches and antimicrobial resistances in blackwater anaerobic digesters, Water Res., 2021, 117035 CrossRef CAS PubMed.
- B. Christgen, Y. Yang, S. Z. Ahammad, B. Li, D. C. Rodriquez, T. Zhang and D. W. Graham, Metagenomics Shows That Low-Energy Anaerobic−Aerobic Treatment Reactors Reduce Antibiotic Resistance Gene Levels from Domestic Wastewater, Environ. Sci. Technol., 2015, 49(4), 2577–2584 CrossRef CAS.
- D. Calderón-Franco, M. C. M. van Loosdrecht, T. Abeel and D. G. Weissbrodt, Free-floating extracellular DNA: Systematic profiling of mobile genetic elements and antibiotic resistance from wastewater, Water Res., 2021, 189, 116592 CrossRef PubMed.
- M. A. Stokholm-Bjerregaard, A. A. Hansen, H. Strandbæk, I. Vølund, D. Thornberg, L. Hughes and P. H. Nielsen, Intelligent and dynamic control of optimal WWTP operation from microbial sequencing, in IWA World Water Congress & Exhibition, ed. IWA, IWA Publishing, Copenhagen, Denmark, 2021 Search PubMed.
- C. Cimon, P. Kadota and C. Eskicioglu, Effect of biochar and wood ash amendment on biochemical methane production of wastewater sludge from a temperature phase anaerobic digestion process, Bioresour. Technol., 2020, 297, 122440 CrossRef CAS PubMed.
- D. J. Batstone, J. Keller, I. Angelidaki, S. V. Kalyuzhnyi, S. G. Pavlostathis, A. Rozzi, W. T. Sanders, H. Siegrist and V. A. Vavilin, The IWA Anaerobic Digestion Model No 1 (ADM1), Water Sci. Technol., 2002, 45(10), 65–73 CrossRef CAS.
- H. Carrère, C. Dumas, A. Battimelli, D. J. Batstone, J. P. Delgenès, J. P. Steyer and I. Ferrer, Pretreatment methods to improve sludge anaerobic degradability: A review, J. Hazard. Mater., 2010, 183(1–3), 1–15 CrossRef PubMed.
- Y. Chen, J. J. Cheng and K. S. Creamer, Inhibition of anaerobic digestion process: A review, Bioresour. Technol., 2008, 99(10), 4044–4064 CrossRef CAS PubMed.
- J. B. Van Lier, A. Tilche, B. K. Ahring, H. Macarie, R. Moletta, M. Dohanyos, L. W. Hulshoff Pol, P. Lens and W. Verstraete, New perspectives in anaerobic digestion, Water Sci. Technol., 2001, 43(1), 1–18 CrossRef CAS.
- W. Sarni, C. White, R. Webb, K. Cross and R. Glotzbach, Digital Water: Industry leaders chart the transformation journey, IWA, London, UK, 2019, p. 43 Search PubMed.
- Key reference: D. G. Weissbrodt, M. K. H. Winkler and G. F. Wells, Responsible science, engineering and education for water resource recovery and circularity, Environ. Sci.: Water Res. Technol., 2020, 6(8), 1952–1966 RSC.
- M. D. Besmer, D. G. Weissbrodt, B. E. Kratochvil, J. A. Sigrist, M. S. Weyland and F. Hammes, The feasibility of automated online flow cytometry for in-situ monitoring of microbial dynamics in aquatic ecosystems, Front. Microbiol., 2014, 5, 265 Search PubMed.
- P. H. Nielsen, OnlineDNA - Optimerede miljøteknologiske systemer med online overvågning af mikrobielle samfund, Aalborg University, 2017 Search PubMed.
- B. Guo, Z. Sheng and Y. Liu, Evaluation of influent microbial immigration to activated sludge is affected by different-sized community segregation, npj Clean Water, 2021, 4(1), 20 CrossRef.
- Key reference: D. Frigon and G. Wells, Microbial immigration in wastewater treatment systems: analytical considerations and process implications, Curr. Opin. Biotechnol., 2019, 57, 151–159 CrossRef CAS.
- P. Roots, F. Sabba, A. F. Rosenthal, Y. Wang, Q. Yuan, L. Rieger, F. Yang, J. A. Kozak, H. Zhang and G. F. Wells, Integrated shortcut nitrogen and biological phosphorus removal from mainstream wastewater: process operation and modeling, Environ. Sci.: Water Res. Technol., 2020, 6(3), 566–580 RSC.
- Y. D. Scherson, A. Roa, G. Darling and C. S. Criddle, Sidestream Treatment with Energy Recovery from Nitrogen Waste: The Coupled Aerobic-anoxic Nitrous Decomposition Operation (CANDO), Proceedings of the Water Environment Federation, 2014, 2014(9), 1114–1125 CrossRef.
- P. Roots, Y. Wang, A. F. Rosenthal, J. S. Griffin, F. Sabba, M. Petrovich, F. Yang, J. A. Kozak, H. Zhang and G. F. Wells, Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor, Water Res., 2019, 157, 396–405 CrossRef CAS PubMed.
- J. S. Griffin and G. F. Wells, Regional synchrony in full-scale activated sludge bioreactors due to deterministic microbial community assembly, ISME J., 2017, 11(2), 500–511 CrossRef CAS PubMed.
- G. F. Wells, H. D. Park, B. Eggleston, C. A. Francis and C. S. Criddle, Fine-scale bacterial community dynamics and the taxa-time relationship within a full-scale activated sludge bioreactor, Water Res., 2011, 45(17), 5476–5488 CrossRef CAS.
- I. D. Ofiteru, M. Lunn, T. P. Curtis, G. F. Wells, C. S. Criddle, C. A. Francis and W. T. Sloan, Combined niche and neutral effects in a microbial wastewater treatment community, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(35), 15345–15350 CrossRef CAS.
- C. Ronda, S. P. Chen, V. Cabral, S. J. Yaung and H. H. Wang, Metagenomic engineering of the mammalian gut microbiome in situ, Nat. Methods, 2019, 16(2), 167–170 CrossRef CAS PubMed.
- Key reference: L. Corominas, M. Garrido-Baserba, K. Villez, G. Olsson, U. Cortés and M. Poch, Transforming data into knowledge for improved wastewater treatment operation: A critical review of techniques, Environ. Model. Softw., 2018, 106, 89–103 CrossRef.
- K. Villez, J. Billeter and D. Bonvin, Incremental parameter estimation under Rank-Deficient measurement conditions, Processes, 2019, 7(2), 75 CrossRef.
- D. Wang, S. Thunéll, U. Lindberg, L. Jiang, J. Trygg, M. Tysklind and N. Souihi, A machine learning framework to improve effluent quality control in wastewater treatment plants, Sci. Total Environ., 2021, 784, 147138 CrossRef CAS PubMed.
- F. Li, A. Amaral and P. A. Vanrolleghem, in An essential tool for WRRF modelling: A realistic and complete influent generator for flow rate and water quality based on machine learning, 93rd Water Environment Federation Technical Exhibition and Conference 2020, WEFTEC 2020, 2020, pp. 303–309 Search PubMed.
- M. Sweeney and J. Kabouris, Modeling, instrumentation, automation, and optimization of water resource recovery facilities (2019) DIRECT, Water Environ. Res., 2020, 92(10), 1499–1503 CrossRef CAS.
- L. J. Müller, A. Kätelhön, M. Bachmann, A. Zimmermann, A. Sternberg and A. Bardow, A Guideline for Life Cycle Assessment of Carbon Capture and Utilization, Front. Energy Res., 2020, 8, 15 CrossRef.
- L. Corominas, D. M. Byrne, J. S. Guest, A. Hospido, P. Roux, A. Shaw and M. D. Short, The application of life cycle assessment (LCA) to wastewater treatment: A best practice guide and critical review, Water Res., 2020, 184, 116058 CrossRef CAS PubMed.
- N. van Linden, R. Shang, G. Stockinger, B. Heijman and H. Spanjers, Separation of natural organic matter and sodium chloride for salt recovery purposes in zero liquid discharge, Water Resour. Ind., 2020, 23, 100117 CrossRef.
- T. Inaba, T. Hori, H. Aizawa, Y. Sato, A. Ogata and H. Habe, Microbiomes and chemical components of feed water and membrane-attached biofilm in reverse osmosis system to treat membrane bioreactor effluents, Sci. Rep., 2018, 8(1), 16805 CrossRef PubMed.
- T. Møretrø and S. Langsrud, Residential Bacteria on Surfaces in the Food Industry and Their Implications for Food Safety and Quality, Compr. Rev. Food Sci. Food Saf., 2017, 16(5), 1022–1041 CrossRef.
- E. Hansen and M. A. S. Rodrigues, Aquim, P. M. d., Wastewater reuse in a cascade based system of a petrochemical industry for the replacement of losses in cooling towers, J. Environ. Manage., 2016, 181, 157–162 CrossRef CAS PubMed.
- A. Friedlander, S. Nir, M. Reches and M. Shemesh, Preventing Biofilm Formation by Dairy-Associated Bacteria Using Peptide-Coated Surfaces, Front. Microbiol., 2019, 10, 1405 CrossRef PubMed.
- M. Jafari, M. Vanoppen, J. M. C. van Agtmaal, E. R. Cornelissen, J. S. Vrouwenvelder, A. Verliefde, M. C. M. van Loosdrecht and C. Picioreanu, Cost of fouling in full-scale reverse osmosis and nanofiltration installations in the Netherlands, Desalination, 2020, 500, 114865 CrossRef.
- D. Pichardo-Romero, Z. P. Garcia-Arce, A. Zavala-Ramírez and R. Castro-Muñoz, Current Advances in Biofouling Mitigation in Membranes for Water Treatment: An Overview, Processes, 2020, 8(2), 182 CrossRef CAS.
- P. Desmond, E. Morgenroth and N. Derlon, Physical structure determines compression of membrane biofilms during Gravity Driven Membrane (GDM) ultrafiltration, Water Res., 2018, 143, 539–549 CrossRef CAS PubMed.
- H. Ozgun, R. K. Dereli, M. E. Ersahin, C. Kinaci, H. Spanjers and J. B. van Lier, A review of anaerobic membrane bioreactors for municipal wastewater treatment: Integration options, limitations and expectations, Sep. Purif. Technol., 2013, 118, 89–104 CrossRef CAS.
- I. Pinel, L. H. Kim, V. R. Proença Borges, N. M. Farhat, G. J. Witkamp, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Effect of phosphate availability on biofilm formation in cooling towers, Biofouling, 2020, 36, 800–815 CrossRef CAS.
- I. Pinel, D. H. Moed, J. S. Vrouwenvelder and M. C. M. van Loosdrecht, Bacterial community dynamics and disinfection impact in cooling water systems, Water Res., 2020, 172, 115505 CrossRef CAS.
- L. Olabode, A. Greenberg, M. Grippo, A. Fore and G. Rijal, Chicago Area Waterway System Microbiome Study: An understanding of the microbial community structure — the microbiome — of urban waters could provide insight into the human health risks associated with recreational use of these waters, Adv. Water Resour., 2020, 30(4), 20–24 Search PubMed.
- Key reference: E. I. Prest, D. G. Weissbrodt, F. Hammes, M. C. van Loosdrecht and J. S. Vrouwenvelder, Long-Term Bacterial Dynamics in a Full-Scale Drinking Water Distribution System, PLoS One, 2016, 11(10), e0164445 CrossRef CAS PubMed.
- Key reference: A. J. Pinto, J. Schroeder, M. Lunn, W. Sloan and L. Raskin, Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome, MBio, 2014, 5(3), e01135-14 CrossRef PubMed.
- T. Klein, D. Zihlmann, N. Derlon, C. Isaacson, I. Szivák, D. G. Weissbrodt and W. Pronk, Biological control of biofilms on membranes by metazoans, Water Res., 2016, 88, 20–29 CrossRef CAS.
- R. S. Kantor, S. E. Miller and K. L. Nelson, The Water Microbiome Through a Pilot Scale Advanced Treatment Facility for Direct Potable Reuse, Front. Microbiol., 2019, 10, 993 CrossRef PubMed.
- C. R. Proctor and F. Hammes, Drinking water microbiology—from measurement to management, Curr. Opin. Biotechnol., 2015, 33, 87–94 CrossRef CAS PubMed.
- A. Bruno, A. Sandionigi, M. Bernasconi, A. Panio, M. Labra and M. Casiraghi, Changes in the Drinking Water Microbiome: Effects of Water Treatments Along the Flow of Two Drinking Water Treatment Plants in a Urbanized Area, Milan (Italy), Front. Microbiol., 2018, 9, 2557 CrossRef PubMed.
- B. W. Stamps, M. B. Leddy, M. H. Plumlee, N. A. Hasan, R. R. Colwell and J. R. Spear, Characterization of the Microbiome at the World's Largest Potable Water Reuse Facility, Front. Microbiol., 2018, 9, 2435–2435 CrossRef.
- P. Ji, J. Parks, M. Edwards and A. Pruden, Impact of Water Chemistry, Pipe Material and Stagnation on the Building Plumbing Microbiome, PLoS One, 2015, 10, e0141087 CrossRef PubMed.
- Key reference: Y. Zhang and W.-T. Liu, The application of molecular tools to study the drinking water microbiome – Current understanding and future needs, Crit. Rev. Environ. Sci. Technol., 2019, 49(13), 1188–1235 CrossRef CAS.
- N. M. Hull, F. Ling, A. J. Pinto, M. Albertsen, H. G. Jang, P.-Y. Hong, K. T. Konstantinidis, M. LeChevallier, R. R. Colwell and W.-T. Liu, Drinking Water Microbiome Project: Is it Time?, Trends Microbiol., 2019, 27(8), 670–677 CrossRef CAS.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |