Open Access Article
Open Access ArticleA novel method for PEGylation of chitosan nanoparticles through photopolymerization†
Ugur Bozuyuk‡§
 ,
Ipek S. Gokulu‡,
Nihal Olcay Dogan
,
Ipek S. Gokulu‡,
Nihal Olcay Dogan and
Seda Kizilel
and
Seda Kizilel *
*
Chemical and Biological Engineering, Koç University, Sariyer, Istanbul 34450, Turkey. E-mail: skizilel@ku.edu.tr
First published on 7th May 2019
Abstract
An ultrafast and convenient method for PEGylation of chitosan nanoparticles has been established through a photopolymerization reaction between the acrylate groups of PEG and methacrylated-chitosan nanoparticles. The nanoparticle characteristics under physiological pH conditions were optimized through altered PEG chain length, concentration and duration of UV exposure. The method developed here has potential for clinical translation of chitosan nanoparticles. It also allows for the scalable and fast synthesis of nanoparticles with colloidal stability.
The latest progress regarding the framework of bioengineered nanoparticles has led to the development of new biodegradable nanoparticles for potential treatment purposes.1–6 Recent methods in nanomedicine have leaned towards developing colloidally stable, biocompatible nanoparticles which can overcome biological barriers without causing any harm to living tissues. Accordingly, the surface charge and size optimization of such particles has gained great importance as they are major factors for the stability of the system.7
The search for bioadhesive nanoparticles that have potential as clinically viable materials extends to a wide scope in the field. One such material is chitosan, which has an application range including food, cosmetics and biomedicine.8–11 As this material is known for its biocompatible,12 biodegradable13 and nontoxic nature, it becomes an expedient choice to be used for therapeutic purposes such as drug or gene delivery.14–16 Moreover, the reaction of primary amines attached to chitosan backbone permit functionalization of this material with photosensitive groups. One of those functionalization is the addition reaction of the amino groups with methacrylic anhydride which produces photosensitive methacrylamide groups.17,18 This modification of chitosan chains allows for photocrosslinking of this adhesive monomer in the presence of a photo initiator under UV light to form a gel.19
Multiple methods are present to synthesize chitosan nanoparticles such as reverse micellar,20 emulsification solvent diffusion,21 and ionotropic gelation method.22,23 Chitosan nanoparticles have been synthesized at acidic pH due to the presence of negatively charged sodium tripolyphosphate (TPP) and cationic chitosan chains, within a short time at room temperature through ionic gelation.14 Majority of research that involves the synthesis of these nanoparticles consider the size and zeta potential of chitosan nanoparticles between pH 4.0 and 5.5, where these methods do not characterize the properties at physiological conditions regardless of this major applicability constraint.24,25 To overcome this obstacle, we have developed a unique PEGylation approach which can be utilized for wide ranges of pH without compromising the solution stability of chitosan. For this reason, PEGylation may be considered as a critical route for advancing chitosan nanoparticles as biomedical tools and making the particle system colloidally stable.26 There are various approaches used for the PEGylation of chitosan nanoparticles such as the PEGylation of chitosan chains prior to nanoparticle synthesis.27,28 However, these existing methods used for direct chemical conjugation of PEG to chitosan are time consuming, require complex reactions and equipment. In our work, we address two fundamental issues for the utilization of chitosan nanoparticles in biomedicine: (i) optimization of chitosan nanoparticle properties at physiological pH, and (ii) development of a novel PEGylation method for chitosan nanoparticles for long term feasibility and large-scale synthesis.
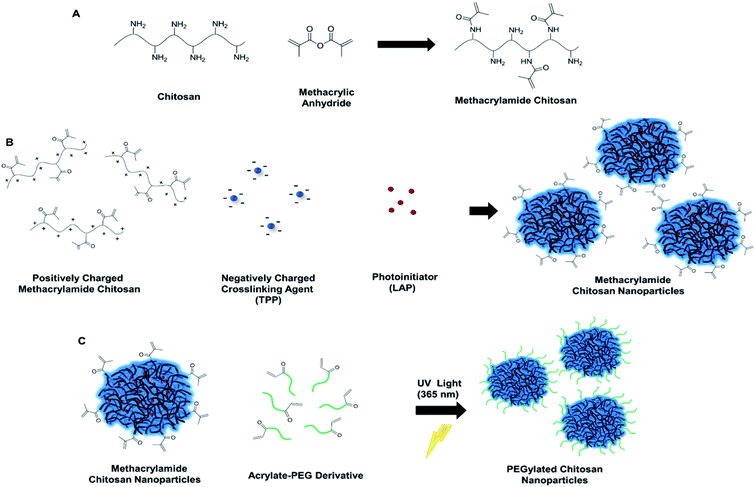
The novel method we introduce here for the PEGylation of chitosan overcomes the limitations associated with the use of chitosan nanoparticles in biomedicine. We prepare ionically crosslinked and PEGylated chitosan nanoparticles through simultaneous addition of photoinitiator with TPP and acrylate-PEG prior to UV light exposure at 365 nm, which promotes covalent bond formation between the acrylate groups of PEG and methacrylamide chitosan nanoparticles (CSMA) (Fig. 1). Methacrylamide chitosan was synthesized according to previously described protocol.17–19 (ESI Experimental section 2†). Synthesis of methacrylamide chitosan is a scalable one-step process and allows for the large amounts of product formation. Two groups of methacrylamide chitosan chains were prepared for the optimization of degree of methacrylation during nanoparticle formation: chitosan functionalized with (1) 50 μL and (2) 100 μL methacrylic anhydride (MA) per 30 mg of chitosan. Aggregation was observed inside the CSMA solution with the higher methacrylation degree (100 μL MA to 30 mg chitosan) for different groups at pH 4.7 (Table S1†). Hence, we continued our experiments with the methacrylamide chitosan polymer that has lower degree of methacrylation. It was possible to obtain nanoparticles with high colloidal stability using the less methacrylated-chitosan polymer with sub 100 nm size and positive charge at pH 4.7 (Table 1, Negative control 1).
| Sample name | PEG MW (kDa) | PEG molea (μmole) | pH | UV | Observed size (nm) | Polydispersity index | Observed zeta potential (mV) |
|---|---|---|---|---|---|---|---|
| a Number of PEG mole per 25 mg of chitosan nanoparticle solution.b Aggregated. | |||||||
| Negative control 1 | — | — | 4.7 | No | 90.75 ± 0.56 | 0.383 ± 0.05 | 32.3 ± 4.34 |
| Negative control 2 | — | — | 7.4 | No | Agg.b | Agg.b | Agg.b |
| 1 | 5 | 4 | 4.7 | Yes | 83.97 ± 1.00 | 0.297 ± 0.08 | 32.2 ± 3.80 |
| 2 | 5 | 4 | 7.4 | Yes | 136.30 ± 6.85 | 0.118 ± 0.01 | 6.01 ± 3.14 |
| 3 | 5 | 7 | 4.7 | Yes | 88.38 ± 0.56 | 0.390 ± 0.28 | 31.8 ± 6.94 |
| 4 | 5 | 7 | 7.4 | Yes | 120.06 ± 6.91 | 0.230 ± 0.02 | 4.23 ± 3.36 |
| 5 | 5 | 10 | 4.7 | Yes | 89.31 ± 0.80 | 0.411 ± 0.01 | 31.9 ± 4.41 |
| 6 | 5 | 10 | 7.4 | Yes | Agg.b | Agg.b | Agg.b |
| 7 | 10 | 4 | 4.7 | Yes | 91.93 ± 1.33 | 0.424 ± 0.01 | 30.7 ± 3.82 |
| 8 | 10 | 4 | 7.4 | Yes | 189.56 ± 11.87 | 0.145 ± 0.01 | 5.64 ± 3.20 |
| 9 | 10 | 7 | 4.7 | Yes | 83.55 ± 4.03 | 0.466 ± 0.01 | 30.2 ± 4.05 |
| 10 | 10 | 7 | 7.4 | Yes | Agg.b | Agg.b | Agg.b |
| 11 | 10 | 10 | 4.7 | Yes | 60.82 ± 0.10 | 0.496 ± 0.01 | 28.6 ± 3.76 |
| 12 | 10 | 10 | 7.4 | Yes | Agg.b | Agg.b | Agg.b |
Degree of methacrylation for the less methacrylated polymer (50 μL MA to 300 mg chitosan) was calculated with 2,4,6-trinitrobenzenesulfonic acid (TNBS) assay, where 20% of amino groups were converted into photosensitive methacrylamide groups. However, this group of methacrylamide chitosan nanoparticles had no colloidal stability and aggregated irreversibly at pH 7.4 (Table 1, Negative control 2). To obtain PEGylated chitosan nanoparticles, we added the photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) to methacrylamide chitosan monomer solutions prior to nanoparticle synthesis. After nanoparticle synthesis, we added acrylate-PEG (5 and 10 kDa molecular weights) into nanoparticle solutions and exposed them to UV light with altered exposure time. Optimization of the photoinitiator concentration inside the CSMA solutions were completed through addition of 6.9 μL or 69 μL of 1% (w/v) LAP solution simultaneously with TPP into the CSMA solutions (2.5 mL TPP, and 10 mL CSMA solution). We first added 69 μL of LAP to the CSMA solution and prepared nanoparticles with subsequent addition of TPP. Then, we exposed this nanoparticle solution to UV light for 1 min without any addition of acrylate-PEG. We observed aggregation of nanoparticles at pH 4.7 (pH of the solution after mixing with CS and TPP) due to crosslinking of nanoparticles with each other as a result of high number of radicalized acrylate groups (ESI, Table S1, Sample 2†). Then, we decreased LAP amount by tenfold, from 69 μL to 6.9 and 1 μL, and observed no aggregation at pH 4.7 with 1 and 10 min of UV light exposure for both groups (Table S1, Samples 4 and 10†). Accordingly, we prepared five different nanoparticle solutions of 12.5 mL which contained 6.9 μL of LAP and 7 μmoles of 5 kDa acrylate-PEG. Next, we exposed these solutions to UV light for altered durations of 0, 1, 5, 10 and 20 minutes. For these groups, no aggregation was observed at pH 4.7. However, for the groups that were exposed to 0, 1, and 5 minutes of UV, aggregations were visible at pH 7.4, probably due to no or insufficient PEGylation reaction. On the other hand, the groups that were exposed to 10 and 20 minutes of UV light did not aggregate at pH 7.4 (ESI, Table S1†). The solutions of these groups of 10 and 20 minutes of UV exposure were observed as clear at pH 7.4, which was a direct indication for successful PEGylation. The group that had 1 μL LAP solution aggregated at pH 7.4 probably due to insufficient PEGylation (ESI, Table S1, Sample 11†). We also investigated the direct effect of LAP initiator to the PEGylation, and the group without initiator aggregated at pH 7.4 (ESI, Table S1, Sample 12†). We measured larger particle sizes for the group that was exposed to UV for 20 minutes, probably due to crosslinking of nanoparticles (Fig. S1 and S2†). From these results, we concluded that 6.9 μL LAP and 10 min UV exposure conditions are optimal for fast and simple PEGylation of nanoparticles.
Thus, this condition was utilized for the following experimental trials. We used 5 kDa and 10 kDa chain lengths of PEG for the PEGylation of chitosan nanoparticles. Six different conditions of 4, 7 and 10 μmoles of 5 kDa and 10 kDa PEG were used per 5 mg of chitosan nanoparticle solution in different trials as was presented in Table 1. Increasing the amount of PEG resulted in alterations in size and zeta potential. These alterations were not proportional, that is, number of acrylate-PEG macromolecules were used in excess compared to the number of methacrylamide groups in nanoparticles even with the condition of lowest number of PEG moles. Thus, increasing the number of PEG molecules caused slight changes in the nanoparticle properties with, non-proportional zeta potential differences. However, there is a difference between comparable 5 and 10 kDa groups, where 10 kDa groups have slightly less zeta potential. This change can be explained as longer PEG chain lengths cause more hindrance effects on the surface of the nanoparticles.27 We were able to obtain nanoparticles with decreasing surface charge at pH 7.4 for 5 kDa groups. This also means that, it is possible to tune the surface charge of nanoparticles to some extent. However, increasing the amount of PEG to decrease surface charge of nanoparticles resulted in aggregation.
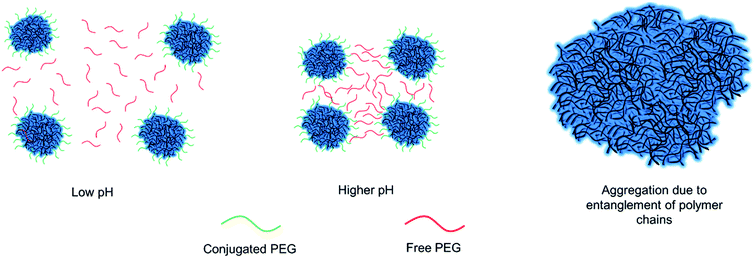
These aggregations can be due to the tangling of free and conjugated PEG on the surface of the nanoparticles which stay closer to each other at increased pH levels due to ionic attraction forces as is shown schematically in Fig. 2. This effect was more obvious in 10 kDa groups since longer chains induces more entanglement in the solution (Table 1). The samples with 4 and 7 μmoles of 5 kDa PEG and 4 μmoles of 10 kDa PEG proved that these respective conditions were optimal for biomedical applications, as they remained stable under physiological pH (Table 1). Furthermore, cytotoxicity of PEGylated chitosan nanoparticles was carried out using HEK293-T cells via CellTiter-Glo Luminescent Cell Viability Assay. Cells were treated with altered concentrations of PEGylated chitosan nanoparticles (25, 50, 75 μg mL−1) for 48 h. Cell survival was measured as higher than 80% for all groups with the highest dose of nanoparticle concentration (Fig. S3†). These results suggest that PEGylated chitosan nanoparticles are non-toxic to HEK293-T cells at high doses.
Presence of PEG in the PEGylated chitosan nanoparticles was confirmed via FTIR analysis and results are shown in Fig. S4.† PEGylated chitosan nanoparticles were dialyzed to eliminate the unreacted PEG macromolecules. FT-IR spectrum of PEGylated chitosan nanoparticles retained C–H stretches around 2800 cm−1 and amine stretches around 1639 cm−1, which further confirmed PEGylation of nanoparticles. Moreover, scanning electron microscopy (SEM) image was examined to visualize these nanoparticles as presented in Fig. S5–S7.† The particle characterization results indicated an increasing trend in particle sizes and a decreasing trend in observed zeta potentials with increasing molecular weight of PEG due to the increased number of conjugated PEG chains on the surface of the nanoparticles.
In summary, a new and simple PEGylation method has been introduced in this study, through photopolymerization of functional photosensitive groups of chitosan. This method has shown to be significant as it is achieved within minutes and does not involve complex equipment compared to the existing PEGylation of chitosan methods. The results suggest that particles synthesized using this approach are non-toxic and colloidally stable under physiological pH condition. The optimization of nanoparticle characteristics for different UV exposure durations, PEG chain lengths and concentrations has proven that the desired characteristics for PEGylated chitosan nanoparticles can be achieved under different conditions combinations. It has been concluded that the simultaneous increase in PEG chain length and concentration in nanoparticle solution should be avoided as free and conjugated PEG chains might promote chain entanglements with shorter intermolecular distances at physiological pH. This new method can be used for the realistic biomedical applications of chitosan nanoparticles and offers scalable and fast fabrication of colloidally stable nanoparticles.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study is supported by the Scientific and Technological Research Council of Turkey (TUBITAK) under International Support Program (COST Action – European Cooperation in Science and Technology – CA15216, project number: 116M995). Authors would like to thank M. Anwaar Nazeer for his help with FE-SEM experiments. The authors gratefully acknowledge use of the services and facilities of the Koç University Research Center for Surface Science (KUYTAM) and Koç University Research Center for Translational Medicine (KUTTAM), funded by the Presidency of Turkey, Presidency of Strategy and Budget. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Presidency of Strategy and Budget.Notes and references
- M. Ferrari, Nat. Rev. Cancer, 2005, 5, 161 CrossRef CAS PubMed.
- X.-J. Liang, C. Chen, Y. Zhao, L. Jia and P. C. Wang, Curr. Drug Metab., 2008, 9, 697–709 CrossRef CAS.
- R. A. Petros and J. M. DeSimone, Nat. Rev. Drug Discovery, 2010, 9, 615 CrossRef CAS PubMed.
- M. Saboktakin, Adv. Mater. Sci., 2017, 2, 1–14 Search PubMed.
- T. Sun, Y. S. Zhang, B. Pang, D. C. Hyun, M. Yang and Y. Xia, Angew. Chem., Int. Ed., 2014, 53, 12320–12364 CAS.
- L. Zhang, F. Gu, J. Chan, A. Wang, R. Langer and O. Farokhzad, Clin. Pharmacol. Ther., 2008, 83, 761–769 CrossRef CAS PubMed.
- A. Albanese, P. S. Tang and W. C. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- M. Aider, LWT--Food Sci. Technol., 2010, 43, 837–842 CrossRef CAS.
- P. K. Dutta, J. Dutta and V. Tripathi, J. Sci. Ind. Res., 2004, 63, 20–31 CAS.
- M. Rinaudo, Prog. Polym. Sci., 2006, 31, 603–632 CrossRef CAS.
- E. I. Rabea, M. E.-T. Badawy, C. V. Stevens, G. Smagghe and W. Steurbaut, Biomacromolecules, 2003, 4, 1457–1465 CrossRef CAS PubMed.
- S. Rodrigues, M. Dionísio, C. R. López and A. Grenha, J. Funct. Biomater., 2012, 3, 615–641 CrossRef CAS PubMed.
- H. Tan, C. R. Chu, K. A. Payne and K. G. Marra, Biomaterials, 2009, 30, 2499–2506 CrossRef CAS PubMed.
- M. Mohammed, J. Syeda, K. Wasan and E. Wasan, Pharmaceutics, 2017, 9, 53 CrossRef.
- N. Bhattarai, J. Gunn and M. Zhang, Adv. Drug Delivery Rev., 2010, 62, 83–99 CrossRef CAS PubMed.
- K. Roy, H.-Q. Mao, S.-K. Huang and K. W. Leong, Nat. Med., 1999, 5, 387 CrossRef CAS.
- L. M. Yu, K. Kazazian and M. S. Shoichet, J. Biomed. Mater. Res., Part A, 2007, 82, 243–255 CrossRef.
- B. G. Amsden, A. Sukarto, D. K. Knight and S. N. Shapka, Biomacromolecules, 2007, 8, 3758–3766 CrossRef CAS.
- U. Bozuyuk, O. Yasa, I. C. Yasa, H. Ceylan, S. Kizilel and M. Sitti, ACS Nano, 2018, 12, 9617–9625 CrossRef CAS PubMed.
- F. Brunel, L. Véron, L. David, A. Domard and T. Delair, Langmuir, 2008, 24, 11370–11377 CrossRef CAS PubMed.
- P. Khachane and M. Nagarsenker, Pharmazie, 2011, 66, 334–338 CAS.
- Y. Dong, W. K. Ng, S. Shen, S. Kim and R. B. Tan, Carbohydr. Polym., 2013, 94, 940–945 CrossRef CAS.
- K. Divya and M. Jisha, Environ. Chem. Lett., 2018, 16, 101–112 CrossRef CAS.
- A. Makhlof, Y. Tozuka and H. Takeuchi, Eur. J. Pharm. Sci., 2011, 42, 445–451 CrossRef CAS.
- M. Malhotra, C. Lane, C. Tomaro-Duchesneau, S. Saha and S. Prakash, Int. J. Nanomed., 2011, 6, 485 CAS.
- Y. Zhang, J. Chen, Y. Zhang, Y. Pan, J. Zhao, L. Ren, M. Liao, Z. Hu, L. Kong and J. Wang, Biotechnol. Appl. Biochem., 2007, 46, 197–204 CrossRef CAS PubMed.
- U. Bozuyuk, N. O. Dogan and S. Kizilel, ACS Appl. Mater. Interfaces, 2018, 10, 33945–33955 CrossRef CAS PubMed.
- L. Casettari, D. Vllasaliu, E. Castagnino, S. Stolnik, S. Howdle and L. Illum, Prog. Polym. Sci., 2012, 37, 659–685 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00780f |
| ‡ The authors have equal authorship. |
| § Current address: Physical Intelligence Department, Max Planck Institute for Intelligent Systems, Heisenbergstraβe 3, 70569 Stuttgart, Germany. |
| This journal is © The Royal Society of Chemistry 2019 |