Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Highly selective microglial uptake of ceria–zirconia nanoparticles for enhanced analgesic treatment of neuropathic pain†
Boomin
Choi‡
a,
Min
Soh‡
 bc,
Yelina
Manandhar‡
a,
Dokyoon
Kim
d,
Sang Ihn
Han
bc,
Seungmin
Baik
bc,
Kwangsoo
Shin
bc,
Sagang
Koo
bc,
Hyek Jin
Kwon
bc,
Giho
Ko
bc,
Junyoung
Oh
a,
Heehong
Hwang
a,
Taeghwan
Hyeon
bc,
Yelina
Manandhar‡
a,
Dokyoon
Kim
d,
Sang Ihn
Han
bc,
Seungmin
Baik
bc,
Kwangsoo
Shin
bc,
Sagang
Koo
bc,
Hyek Jin
Kwon
bc,
Giho
Ko
bc,
Junyoung
Oh
a,
Heehong
Hwang
a,
Taeghwan
Hyeon
 *bc and
Sung Joong
Lee
*bc and
Sung Joong
Lee
 *a
*a
aDepartment of Neuroscience and Physiology, Dental Research Institute, School of Dentistry, Seoul National University, Seoul 08826, Republic of Korea. E-mail: sjlee87@snu.ac.kr
bCenter for Nanoparticle Research, Institute for Basic Science (IBS), Seoul 08826, Republic of Korea
cSchool of Chemical and Biological Engineering, and Institute of Chemical Processes, Seoul National University, Seoul 08826, Republic of Korea. E-mail: thyeon@snu.ac.kr
dDepartment of Bionano Engineering and Bionanotechnology, Hanyang University, Ansan 15588, Republic of Korea
First published on 2nd September 2019
Abstract
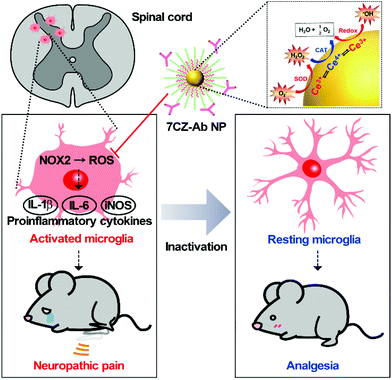
Neuropathic pain is a chronic and pathological pain caused by injury or dysfunction in the nervous system. Pro-inflammatory microglial activation with aberrant reactive oxygen species (ROS) generation in the spinal cord plays a critical role in the development of neuropathic pain. However, the efficacy of current therapeutic methods for neuropathic pain is limited because only neurons or neural circuits involved in pain transmission are targeted. Here, an effective strategy to treat pain hypersensitivity using microglia-targeting ceria–zirconia nanoparticles (CZ NPs) is reported. The CZ NPs are coated with microglia-specific antibodies to promote their delivery to microglia, and thus to improve their therapeutic efficacy. The targeted delivery facilitates the elimination of both pro-inflammatory cytokines and ROS in microglia, enabling the rapid and effective inhibition of microglial activation. As a result, greatly ameliorated mechanical allodynia is achieved in a spinal nerve transection (SNT)-induced neuropathic pain mouse model, proving the potent analgesic effect of the microglia-targeting CZ NPs. Given the generality of the approach used in this study, the microglia-targeting CZ NPs are expected to be useful for the treatment of not only neuropathic pain but also other neurological diseases associated with the vicious activation of microglia.
Introduction
Neuropathic pain is a type of pathological pain caused by injury or dysfunction in the nervous system. Clinical symptoms of this devastating disease are characterized by spontaneous pain, pain perception against non-noxious stimuli (allodynia), and exaggerated pain perception against noxious stimuli (hyperalgesia). It is well-known that pro-inflammatory activation of spinal cord microglia, innate immune cells in the central nervous system (CNS), is one of the key etiological factors in the pathogenesis of neuropathic pain.1,2 Signals derived from injured nerves activate microglia at the spinal dorsal horn and induce subsequent expression of pro-inflammatory cytokines such as interleukin 1 beta (IL-1β), interleukin 6 (IL-6), and tumor necrosis factor-alpha (TNF-α). This inflammatory milieu, in turn, sensitizes the pain-transmitting neural circuits, resulting in pain sensitization at the spinal cord level, the so-called central sensitization.3–5 Given these findings, modulating neuro-inflammation in the spinal cord after nerve injury by targeting microglial cells can be an effective strategy to prevent or resolve the pathological changes that develop chronic pain. Previous studies have shown that spinal nerve transection (SNT) injury induces pro-inflammatory microglial activation in the spinal dorsal horn, which in turn leads to pain central sensitization and neuropathic pain.6 Furthermore, nicotinamide adenine dinucleotide phosphate (NADPH) oxidase 2 (Nox2)-derived reactive oxygen species (ROS) production plays a critical role in the nerve injury-induced pro-inflammatory spinal microglial activation and subsequent pain sensitization.6–8 Since Nox2 generates superoxide anions which can be converted to other ROS such as hydrogen peroxide,9 reducing the microglial ROS level is significant in the intervention against the pathogenesis of neuropathic pain and other CNS diseases (Scheme 1).6,10–12Recently, nanomaterials have been widely used in a variety of biomedical applications due to their interesting biological properties.13–20 Despite such achievements, however, promoting the therapeutic efficacy of nanomaterials has been a major consideration for successful application since therapeutic nanomaterials are still far from clinical translation. Nevertheless, endeavors in developing ceria nanoparticles (NPs) as an effective therapeutic agent have demonstrated the promising potential for the treatment of diseases associated with abrupt surges of ROS.21–27 Reversible redox switching between Ce3+ and Ce4+ ions in ceria NPs enables the NPs to scavenge ROS for a prolonged period.28–30 Moreover, since it was revealed that Ce3+ ions are more important than Ce4+ ions in removing ROS and subsequent amelioration of inflammatory diseases, systems to sustain higher Ce3+ contents in ceria NPs were designed by inserting dopant ions.31–36 For example, ceria–zirconia NPs outperform ceria NPs in terms of therapeutic efficacy in a sepsis model by showing a greater activity toward the elimination of superoxide anions (O2−) and hydroxyl radicals (˙OH), which are the main culprits in the pathogenesis of many inflammatory diseases.37 Meanwhile, approaches for the targeted delivery of nano-therapeutics are important to promote the efficacy of drugs while avoiding side-effects.38–42 The adequate delivery strategy should accompany the therapeutic ceria-based NPs so that they are delivered specifically to the target cells or tissues and contained within the confined area of the disease only.23,43–46 The rational design of such a strategy would result in a higher recovery rate for patients.
Experimental
Materials
Cerium(III) acetylacetonate hydrate, zirconium(IV) acetylacetonate hydrate, fluorescein isothiocyanate (FITC), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDCI), N-hydroxysuccinimide (NHS), triethylamine (TEA) and a superoxide dismutase (SOD) assay kit were purchased from Sigma-Aldrich Inc. (St Louis, Missouri, USA). Oleylamine (approximate C18-content of 80–90%) was purchased from Acros Organics (Geel, Belgium). Acetone (99%) and chloroform (99%) were purchased from Samchun Chemicals (Seoul, Korea). 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (mPEG(2000)-PE) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (DSPE-PEG(2000)-amine) were purchased from Avanti Polar Lipids Inc. (Alabaster, Alabama, USA). Purified and FITC anti-mouse/human CD11b antibodies (Ab and FITC-Ab, respectively) were purchased from BioLegend (San Diego, California, USA). An Amplex® red hydrogen peroxide/peroxidase assay kit was purchased from Molecular Probes, Inc. (Eugene, Oregon, USA). An OxiSelect™ hydroxyl radical antioxidant capacity (HORAC) activity assay kit was purchased from Cell Biolabs, Inc. (San Diego, California, USA).Synthesis of 2 nm Ce0.7Zr0.3O2 nanoparticles (7CZ NPs)
In the same synthesis process as previously described,37 0.36 mg of cerium(III) acetylacetonate hydrate and 0.14 mg of zirconium(IV) acetylacetonate hydrate were added to 15 mL of oleylamine. The mixture was first sonicated for 10 min at 20 °C and then heated up to 80 °C with a heating rate of 2 °C min−1. The mixture was reacted at 80 °C for 1 day and then cooled down to 20 °C. The product was washed with acetone (100 mL) and collected by centrifugation at 5000 rpm several times. The resulting 7CZ NPs were dispersed in chloroform with a final concentration of 10 mg mL−1.Conjugation of phospholipid–polyethylene glycol-FITC
6 mg of DSPE-PEG(2000)-amine and 0.6 mg of fluorescein isothiocyanate were dispersed in 0.6 ml of chloroform. The mixture was heated and kept at 40 °C for 4 h under stirring. After the 4 h reaction, FITC was covalently conjugated to the amine-group in PEG.Conjugation of CD11b antibody to NHS-ester
In the reaction of EDC coupling, 1.5 mg of CD11b or FITC-CD11b antibody was added to the mixed solution of 30 mg of EDCI, 18 mg of NHS-ether, 60 μl of TEA and 9 ml of deionized (DI) water. The mixture was shaken for 2 h at room temperature. After the 2 h reaction, the carboxyl acid group of the CD11b antibody was covalently conjugated to the NHS-ester as an amine-reactive intermediate.Synthesis of phospholipid–polyethylene glycol-capped 7CZ, 7CZ-FITC NPs
To make water dispersible 7CZ NPs, 1.5 ml of 7CZ NPs dispersed in chloroform (10 mg mL−1) was mixed with 4.5 mL of a mixture of PEG(2000) in chloroform (10 mg ml−1, with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of mPEG(2000)-PE to DSPE-PEG(2000)-amine). In the case of 7CZ-FITC NPs, the same amount of 7CZ NPs dispersed in chloroform was mixed with 4.5 mL of a mixture of PEG(2000) and PEG(2000)-FITC as prepared in chloroform (10 mg ml−1, with a 10
1 ratio of mPEG(2000)-PE to DSPE-PEG(2000)-amine). In the case of 7CZ-FITC NPs, the same amount of 7CZ NPs dispersed in chloroform was mixed with 4.5 mL of a mixture of PEG(2000) and PEG(2000)-FITC as prepared in chloroform (10 mg ml−1, with a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio of mPEG(2000)-PE to DSPE-PEG(2000)-amine to DSPE-PEG(2000)-FITC). Each sample was treated using a rotary evaporator and a vacuum oven at 70 °C for 2 h to remove chloroform thoroughly. The resultant mixture was then dispersed in 5 ml of DI water to form a transparent colloidal suspension. The residues of phospholipid–PEG were removed by filtration using a 0.4 μm filter and ultracentrifugation several times. The purified product of each sample was kept in DI water.
2 ratio of mPEG(2000)-PE to DSPE-PEG(2000)-amine to DSPE-PEG(2000)-FITC). Each sample was treated using a rotary evaporator and a vacuum oven at 70 °C for 2 h to remove chloroform thoroughly. The resultant mixture was then dispersed in 5 ml of DI water to form a transparent colloidal suspension. The residues of phospholipid–PEG were removed by filtration using a 0.4 μm filter and ultracentrifugation several times. The purified product of each sample was kept in DI water.
Conjugation of CD11b antibody to 7CZ or 7CZ-FITC NPs
To conjugate CD11b or FITC-CD11b antibody to 7CZ or 7CZ-FITC NPs, respectively, the prepared amount of the water-dispersed sample (7CZ or 7CZ-FITC NPs) was added to the intermediate of the antibody–NHS ester mixture prepared in DI water. The mixture including both NPs and the antibody was stirred using a magnetic bar at room temperature for 12 h. Next, the reaction product was repeatedly washed by removing the supernatant of the sample after each ultracentrifugation. The purified sample of 7CZ-Ab or 7CZ-FITC-Ab NPs was finally dispersed in DI water.Characterization of 7CZ and/or 7CZ-Ab NPs
Transmission electron microscopy (TEM) and scanning transmission electron microscopy (STEM) at 200 kV (JEM-2100f, JEOL, Japan) were used to analyze 7CZ NPs by releasing a droplet of the sample dispersion onto a carbon-coated copper grid. The diffusion time and diffusion coefficient of the samples were analyzed using a fluorescence correlation spectrometer equipped with a confocal microscope (LSM 780 NLO, Carl Zeiss, Germany). Hydrodynamic diameters and zeta potentials of the samples were obtained by dynamic light scattering (DLS) measurements using a Zetasizer Nano-ZS system (Malvern Instruments, Inc., UK). X-ray diffraction (XRD) patterns of the sample were obtained with a diffractometer (New D8 Advance, Bruker, Germany). Phase identification was performed using JCPDS-ICDD 2000 software. X-ray photoelectron spectroscopy (XPS) analysis of the sample was performed using an XP spectrometer (AXIS-HSi, Kratos, UK). Each peak was fitted by CasaXPS software. Energy-dispersive X-ray spectroscopy (EDS) analysis was performed with a single drift detector (X-MaxN, Oxford Instruments, UK). AZtecTEM software was used to analyze the atomic content of the sample. Analysis for elemental confirmation and concentration of the sample was performed using an inductively coupled plasma atomic emission spectrometer (ICP-AES; ICPS-1000IV, Shimadzu, Japan). Assays for ROS determination were performed with a multiple plate reader (Victor X4, PerkinElmer, USA) by measuring light absorbance or fluorescence from the samples.SOD mimetic activity assay
All assays for ROS determination proceeded in almost the same way as previously described.37 The SOD assay kit (Sigma-Aldrich, USA) was used to assess the superoxide anion scavenging activity. First, 20 μL of each sample with a final concentration of 0.1 mM was added to 160 μL of a 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium sodium salt (WST-1) working solution. Then, 20 μL of a xanthine oxidase solution as a superoxide anion generator was added to each microplate well. After incubating at 37 °C for 20 min, the absorbance of each well at 450 nm was measured using a multiple plate reader (Victor X4). Since the absorbance is proportional to the amount of superoxide anion, the inhibition rate of the superoxide was calculated by quantifying the reduction in color development.Catalase (CAT) mimetic activity assay
The Amplex® red hydrogen peroxide/peroxidase assay kit (Molecular Probes, Inc., USA) was used to assess the quenching activities of hydrogen peroxide. Amplex® red reagent (10-acetyl-3,7-dihydroxyphenoxazine) reacts with H2O2, in combination with horseradish peroxide (HRP), to produce a red fluorescent oxidation product, resorufin. The fluorescence of resorufin (excitation and emission maximum at 571 and 585 nm, respectively) represents the H2O2 level in a solution. First, 10 μL of each sample with a final concentration of 0.1 mM was mixed with 40 μL of a H2O2 solution with a final concentration of 5 μM in each microplate well. After pre-incubating for 20 minutes, 50 μL of the Amplex® Red reagent/HRP working solution was added to each well, and the samples were then protected from light and incubated at 25 °C for 30 min. Then, the fluorescence was measured using a multiple plate reader (Victor X4).Hydroxyl radical antioxidant capacity (HORAC) activity assay
The HORAC assay kit (Cell Biolabs, Inc., USA) was used to assess the hydroxyl radical scavenging activity. First, 20 μL of each sample with a final concentration of 0.1 mM was added to 140 μL of the fluorescent probe. After incubating at 25 °C for 30 min, 20 μL of a hydroxyl initiator and 20 μL of Fenton reagent were added to each microplate well to generate hydroxyl radicals. After shaking for 15 s and incubating at 25 °C for 20 min, the fluorescence was then measured using a multiple plate reader (Victor X4).Animals
Male C57BL/6 mice aged 7–10 weeks were purchased from DBL, South Korea for experimental use. Four to five mice were housed in a plastic cage with standard bedding. They had access to food and water ad libitum. They were accommodated at a constant room temperature of 23 °C ± 2 °C and a 12 h dark/light cycle. All surgical and experimental procedures were approved by the Institutional Animal Care and Use Committee at Seoul National University. The animal treatments were performed in agreement with the guidelines of the International Association for the Study of Pain.SNT-induced neuropathic pain model
Mice were anesthetized with an intraperitoneal (i.p.) injection of 2% avertin, and SNT was induced by transecting the L5 spinal nerve as described previously.6 Briefly, an incision was made in the skin from the spinal processes at the L4 to L2 level. The paraspinal muscles were separated and the L6 transverse process was partially removed. The right L5 spinal nerve was exposed and carefully transected with small scissors. Then, 10% povidone–iodine topical solution was applied to the site of incision and the surgical site was closed with surgical staples. Sham-operated mice were subjected to the removal of the L6 transverse process. Sterile procedures were used throughout the surgery to prevent infection and to minimize the influence of inflammation.Intrathecal injection
For the administration of 7CZ, 7CZ-Ab, or saline, mice were injected with avertin (2%) for anesthesia. After shaving the back of the mice, nanoparticles (10 μg per 5 μl) or saline was injected using a 10 μl Hamilton syringe (Hamilton Company, Reno, NV, USA) with a 30-gauge one-half-inch needle into subarachnoid space; a slight tail-flick denoted appropriate administration of the test compounds.Behavioral analysis
All animal experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee, Seoul National University. A 50% withdrawal threshold was measured using a set of von Frey filaments (0.02–4 g, Stoelting, IL, USA), following an up-down method.47 Mice were placed in a cage with a wire mesh bottom which allowed full access to the paws. Nocifensive behaviors were evoked by a light touch of the filaments to the plantar right hind paw with sufficient force to cause slight buckling against the paw. The paws were touched with a series of von Frey filaments in an ascending order of strength, in intervals allowing for the resolution of behavioral response to the previous stimuli. A positive response was noted if the mice flinched, sharply withdrew or licked the paw (ESI video†). In the up-down method, when there was a lack of response to a filament, a next higher filament was used, while with a positive response, a lower filament was used. Once a threshold approach was denoted by a change in the response, another four von Frey presentations were done and the paw withdrawal threshold (PWT) was calculated by combining the value of the final von Frey filament used with an adjustment factor based on the response pattern of the animal.Primary glial cells and microglia culture
One-day-old C57BL/6 mice pups were used to culture primary glia using a procedure previously described.48 After removing the meninges from the cerebral hemisphere, tissue was dissociated into a single-cell suspension through gentle repetitive pipetting. Cells were cultured in Dulbecco's Modified Eagle's medium supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
×![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NEAA, and 1
NEAA, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
×![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) antibiotic/antimycotic in 75 cm2 flasks at 37 °C in a 5% CO2 incubator, and the medium was changed every five days. To isolate microglia from the mixed glia cells, the flasks were kept in a rotating shaker at 250 rpm for 3 h and then tapped firmly 15–20 days after culturing. The floating cells were collected and plated on poly-D-lysine (PDL)-coated glass coverslips in a four-well plate with 2 × 104 cells per well. After 15 min, the media were replaced with fresh media in order to eliminate unbound non-microglial cells and debris.
antibiotic/antimycotic in 75 cm2 flasks at 37 °C in a 5% CO2 incubator, and the medium was changed every five days. To isolate microglia from the mixed glia cells, the flasks were kept in a rotating shaker at 250 rpm for 3 h and then tapped firmly 15–20 days after culturing. The floating cells were collected and plated on poly-D-lysine (PDL)-coated glass coverslips in a four-well plate with 2 × 104 cells per well. After 15 min, the media were replaced with fresh media in order to eliminate unbound non-microglial cells and debris.
Real-time RT-PCR
The cDNA was synthesized using total RNA from mouse spinal cord tissue or cells cultured in vitro. The reverse transcription mixture consisted of 2 μg of total RNA, oligo-dT, M-MLV, RNase inhibitor, DTT, and RT-PCR buffer and was synthesized at 37 °C for 1 h. Real-time RT-PCR was performed using SYBR Green PCR Master Mix and ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA) as described previously.49 The following PCR primer sequences were used: GAPDH forward, 5′-AGG TCA TCC CAG AGC TGA ACG-3′; GAPDH reverse, 5′-CAC CCT GTT GCT GTA GCC GTA-3′; iNOS forward, 5′-GGC AAA CCC AAG GTC TAC GTT-3′; iNOS reverse, TCG CTC AAG TTC AGC TTG GT; IL-1β forward, 5′-GTG CTG TCG GAC CCA TAT GA-3′; IL-1β reverse, 5′-TTG TCG TTG CTT GGT TCT CC-3′; IL-6 forward, 5′-CCA CGA TTT CCC AGA GAA CAT-3′; and IL-6 reverse, 5′-TCC ATC CAG TTG CCT TCT TGG-3′. The mRNA level for each gene was normalized to the mRNA level of the GAPDH gene and presented as fold induction. Fold induction was calculated using the 2-ΔΔCT method, as described previously.50 All real-time RT-PCR experiments were performed at least three times, and the mean ± standard errors of the mean (SEM) values were reported unless otherwise noted.Flow cytometry analysis
The spinal cord tissue from lumbar 1 to 6 was removed and homogenized mechanically to a single cell suspension. For in vitro experiments, the mixed glial cells were detached using 0.25% trypsin with 3 min incubation at 37 °C and collected. Cells were washed with ice-cold PBS and 2% FBS and incubated with Fc BlockerTM (BD Bioscience, San Jose, CA, USA) for 10 min at 4 °C prior to staining. Then, cells stained with CD11b-APC (Biolegend Inc., San Diego, CA, USA) were analyzed with a BD FACSVERSE flow cytometer (BD Bioscience) to measure the CD11b+ microglia population. Likewise, cells stained with ACSA A-2-PE (Miltenyi Biotec, Bergisch Gladbach, Germany) were analyzed to measure the ACSA-2+/GLAST+ astrocyte population. Cells stained with Thy-1.2 violet (Biolegend Inc.) were analyzed to measure Thy-1+ neurons. Data were acquired and analyzed using BD FACSuite v1.2 (BD Biosciences).Immunofluorescence
Immunostaining was carried out using previously established protocols.51 The spinal cord sections were incubated in a blocking solution (5% normal goat serum, 2% bovine serum albumin (BSA), and 0.1% Triton X-100) for 1 h at room temperature (RT). Sections were then incubated overnight at 4 °C with primary antibody for rabbit-anti-Iba1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Wako, Osaka, Japan) or mouse-anti-8-OHG (1
1000; Wako, Osaka, Japan) or mouse-anti-8-OHG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Abcam, Cambridge, UK). After rinsing in 0.1 M PBS, the sections were incubated for 1 h at RT with a mixture of Cy3-conjugated secondary antibodies (1
200; Abcam, Cambridge, UK). After rinsing in 0.1 M PBS, the sections were incubated for 1 h at RT with a mixture of Cy3-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Jackson ImmunoResearch) and mounted with VectaShield medium (Vector Labs, Burlingame, CA, USA). Fluorescence images were obtained using a confocal microscope (LSM800; Carl Zeiss, Oberkochen, Germany). For the quantification of immunoreactivity, stained sections from the mouse spinal cord (L4–6 region) were taken and quantified using LSM 800 software. For immunocytochemistry (ICC), pure microglial cells were seeded onto a PDL-coated cover glass (2 × 104 cells per 12 mm2). After 2 days of proper attachment and growth, the cells were treated with FITC-conjugated nanoparticles. For ROS detection, cells were labeled with 10 μM CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichloro-dihydrofluorescein diacetate, acetyl ester) at 37 °C for 40 min. Tert-butyl hydroperoxide was treated for 1 h at 37 °C followed by DPBS washing, then fluorescence was detected by using a microplate reader (SPARK 10 M, TECAN, Austria). To get the image, cells were fixed in 2% PFA in 0.1 M PBS (pH 7.4) for 15 min. The antibody treatment protocol is same as histochemistry. Fluorescence images were obtained using a fluorescence microscope (Digital Inverted Fluorescence Microscope; Nikon, Tokyo, Japan).
200; Jackson ImmunoResearch) and mounted with VectaShield medium (Vector Labs, Burlingame, CA, USA). Fluorescence images were obtained using a confocal microscope (LSM800; Carl Zeiss, Oberkochen, Germany). For the quantification of immunoreactivity, stained sections from the mouse spinal cord (L4–6 region) were taken and quantified using LSM 800 software. For immunocytochemistry (ICC), pure microglial cells were seeded onto a PDL-coated cover glass (2 × 104 cells per 12 mm2). After 2 days of proper attachment and growth, the cells were treated with FITC-conjugated nanoparticles. For ROS detection, cells were labeled with 10 μM CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichloro-dihydrofluorescein diacetate, acetyl ester) at 37 °C for 40 min. Tert-butyl hydroperoxide was treated for 1 h at 37 °C followed by DPBS washing, then fluorescence was detected by using a microplate reader (SPARK 10 M, TECAN, Austria). To get the image, cells were fixed in 2% PFA in 0.1 M PBS (pH 7.4) for 15 min. The antibody treatment protocol is same as histochemistry. Fluorescence images were obtained using a fluorescence microscope (Digital Inverted Fluorescence Microscope; Nikon, Tokyo, Japan).
MTS assay
Mixed glial cells were grown on a 96-well plate with 3 × 104 cells per well after which they were treated with a cytotoxic reagent for 6 and 24 h. Then, 20 μl of MTS reagent (Promega, Madison, WI, USA) was added directly into the cell culture media and incubated at 37 °C for 1 h. Reduction of the MTS tetrazolium compound by viable cells to generate the formazan product soluble in the serum-free media was quantified by measuring the absorbance at 492 nm using an EMax® Plus Microplate Reader (Molecular Devices, San Jose, CA, USA). Absorbances obtained from samples treated with cytotoxic reagents were compared to absorbance in control samples to obtain the percentage of cell viability.Statistical analysis
All data are presented as the mean value with a standard error of the mean (SEM). Differences between groups for behavior tests were determined by the PASW statistical program (SPSS Inc.) with the LSD test. One-way ANOVA determined differences between groups for other data with a Newman–Keuls multiple comparison test. p < 0.05 was considered statistically significant.Results and discussion
Preparation, characterization and ROS scavenging performance of 7CZ and 7CZ-Ab NPs
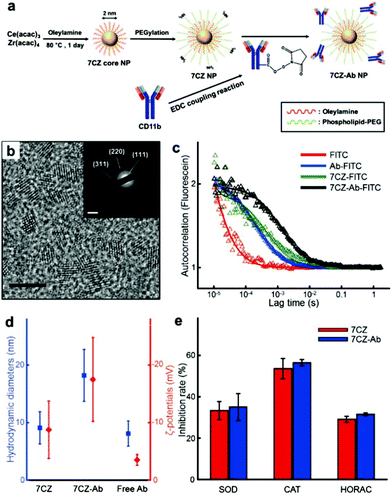
To obtain ceria–zirconia NPs (Ce0.7Zr0.3O2; 7CZ NPs) functionalized with the microglia-targeting antibodies, 7CZ NPs are first synthesized via a non-hydrolytic sol–gel reaction under mild conditions.37 The 7CZ NPs are further processed into CD11b antibody-conjugated 7CZ NPs (7CZ-Ab NPs) by the formation of stable amide bonds between the antibodies and the phospholipid–polyethylene glycol (PEG) shell on the NPs (Fig. 1a). High-resolution and scanning transmission electron microscopy (HRTEM and STEM, respectively) images show the discrete and well-defined lattice structure of the 2 nm-sized uniform 7CZ NP cores (Fig. 1b and Fig. S1†). The selected area electron diffraction (SAED) and X-ray diffraction (XRD) data indicate the cubic fluorite structure of the 7CZ NPs (Fig. 1b; inset, Fig. S2†). Owing to the increase in tetragonality by the insertion of Zr4+ ions to the ceria NPs, slight peak shifts of the 7CZ NPs in the XRD pattern are observed. Our synthesis method enables the 7CZ NPs to have a solid-solution form with a high Ce3+/Ce4+ ratio (the ratio of Ce3+ ions in 7CZ NPs: 52.6%), as confirmed by X-ray photoelectron spectroscopy (XPS) analysis (Fig. S3a†). To make the NPs water-dispersible, the surface of the 7CZ NPs is encapsulated with PEG, which provides not only restrained adsorption of proteins but also functional groups for further conjugation.52,53 After the PEGylation and subsequent functionalization with the antibodies, we examined the antibody attached to the NPs using both fluorescence correlation spectroscopy (FCS) and dynamic light scattering (DLS) analyses. In the FCS data, 7CZ-Ab NPs present an increased diffusion time and decreased diffusion coefficient compared to those of free antibodies and 7CZ NPs (Fig. 1c and Table S1†). The number of antibodies on a single 7CZ NP is 3.29 and they occupy 70% of the monolayer area for the antibody (ESI†). In addition, DLS measurements show that the hydrodynamic diameters (HD) and the ζ-potential values of the NPs are 9.1 nm and −8.8 mV for the 7CZ NPs, and 18.2 nm and −17.5 mV for the 7CZ-Ab NPs, respectively (Fig. 1d and Fig. S5†). Taken together, successful conjugation of the antibodies on the NPs is shown. To see whether the antibody conjugation affects the ROS scavenging activities of the NPs aside from cell-living conditions, we performed superoxide dismutase (SOD)-mimetic, catalase (CAT)-mimetic, and hydroxyl radical antioxidant capacity (HORAC) activity assays in aqueous media. In all of the assays, the antibody conjugation onto the NPs does not hinder the catalytic activities of the NPs to scavenge ROS, as 7CZ-Ab and 7CZ NPs show comparable levels of performance (Fig. 1e). These results also augment the notion that the elimination of ROS is credited to the core NPs rather than the accompanying antibodies.In vitro microglia-specific delivery of 7CZ-Ab NPs in primary glial cells
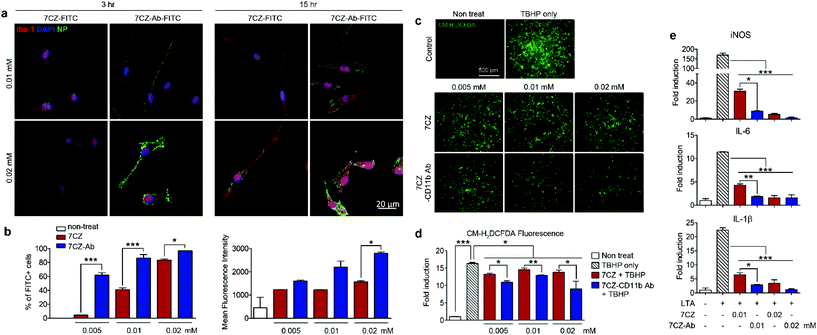
To test and compare microglial uptake of the 7CZ and 7CZ-Ab NPs, two concentrations of fluorescein isothiocyanate (FITC)-conjugated NPs, 0.01 and 0.02 mM, were treated to primary pure microglial culture for 3 and 15 h, after which the FITC signal in the cell populations was detected using both immunocytochemistry (ICC) and flow cytometry analysis. Higher FITC signals from 7CZ-Ab-FITC-treated cells for both concentrations and time points in ICC analysis suggest a greater microglia uptake efficiency of the 7CZ-Ab NPs than that of 7CZ NPs (Fig. 2a). Furthermore, while the FITC signal is not seen in the microglial cells at 3 h after the treatment with 7CZ NPs, it is observed in the membrane of the microglial cells after treating with the 7CZ-Ab NPs for both concentrations under confocal microscopy. 15 h after treatment with the NPs, the whole cytosol of microglia is filled with FITC+ dots for both concentrations of 7CZ-Ab NPs, whereas a few dots of the FITC signal are detected only after treatment with 0.02 mM 7CZ NPs. These results show that 7CZ-Ab NPs attach to the microglial cell membrane and internalize into the cell faster than 7CZ NPs. Accordingly, flow cytometry analysis also reveals a significant increase in the uptake of 7CZ-Ab-FITC NPs than 7CZ-FITC NPs by CD11b-positive microglial cells (Fig. 2b). The percentage of the microglia cells positive for FITC after treatment with 0.005 mM 7CZ-Ab-FITC NPs is approximately 60% at 3 h, while less than 5% are positive after treatment with 7CZ-FITC NPs. At the NP treatment concentration of 0.01 mM, 7CZ-Ab-FITC NPs are taken up by more than 80% of microglia compared to the 7CZ-FITC NP uptake of around 40% microglia (Fig. 2b). Although the FITC-positive populations of microglial cells are similar at the higher concentration (0.02 mM), the mean fluorescence intensity (MFI) of the 7CZ-Ab-FITC NP-treated cells is much higher than that of the 7CZ-FITC NP-treated cells, indicating a higher 7CZ-Ab-FITC NP uptake by microglia at the higher concentration as well. The enhanced uptake of 7CZ by CD11b antibody conjugation was further demonstrated by inductively coupled plasma-mass spectroscopy (ICP-MS) analysis (Fig. S6†). The intracellular concentration of 7CZ-NPs is much higher when microglia are incubated with CD11b Ab-conjugated 7CZ NPs compared to isotype IgG Ab-conjugated 7CZ NPs. Taken together, these data show that CD11b antibody conjugation to 7CZ NPs increases the targeting capacity of the NPs to microglia and enhances their microglial uptake efficiency.In vitro antioxidant and anti-inflammatory effects of 7CZ and 7CZ-Ab NPs in primary glial cells
Pro-inflammatory mediators induced in the activated spinal microglia, such as IL-1β, IL-6, and nitric oxide (NO), contribute to the development of neuropathic pain.54–56 In addition, ROS are implicated in the induction of pro-inflammatory gene expression.57 First, we tested the anti-oxidant effects of 7CZ and 7CZ-Ab in vitro. For this purpose, we used mixed glia instead of pure microglia since they better recapitulate the in vivo microenvironment.58,59 The treatment with tert-butyl hydroperoxide (TBHP), an exogenous oxidative stress inducer increased intracellular ROS levels in primary glial cells, which were measured by CM-H2DCFDA fluorescence (Fig. 2c). When treated with 7CZ-Ab, the TBHP-induced ROS production is significantly reduced (Fig. 2c and d). Upon quantification of the fluorescence levels, the inhibitory effects on TBHP-induced ROS production are more pronounced after 7CZ-Ab pretreatment than 7CZ pretreatment (Fig. 2d). Furthermore, we investigated whether 7CZ and 7CZ-Ab NPs inhibit the expression of pain-mediating genes in primary glial cells in vitro. Mixed glial cells were either treated with lipoteichoic acid (LTA) (1 μg ml−1), a toll-like receptor 2 (TLR2) agonist,60 or co-treated with either LTA + 7CZ NPs or LTA + 7CZ-Ab NPs at two different NP concentrations (0.01 and 0.02 mM) for 15 h. Upon the LTA treatment, mRNA expression of inducible nitric oxide synthase (iNOS), IL-6, and IL-1β increases by 169-, 12- and 22-fold, respectively (Fig. 2e). Co-treatment with either 7CZ NPs or 7CZ-Ab NPs results in a significant decrease of transcription levels in a dose-dependent manner. This inhibitory effect is more pronounced with the treatment of 7CZ-Ab NPs than 7CZ NPs as demonstrated by the significantly higher reduction rate of the cytokines and iNOS with the 7CZ-Ab NP treatment. The LTA-induced mRNA expression levels of iNOS, IL-6, and IL-1β are reduced by 95, 86, and 91% with the treatment of 0.01 mM 7CZ-Ab NPs, respectively, while they are reduced only by 82, 63, and 71% with the 7CZ NPs of the same concentration. These data show that 7CZ-Ab NPs are more potent in inhibiting pain-mediating gene expression in microglia than 7CZ NPs. The cytotoxicity of the 7CZ NPs and 7CZ-Ab NPs to glial cells was assessed by MTS assay (Fig. S6†). No significant cytotoxicity is detected with the treatment of up to 0.04 mM concentrations of 7CZ NPs and 7CZ-Ab NPs. In addition, there is no significant difference in cell toxicity between the 7CZ NPs and 7CZ-Ab NPs.In vivo delivery of intrathecally injected 7CZ and 7CZ-Ab NPs
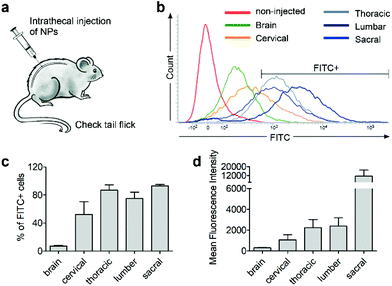
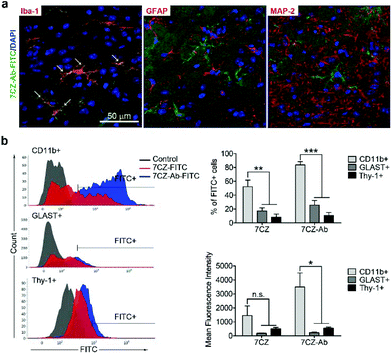
Next, we investigated whether CD11b antibody conjugation could result in an increased uptake of the 7CZ NPs by microglia and confer them specificity to microglia in vivo. Firstly, we checked whether or not intrathecal administration of 7CZ NPs can reach the spinal cord cells (Fig. 3a). After 24 h of FITC-conjugated 7CZ NP administration, although NPs are broadly delivered to the area from the brain to the sacral of the spinal cord, more than 80% of cells in the thoracic to sacral, ∼50% in the cervical, and only ∼7% of the brain cells are positive for FITC (Fig. 3c and d). To check the uptake of 7CZ-Ab NPs in vivo, the NPs were administered in the same way into the spinal canal of mice, and then the spinal cords were analyzed via immunohistochemistry (IHC) after 24 h of injection. The lumbar segment 4–6 (L4–L6) tissue samples were immunostained with cell type-specific antibodies and localization of the FITC signal was observed. As seen in Fig. 4a, the FITC signal mainly co-localizes with ionized calcium binding adaptor molecule 1 (Iba-1)-positive microglia but not with glial fibrillary acidic protein (GFAP)-positive astrocytes and microtubule-associated protein 2 (MAP-2)-positive neurons. For further confirmation, FITC-positive cells in the spinal cord tissue were analyzed by flow cytometry after 24 h of 7CZ-FITC NP or 7CZ-CD11b Ab-FITC NP administration. Upon characterization of the 7CZ Ab-FITC-positive cells by using cell type-specific markers, the FITC signal is detected in 84% of CD11b-positive microglia, 26% of glutamate aspartate transporter (GLAST)-positive astrocytes, and 11% Thy-1-positive neurons (Fig. 4b) for each cell type. A significantly greater percentage of microglial cells shows an uptake of 7CZ-Ab NPs in comparison with astrocytes and neurons. Moreover, the MFI of microglial cells is significantly higher than those of astrocytes and neurons. The significant difference in uptake profiles of 7CZ-Ab NPs between CD11b-positive microglia and other cell types denotes the specificity of the NPs to microglia as a result of the CD11b antibody conjugation to the 7CZ NPs.Analgesic effects of 7CZ and 7CZ-Ab NPs in the SNT-induced neuropathic pain mouse model
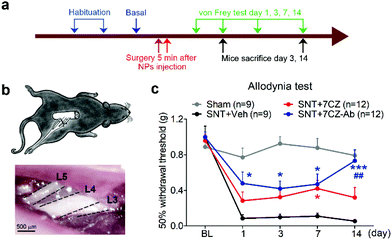
To test if microglia targeting by 7CZ-Ab NPs has a better analgesic effect, we compared the susceptibility of 7CZ NP- or 7CZ-Ab NP-treated mice to nerve injury-induced pain hypersensitivity. After 2 days of habituation on the von Frey test apparatus and evaluation of the basal level, the saline or the NPs were intrathecally injected 5 min before surgery, and then the SNT injury was performed by transecting L5 spinal nerve (Fig. 5a and b). The SNT mice injected with saline display an increased sensitivity to mechanical stimuli as measured by the von Frey test. The paw withdrawal threshold (PWT) to mechanical stimuli decreases from around 0.99 to less than 0.1 g at one-day post injury (Fig. 5c). The threshold remains below 0.1 g for two weeks. In 7CZ NP-treated mice, the PWT increases to 0.28, 0.32, 0.42, and 0.32 g at 1, 3, 7, and 14 days post-injection, respectively indicating a moderate reduction in mechanical allodynia. In the 7CZ-Ab NP-treated mice, where the same molar concentration of the NPs is administered intrathecally, the PWT increases to 0.48, 0.42, 0.47, and 0.73 g at 1, 3, 7, and 14 days post-injection, respectively, indicating that the 7CZ-Ab NPs have a much stronger analgesic effect in comparison with the 7CZ NPs. A significant increase in the withdrawal threshold after treatment with the 7CZ-Ab NPs in comparison with the 7CZ NPs (day 14; p-value = 0.006) confirms their greater therapeutic effect on neuropathic pain treatment, which is mostly due to their augmented selectivity towards microglia.In vivo attenuation of microglial activation and oxidative stress by 7CZ and 7CZ-Ab NPs
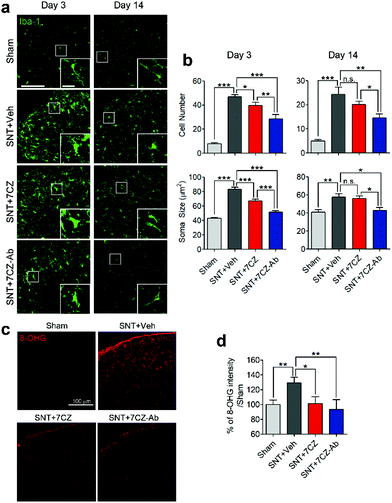
To evaluate the inhibitory effects of 7CZ NPs and 7CZ-Ab NPs on spinal cord microglia activation in vivo, microglia cell number and soma size, two key features of microglia activation,61,62 were measured in L5 spinal cord sections of sham and SNT-injured mice after the injection of saline, 7CZ NPs, or 7CZ-Ab NPs (Fig. 6a). Both microglia cell number and soma size are increased upon SNT. The increase in microglia cell number and soma size observed 3 days post-SNT surgery is significantly reduced by 7CZ and 7CZ-Ab treatment (Fig. 6b). Between these, the inhibitory effects are much stronger in 7CZ-Ab-injected than 7CZ-injected mice (38.7% vs. 15.2% in cell number; 39.7% vs. 19.5% in soma size). At 14 days post-SNT surgery, inhibitory effects are observed only in the 7CZ-Ab-injected group (39.4% in cell number; 25.5% in soma size), not in the 7CZ-injected group (Fig. 6b). To investigate the protective effects of the NPs against oxidative stress, we assessed non-mitochondrial ROS production by using 8-hydroxyguanine (8-OHG) antibody, which detects oxidized nucleic acid resulting from cellular ROS damage. Three days after the SNT surgery, the 8-OHG-immunoreactive signals in the dorsal horn area of SNT-injured mice increase by 29% in comparison with the sham group. In 7CZ NP- and 7CZ-Ab NP-treated mice, the mean intensity decreases to a greater extent in the 7CZ-Ab NP-treated mice (36%, p-value = 0.010) than in the 7CZ-NP-treated mice (28%, p-value = 0.038) compared to the SNT + Veh group (Fig. 6b). Collectively, 7CZ-Ab NPs inhibit SNT-induced spinal cord microglia activation and ROS accumulation more efficiently than 7CZ NPs in vivo.Conclusions
In conclusion, strategies for enhanced therapeutic efficacy against neuropathic pain are achieved by both improved catalytic performance and delivery efficiency of 7CZ-Ab NPs. Greater uptake of the NPs by microglial cells extenuates the vicious activation of microglia by reducing pro-inflammatory gene expression, whereas the respective dose of 7CZ NPs is insufficient to do the same. In accordance with our in vitro data, conferring a microglial specificity on the 7CZ-Ab NPs to target the damaged tissues of allodynic mice attenuates microglial activation and oxidative damage, thus concomitantly inhibiting mechanical allodynia to a greater extent in vivo, compared to the non-targeted 7CZ NPs. These results suggest that the proper combination of therapeutic approaches in the regulation of microglia increases the beneficial effects to treat diseases related to the burst activation of microglia.Author contributions
Taeghwan Hyeon and Sung Joong Lee conceived and designed the experiments. Boomin Choi, Min Soh and Yelina Manandhar performed the experiments and analyzed the data with equal contribution. Taeghwan Hyeon, Sung Joong Lee, Boomin Choi, Min Soh and Yelina Manandhar wrote and refined the article. Dokyoon Kim and Hyek Jin Kwon supervised the acquisition of results. Sang Ihn Han, Seungmin Baik, Kwangsoo Shin, Sagang Koo and Giho Ko performed the experiments of nanoparticle synthesis and analysis. Junyoung Oh and Heehong Hwang performed the biological experiments.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Institute for Basic Science (IBS) in Korea (grant number IBS-R006-D1) and grants from the National Research Foundation of Korea (NRF-2016M3C7A1905074).Notes and references
- S. Beggs, T. Trang and M. W. Salter, Nat. Neurosci., 2012, 15, 1068–1073 CrossRef CAS PubMed.
- K. Inoue and M. Tsuda, Glia, 2009, 57, 1469–1479 CrossRef PubMed.
- M. Tsuda, S. Beggs, M. W. Salter and K. Inoue, Glia, 2013, 61, 55–61 CrossRef PubMed.
- T. Trang, S. Beggs and M. W. Salter, Exp. Neurol., 2012, 234, 354–361 CrossRef CAS PubMed.
- F. Ferrini, T. Trang, T. A. Mattioli, S. Laffray, T. Del'Guidice, L. E. Lorenzo, A. Castonguay, N. Doyon, W. Zhang, A. G. Godin, D. Mohr, S. Beggs, K. Vandal, J. M. Beaulieu, C. M. Cahill, M. W. Salter and Y. De Koninck, Nat. Neurosci., 2013, 16, 183–192 CrossRef CAS PubMed.
- D. Kim, B. You, E. K. Jo, S. K. Han, M. I. Simon and S. J. Lee, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14851–14856 CrossRef CAS PubMed.
- J. Haslund-Vinding, G. McBean, V. Jaquet and F. Vilhardt, Br. J. Pharmacol., 2017, 174, 1733–1749 CrossRef CAS PubMed.
- J. Park, J. S. Min, B. Kim, U. B. Chae, J. W. Yun, M. S. Choi, I. K. Kong, K. T. Chang and D. S. Lee, Neurosci. Lett., 2015, 584, 191–196 CrossRef CAS PubMed.
- J. K. Andersen, Nat. Med., 2004, 10(Suppl), S18–S25 CrossRef PubMed.
- X. Gao, H. K. Kim, J. M. Chung and K. Chung, Pain, 2007, 131, 262–271 CrossRef CAS PubMed.
- S. K. Rajendrakumar, V. Revuri, M. Samidurai, A. Mohapatra, J. H. Lee, P. Ganesan, J. Jo, Y. K. Lee and I. K. Park, Nano Lett., 2018, 18, 6417–6426 CrossRef CAS PubMed.
- S. R. Cerqueira, J. M. Oliveira, N. A. Silva, H. Leite-Almeida, S. Ribeiro-Samy, A. Almeida, J. F. Mano, N. Sousa, A. J. Salgado and R. L. Reis, Small, 2013, 9, 738–749 CrossRef CAS PubMed.
- D. Liu, C. Poon, K. Lu, C. He and W. Lin, Nat. Commun., 2014, 5, 4182 CrossRef CAS PubMed.
- X. Duan, C. Chan, N. Guo, W. Han, R. R. Weichselbaum and W. Lin, J. Am. Chem. Soc., 2016, 138, 16686–16695 CrossRef CAS PubMed.
- Y. Liu, X. Yang, Z. Huang, P. Huang, Y. Zhang, L. Deng, Z. Wang, Z. Zhou, Y. Liu, H. Kalish, N. M. Khachab, X. Chen and Z. Nie, Angew. Chem., Int. Ed., 2016, 55, 15297–15300 CrossRef CAS PubMed.
- Y. Liu, K. Ai, X. Ji, D. Askhatova, R. Du, L. Lu and J. Shi, J. Am. Chem. Soc., 2017, 139, 856–862 CrossRef CAS PubMed.
- L. S. Lin, J. Song, L. Song, K. Ke, Y. Liu, Z. Zhou, Z. Shen, J. Li, Z. Yang, W. Tang, G. Niu, H. H. Yang and X. Chen, Angew. Chem., Int. Ed., 2018, 57, 4902–4906 CrossRef CAS PubMed.
- Z. Zhou, J. Song, L. Nie and X. Chen, Chem. Soc. Rev., 2016, 45, 6597–6626 RSC.
- Y. Lee, H. Kim, S. Kang, J. Lee, J. Park and S. Jon, Angew. Chem., Int. Ed., 2016, 55, 7460–7463 CrossRef CAS PubMed.
- W. Cai, D. W. Shin, K. Chen, O. Gheysens, Q. Cao, S. X. Wang, S. S. Gambhir and X. Chen, Nano Lett., 2006, 6, 669–676 CrossRef CAS PubMed.
- J. Chen, S. Patil, S. Seal and J. F. McGinnis, Nat. Nanotechnol., 2006, 1, 142–150 CrossRef CAS PubMed.
- C. K. Kim, T. Kim, I. Y. Choi, M. Soh, D. Kim, Y. J. Kim, H. Jang, H. S. Yang, J. Y. Kim, H. K. Park, S. P. Park, S. Park, T. Yu, B. W. Yoon, S. H. Lee and T. Hyeon, Angew. Chem., Int. Ed., 2012, 51, 11039–11043 CrossRef CAS PubMed.
- H. J. Kwon, M. Y. Cha, D. Kim, D. K. Kim, M. Soh, K. Shin, T. Hyeon and I. Mook-Jung, ACS Nano, 2016, 10, 2860–2870 CrossRef CAS PubMed.
- K. L. Heckman, W. DeCoteau, A. Estevez, K. J. Reed, W. Costanzo, D. Sanford, J. C. Leiter, J. Clauss, K. Knapp, C. Gomez, P. Mullen, E. Rathbun, K. Prime, J. Marini, J. Patchefsky, A. S. Patchefsky, R. K. Hailstone and J. S. Erlichman, ACS Nano, 2013, 7, 10582–10596 CrossRef CAS PubMed.
- Q. Bao, P. Hu, Y. Xu, T. Cheng, C. Wei, L. Pan and J. Shi, ACS Nano, 2018, 12, 6794–6805 CrossRef CAS PubMed.
- H. J. Kwon, D. Kim, K. Seo, Y. G. Kim, S. I. Han, T. Kang, M. Soh and T. Hyeon, Angew. Chem., Int. Ed., 2018, 57, 9408–9412 CrossRef CAS PubMed.
- F. Zeng, Y. Wu, X. Li, X. Ge, Q. Guo, X. Lou, Z. Cao, B. Hu, N. J. Long, Y. Mao and C. Li, Angew. Chem., Int. Ed., 2018, 57, 5808–5812 CrossRef CAS PubMed.
- S. M. Hirst, A. S. Karakoti, R. D. Tyler, N. Sriranganathan, S. Seal and C. M. Reilly, Small, 2009, 5, 2848–2856 CrossRef CAS PubMed.
- I. Celardo, J. Z. Pedersen, E. Traversa and L. Ghibelli, Nanoscale, 2011, 3, 1411–1420 RSC.
- S. Das, J. M. Dowding, K. E. Klump, J. F. McGinnis, W. Self and S. Seal, Nanomedicine, 2013, 8, 1483–1508 CrossRef CAS PubMed.
- C. Korsvik, S. Patil, S. Seal and W. T. Self, Chem. Commun., 2007, 1056–1058, 10.1039/b615134e.
- Y. Xue, Q. Luan, D. Yang, X. Yao and K. Zhou, J. Phys. Chem. C, 2011, 115, 4433–4438 CrossRef CAS.
- I. Celardo, M. De Nicola, C. Mandoli, J. Z. Pedersen, E. Traversa and L. Ghibelli, ACS Nano, 2011, 5, 4537–4549 CrossRef CAS PubMed.
- A. Gupta, S. Das, C. J. Neal and S. Seal, J. Mater. Chem. B, 2016, 4, 3195–3202 RSC.
- S. Fernandez-Garcia, L. Jiang, M. Tinoco, A. B. Hungria, J. Han, G. Blanco, J. J. Calvino and X. Chen, J. Phys. Chem. C, 2016, 120, 1891–1901 CrossRef CAS.
- A. Kumar, S. Babu, A. S. Karakoti, A. Schulte and S. Seal, Langmuir, 2009, 25, 10998–11007 CrossRef CAS PubMed.
- M. Soh, D. W. Kang, H. G. Jeong, D. Kim, D. Y. Kim, W. Yang, C. Song, S. Baik, I. Y. Choi, S. K. Ki, H. J. Kwon, T. Kim, C. K. Kim, S. H. Lee and T. Hyeon, Angew. Chem., Int. Ed., 2017, 56, 11399–11403 CrossRef CAS PubMed.
- J. Yoo, D. Lee, V. Gujrati, N. S. Rejinold, K. M. Lekshmi, S. Uthaman, C. Jeong, I. K. Park, S. Jon and Y. C. Kim, J. Controlled Release, 2017, 246, 142–154 CrossRef CAS PubMed.
- M. Huo, L. Wang, Y. Chen and J. Shi, Nat. Commun., 2017, 8, 357 CrossRef PubMed.
- L. Pan, J. Liu and J. Shi, Chem. Soc. Rev., 2018, 47, 6930–6946 RSC.
- C. Carrillo-Carrion, M. Atabakhshi-Kashi, M. Carril, K. Khajeh and W. J. Parak, Angew. Chem., Int. Ed., 2018, 57, 5033–5036 CrossRef CAS PubMed.
- S. A. Costa, D. Mozhdehi, M. J. Dzuricky, F. J. Isaacs, E. M. Brustad and A. Chilkoti, Nano Lett., 2019, 19, 247–254 CrossRef CAS PubMed.
- M. Colombo, L. Fiandra, G. Alessio, S. Mazzucchelli, M. Nebuloni, C. De Palma, K. Kantner, B. Pelaz, R. Rotem, F. Corsi, W. J. Parak and D. Prosperi, Nat. Commun., 2016, 7, 13818 CrossRef CAS PubMed.
- Y. L. Zhou, L. Zhang, Z. Zhou, W. Liu, Y. Lu, S. He, Y. Cui, Y. Qin and M. Hua, J. Biomed. Nanotechnol., 2018, 14, 2185–2197 CrossRef CAS PubMed.
- S. M. Davis, D. Reichel, Y. Bae and K. R. Pennypacker, Pharm. Res., 2018, 35, 6 CrossRef PubMed.
- S. R. Cerqueira, B. L. Silva, J. M. Oliveira, J. F. Mano, N. Sousa, A. J. Salgado and R. L. Reis, Macromol. Biosci., 2012, 12, 591–597 CrossRef CAS PubMed.
- S. R. Chaplan, F. W. Bach, J. W. Pogrel, J. M. Chung and T. L. Yaksh, J. Neurosci. Methods, 1994, 53, 55–63 CrossRef CAS PubMed.
- S. J. Lee, T. Zhou, C. Choi, Z. Wang and E. N. Benveniste, J. Immunol., 2000, 164, 1277–1285 CrossRef CAS PubMed.
- H. Kim, B. Choi, H. Lim, H. Min, J. H. Oh, S. Choi, J. G. Cho, J. S. Park and S. J. Lee, Mol. Pain, 2017, 13, 1744806917697006 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- I. H. Cho, J. Hong, E. C. Suh, J. H. Kim, H. Lee, J. E. Lee, S. Lee, C. H. Kim, D. W. Kim, E. K. Jo, K. E. Lee, M. Karin and S. J. Lee, Brain, 2008, 131, 3019–3033 CrossRef PubMed.
- P. Del Pino, F. Yang, B. Pelaz, Q. Zhang, K. Kantner, R. Hartmann, N. Martinez de Baroja, M. Gallego, M. Moller, B. B. Manshian, S. J. Soenen, R. Riedel, N. Hampp and W. J. Parak, Angew. Chem., Int. Ed., 2016, 55, 5483–5487 CrossRef CAS PubMed.
- B. Pelaz, P. del Pino, P. Maffre, R. Hartmann, M. Gallego, S. Rivera-Fernandez, J. M. de la Fuente, G. U. Nienhaus and W. J. Parak, ACS Nano, 2015, 9, 6996–7008 CrossRef CAS PubMed.
- P. M. Grace, K. A. Strand, E. L. Galer, D. J. Urban, X. Wang, M. V. Baratta, T. J. Fabisiak, N. D. Anderson, K. Cheng, L. I. Greene, D. Berkelhammer, Y. Zhang, A. L. Ellis, H. H. Yin, S. Campeau, K. C. Rice, B. L. Roth, S. F. Maier and L. R. Watkins, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E3441–E3450 CrossRef CAS PubMed.
- M. Kobayashi, H. Konishi, A. Sayo, T. Takai and H. Kiyama, J. Neurosci., 2016, 36, 11138–11150 CrossRef CAS PubMed.
- Y. W. Wang, X. Zhang, C. L. Chen, Q. Z. Liu, J. W. Xu, Q. Q. Qian, W. Y. Li and Y. N. Qian, Neurosci. Lett., 2017, 647, 85–90 CrossRef CAS PubMed.
- J. Ye, Z. Jiang, X. Chen, M. Liu, J. Li and N. Liu, J. Neurochem., 2017, 142, 215–230 CrossRef CAS PubMed.
- J. V. Welser-Alves, S. J. Crocker and R. Milner, J. Neuroinflammation, 2011, 8, 61 CrossRef CAS PubMed.
- F. Aloisi, G. Penna, J. Cerase, B. Menendez Iglesias and L. Adorini, J. Immunol., 1997, 159, 1604–1612 CAS.
- D. Kim, M. A. Kim, I. H. Cho, M. S. Kim, S. Lee, E. K. Jo, S. Y. Choi, K. Park, J. S. Kim, S. Akira, H. S. Na, S. B. Oh and S. J. Lee, J. Biol. Chem., 2007, 282, 14975–14983 CrossRef CAS PubMed.
- Z. Chen, W. Jalabi, K. B. Shpargel, K. T. Farabaugh, R. Dutta, X. Yin, G. J. Kidd, C. C. Bergmann, S. A. Stohlman and B. D. Trapp, J. Neurosci., 2012, 32, 11706–11715 CrossRef CAS PubMed.
- B. M. Davis, M. Salinas-Navarro, M. F. Cordeiro, L. Moons and L. De Groef, Sci. Rep., 2017, 7, 1576 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9nr02648g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |