Open Access Article
Open Access ArticleAntimicrobial peptide based magnetic recognition elements and Au@Ag-GO SERS tags with stable internal standards: a three in one biosensor for isolation, discrimination and killing of multiple bacteria in whole blood†
Kaisong
Yuan‡
ac,
Qingsong
Mei‡
b,
Xinjie
Guo
a,
Youwei
Xu
d,
Danting
Yang
e,
Beatriz Jurado
Sánchez
 c,
Bingbing
Sheng
a,
Chusheng
Liu
a,
Ziwei
Hu
a,
Guangchao
Yu
f,
Hongming
Ma
f,
Hao
Gao
*a,
Christoph
Haisch
g,
Reinhard
Niessner
g,
Zhengjing
Jiang
*a and
Haibo
Zhou
c,
Bingbing
Sheng
a,
Chusheng
Liu
a,
Ziwei
Hu
a,
Guangchao
Yu
f,
Hongming
Ma
f,
Hao
Gao
*a,
Christoph
Haisch
g,
Reinhard
Niessner
g,
Zhengjing
Jiang
*a and
Haibo
Zhou
 *a
*a
aInstitute of Pharmaceutical Analysis, College of Pharmacy, Jinan University, Guangzhou, Guangdong 510632, China. E-mail: haibo.zhou@jnu.edu.cn; jzjjackson@hotmail.com; tghao@jnu.edu.cn
bSchool of Medical Engineering, Hefei University of Technology, Tunxi road 193, Hefei 230009, China
cDepartment of Analytical Chemistry, Physical Chemistry and Chemical Engineering, University of Alcala, Alcala de Henares E-28871, Madrid, Spain
dShanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai 201210, China
eDepartment of Preventative Medicine, Zhejiang Provincial Key Laboratory of Pathological and Physiological Technology, Medical School of Ningbo University, Ningbo, Zhejiang 315211, China
fThe First Affiliated Hospital of Jinan University, Guangzhou, Guangdong 510632, China
gInstitute of Hydrochemistry and Chair for Analytical Chemistry, Technical University of Munich, Marchioninistr. 17, D-81377, Munich, Germany
First published on 2nd November 2018
Abstract
In this study, a new biosensor based on a sandwich structure has been developed for the isolation and detection of multiple bacterial pathogens via magnetic separation and SERS tags. This novel assay relies on antimicrobial peptide (AMP) functionalized magnetic nanoparticles as “capturing” probes for bacteria isolation and gold coated silver decorated graphene oxide (Au@Ag-GO) nanocomposites modified with 4-mercaptophenylboronic acid (4-MPBA) as SERS tags. When different kinds of bacterial pathogens are combined with the SERS tags, the “fingerprints” of 4-MPBA show corresponding changes due to the recognition interaction between 4-MPBA and different kinds of bacterial cell wall. Compared with the label-free SERS detection of bacteria, 4-MPBA here can be used as an internal standard (IS) to correct the SERS intensities with high reproducibility, as well as a Raman signal reporter to enhance the sensitivity and amplify the differences among the bacterial “fingerprints”. Thus, three bacterial pathogens (Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa) were successfully isolated and detected, with the lowest concentration for each of the strains detected at just 101 colony forming units per mL (CFU mL−1). According to the changes in the “fingerprints” of 4-MPBA, three bacterial strains were successfully discriminated using discriminant analysis (DA). In addition, the AMP modified Fe3O4NPs feature high antibacterial activities, and can act as antibacterial agents with low cellular toxicology in the long-term storage of blood for future safe blood transfusion applications. More importantly, this novel method can be applied in the detection of bacteria from clinical patients who are infected with bacteria. In the validation analysis, 97.3% of the real blood samples (39 patients) could be classified effectively (only one patient infected with E. coli was misclassified). The multifunctional biosensor presented here allows for the simultaneous isolation, discrimination and killing of bacteria, suggesting its high potential for clinical diagnosis and safe blood transfusions.
Introduction
Infections caused by bacterial diseases are a global health threat to the general public and demand the development of fast, sensitive and accurate diagnostic methods.1,2 Traditional methods for pathogen detection fall within three categories: standard plate colony counting, polymerase chain reaction (PCR), and immunology based techniques such as enzyme-linked immunosorbent assays (ELISA).3 Yet, standard plate colony counting always involves separation, identification, culturing and counting, which are complex and time-consuming (typically a few days). Despite PCR and ELISA overcoming such time limitations, such methods require expensive, specialized equipment, complicated sample pretreatment, and even lack the ability to remove interference. To overcome such deficiencies, the development of miniaturized biosensors with much shorter analysis time, higher sensitivity and specificity is currently a hot research topic.4–6Surface enhanced Raman scattering (SERS) has become a subject of interest which may realize the quick, sensitive and effective detection of bacteria.7 The main advantage of the SERS detection of pathogens is the ability to provide sharp, specific fingerprint spectra of the bacteria, making it easy to discriminate among different kinds of bacteria from a mixed sample matrix.8 Recently, Wang et al. have modified 4-mercaptophenylboronic acid (4-MPBA) on an AgNPs@Si chip for the capture and SERS discrimination of E. coli and S. aureus in blood samples. When different kinds of bacteria are captured by the 4-MPBA modified chip, the Raman “fingerprints” of 4-MPBA will show corresponding changes; hence the bacteria can be identified. However, the capture of the bacteria is based on the interaction between boronic acid and the diol group of the saccharide. Thus, blood cells with saccharides will also be captured by the SERS chip due to the poor selectivity of 4-MPBA.9 Though the “fingerprints” help to discriminate bacteria from the interference, as the sample become more complex, the selectivity will be limited. To improve the detection selectively, recognition elements with higher specificity have been introduced to capture bacteria.10–12 They mainly include antibodies,13–15 aptamers,16,17 and antibiotics.18 Antibodies with high specificity suffer from high cost, as well as poor stability under harsh environmental conditions. Conversely, aptamers are good recognition elements with high specificity and good stability. Yet, the aptamers available for the specific capture of bacteria are limited. Antibiotics possess the advantages of low cost, high stability, and specificity for the capture of most bacteria. Nevertheless, as a small molecule, an antibiotic provides few binding sites for bacterial recognition, which may restrict their effective capture.
Antimicrobial peptides (AMPs), which are found in multiple niches in nature and typically consist of 10–40 residues, have several attractive advantages as the bacteria capture element.19,20 They are intrinsically stable in harsh environments, display lower costs and possess a higher density/number of recognition sites for bacteria capture due to the long chain of the peptide.21 Several groups have explored the viability of using AMPs as molecular recognition elements in the impedimetric22,23 or electrogenerated chemiluminescence24 detection of bacteria. Their research has fully testified to the effective capture ability of AMP. To our knowledge, the utilization of AMP as a capture element for the SERS detection of bacteria has not yet been reported.
On the other hand, SERS tags with high sensitivity and stability are also an important factor in bacterial detection. Silver-coated gold nanoparticles (Au@AgNPs) are an excellent SERS substrate which shows higher SERS activity and more uniformity of particle size distribution compared with traditional pure AgNPs and AuNPs.25,26 However, the poor stability of Au@Ag NPs will limit their application. They must be kept at low temperature or in a dark place, have a short period of viability and are unsuitable for further surface modification. Graphene-based nanocomposites will solve this problem. Graphene oxide (GO) is a type of 2-D nanomaterial with a large surface area and good biocompatibility.27 It can stabilize the SERS signals and protect metal nanoparticles from oxidation, endowing this flexible substrate with a long-term stability without decline in SERS activity.28,29 As a result, the combination with GO will stabilize the SERS activity of Au@AgNPs, making the SERS active substrate more durable and will be beneficial for further chemical modification. Another smart strategy for improving SERS stability is to use an IS (such as 4-mercaptopyridine30 or multilayered graphitic magnetic nanocapsules31) to eliminate the influence of uncontrollable aggregation of NPs, which is known to cause significant variations in SERS intensity. Ideally, 4-MPBA is a Raman signal reporter which shows a strong Raman signal and has intrinsic peaks of 1075 cm−1 or 1586 cm−1. It can not only be used as a capture probe to combine with saccharides on bacterial cell walls, as described above,9 but is also a desirable IS to correct the SERS intensities.
Inspired by these elegant points, we herein develop a SERS sandwich strategy for the sensitive detection and discrimination of three different kinds of bacteria directly in blood samples. AMP based Fe3O4NPs were first used in the selective capture and magnetic enrichment of bacteria from the mixture, and they show some superiority over ordinary 4-MPBA, antibodies, aptamers, or antibiotic based capture elements. Hence, other interference from the mixture such as cells or proteins has been effectively removed, as well as there being an increment in sensitivity due to the magnetic enrichment. Au@Ag-GO nanocomposites with high SERS activity and stability have also been fabricated, and further modified with 4-MPBA to act as SERS tags. The 4-MPBA on the SERS tags not only corrected the SERS intensities, but also enhanced the sensitivity and amplified the differences in the “fingerprints”. Hence, such SERS tags would combine with a bacteria@Fe3O4 complex to form a sandwich structure following bacteria capture, and provide strong Raman signals for SERS detection. When different kinds of bacteria combined with 4-MPBA, their SERS “fingerprints” showed corresponding changes. In this way, E. coli, S. aureus and P. aeruginosa were discriminated with LOD of 101 CFU mL−1, respectively. This novel method was further used in the detection of bacteria from clinical patients who were infected with bacteria. Additionally, we demonstrate the potential of AMP modified Fe3O4NPs to inactivate potential bacterial contamination in blood transfusions settings. This biosensor holds considerable promise to act as a multifunctional platform in the simultaneous capture, discrimination and inactivation of bacteria.
Results and discussion
Pathogen detection principle via the SERS sandwich strategy
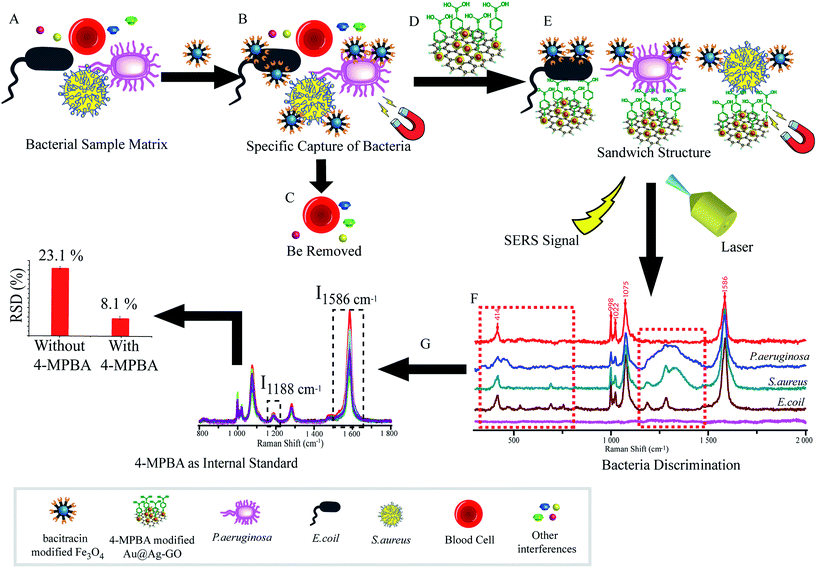
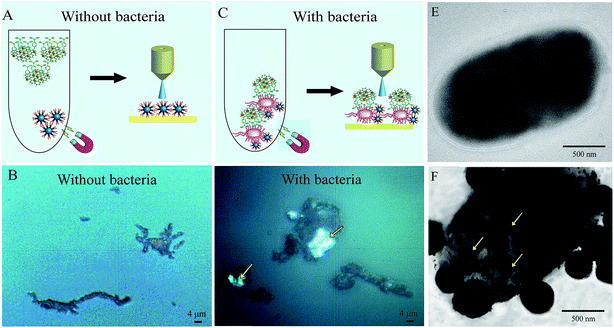
As illustrated in Fig. 1, the SERS sandwich strategy for the detection of bacteria is based on the formation of a sandwich structure of Fe3O4NPs/bacteria/SERS tags. The AMP based Fe3O4NPs were first cultured with a sample matrix, containing the bacteria, blood cells, and other interferences (Fig. 1A). Then AMP modified Fe3O4NPs will specifically bind to the bacteria, and the presence of the Fe3O4NPs will allow for the magnetic separation of the bacteria from the sample matrix (Fig. 1B). In this way, the blood cells or any other interference will be removed (Fig. 1C). Subsequently, SERS tags modified with 4-MPBA as bio-recognition molecules and Raman reporters are added and incubated to form the sandwich structures (Fig. 1D). In this way, the boronic acid group from 4-MPBA will bind to the peptidoglycan from the cell wall of the bacteria. Such interactions are mediated through the interaction of sugar chains or a hydrophilic peptidoglycan layer via a reversible cyclic cis-diol esterification reaction with the boronic acid group (Fig. S1†).32,33 The sandwich structures are then collected via magnetic separation and detected by a Raman spectrophotometer (Fig. 1E). In the following SERS detection, 4-MPBA will act as the Raman reporter molecule to provide a strong Raman signal. When different kinds of bacteria are combined with the SERS tags, the Raman “fingerprints” of 4-MPBA will show corresponding changes, which can be used to discriminate among different kinds of bacteria (Fig. 1F). In addition, peaks of 4-MPBA can be used to correct the SERS intensities (Fig. 1G). Hence, this novel SERS sandwich strategy enabled the highly sensitive detection and specific discrimination of pathogenic bacteria.Fabrication and characterization of Au@Ag-GO nanocomposites
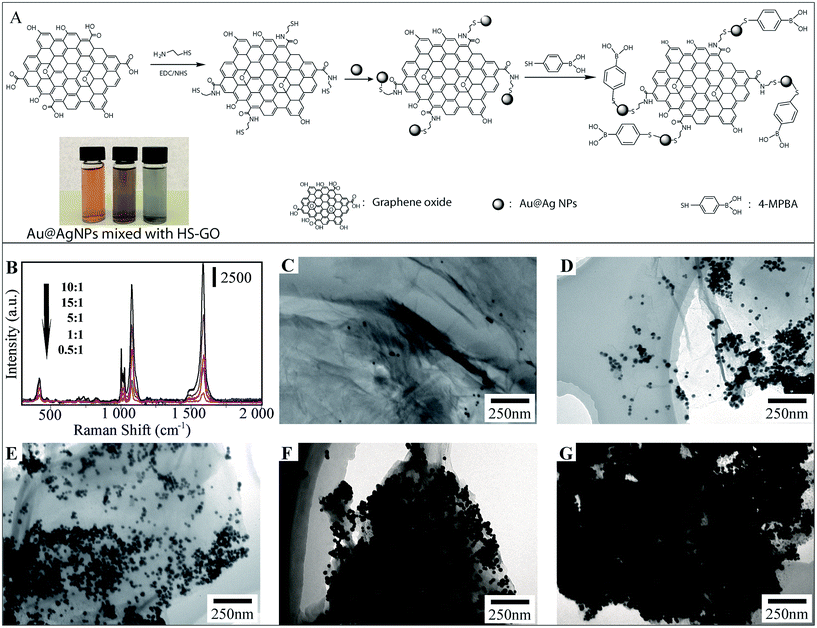
The design and fabrication of 4-MPBA modified Au@Ag-GO SERS tags are illustrated step by step in Fig. 2A, and mainly include the modification of GO with thiol groups, the adsorption of Au@AgNPs onto the GO nanosheet and further modification of Au@Ag-GO with 4-MPBA. Fig. 2A also shows that the color of the Au@AgNPs changes from orange to yellow-green after mixing with thiol modified GO (HS-GO) for 5 min and further turns black after 10 min. This is caused by the thiol induced Au@AgNPs agglomeration, which indicates the successful modification of the thiol group onto the GO surface.Traditional AgNPs with high SERS activity lack uniformity in particle size, resulting in unstable SERS enhancement, while AuNPs with a uniform distribution are poor in SERS activity. The combination of silver and gold into the Au@AgNPs will provide both high and stable SERS activity. TEM image of Fig. S2B† displays the uniform morphology of Au@AgNPs—average diameter of 35 ± 8 nm—compared with the heterogeneous size distribution of AgNPs—average diameter of 40 ± 11 nm—(Fig. S2A†). Fig. S2C† shows the high magnification TEM images of Au@AgNPs with a gold core and a 5 nm thick silver shell. The SERS activities of AuNPs, AgNPs and Au@AgNPs are also evaluated through the Raman spectra of 10−9 M rhodamine 6G (R6G) dispersed in their colloidal solutions. The results show that SERS enhancement is higher in 5 nm Au@AgNPs than in AgNPs or AuNPs (Fig. S3A†). In our previous work, we also studied the SERS enhancement of Au@AgNPs with different shell thicknesses, and the results showed that Au@AgNPs with a 5 nm thick shell show the highest SERS enhancement.34 Hence, Au@AgNPs with a 5 nm thick shell were used in the fabrication of the SERS tags.
The ratio of Au@AgNPs to GO nanosheet exerts a strong influence on the morphology of the Au@Ag-GO nanocomposites. Fig. 2C–G illustrate the TEM images of nanocomposites prepared with an increasing Au@Ag/GO ratio (mL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg) from 0.5
mg) from 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 15
1 to 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. We can see that as the ratio increases, more Au@AgNPs cover the GO surface. When the ratio is 0.5
1. We can see that as the ratio increases, more Au@AgNPs cover the GO surface. When the ratio is 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, few Au@AgNPs can be found on the GO nanosheet. When the ratio is 10
1, few Au@AgNPs can be found on the GO nanosheet. When the ratio is 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Au@AgNPs cover the whole GO nanosheet, forming perfect Au@Ag-GO nanocomposites with the highest SERS activities (Fig. 2B). When the ratio is 15
1, Au@AgNPs cover the whole GO nanosheet, forming perfect Au@Ag-GO nanocomposites with the highest SERS activities (Fig. 2B). When the ratio is 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 many aggregated Au@AgNPs can be found on the GO surface, which in turn decreases such a SERS enhancement effect. Thus, an Au@Ag/GO ratio of 10
1 many aggregated Au@AgNPs can be found on the GO surface, which in turn decreases such a SERS enhancement effect. Thus, an Au@Ag/GO ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 is employed in the fabrication of Au@Ag-GO nanocomposites. The powder XRD patterns (Fig. S3B, point 1.3 in the ESI†) and FTIR (Fig. S3C, point 1.3 in the ESI†) also showed the successful fabrication of 4-MPBA modified SERS tags.
1 is employed in the fabrication of Au@Ag-GO nanocomposites. The powder XRD patterns (Fig. S3B, point 1.3 in the ESI†) and FTIR (Fig. S3C, point 1.3 in the ESI†) also showed the successful fabrication of 4-MPBA modified SERS tags.
Stability and SERS activity of SERS tags
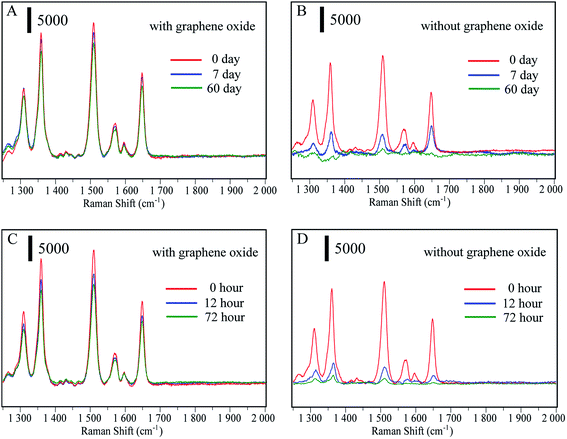
The main advantage of the Au@Ag-GO nanocomposites is the high stability of the SERS signals due to the protection of the reactive Au@AgNPs against oxidation. To confirm this, Au@AgNPs both with and without GO were stored at 4 °C for 0, 7 and 60 days. Then the SERS substrates were mixed with 10−9 R6G solutions to test their SERS activity. Fig. 3A shows that Au@AgNPs with GO still retain high SERS activity after 60 days, while the SERS activity of pure Au@AgNPs (Fig. 3B) declines seriously after only 7 days. When Au@AgNPs with GO and without GO are placed in sunlight for 0, 12, and 72 hours, the Au@Ag-GO nanocomposites (Fig. 3C) also show clearly higher stability than pure Au@AgNPs. Fig. S3D† shows that the corresponding spectra of 4-MPBA on Au@Ag-GO nanocomposites still retain high Raman intensities after lengthy storage. In addition to the high stability, Au@Ag-GO also shows high SERS activity with an enhancement factor (EF) of 1.1 × 108 (please see Fig. S4, point 1.2 in ESI† for the detailed calculation process). Hence, the as-prepared Au@Ag-GO will provide stable and sensitive signals for SERS detection.Another advantage of the combination of GO and Au@AgNPs is the higher adsorptive capacity of 4-MPBA. As a colloid solution, pure Au@AgNPs lack stability and a slight excess of 4-MPBA will lead to aggregation of the colloid. Thus, the modification of 4-MPBA onto the surface of pure Au@AgNPs must be strictly controlled, or the ability to capture bacteria will be limited. In our design, we have mixed the Au@AgNPs with different concentrations of 4-MPBA, and the results (Fig. S5A†) showed that when the concentration of 4-MPBA was in excess of 0.25 μg mL−1, the colloidal solution began to change color. In contrast, the Au@Ag-GO shows high stability even in high concentrations of 4-MPBA. As plotted in Fig. S5B,† the UV absorption of 4-MPBA is obviously reduced after mixing with Au@Ag-GO. A comparison of the adsorbed amount of 4-MPBA between Au@Ag-GO and Au@AgNPs was conducted through measurement of their surface coverage. The calculation process is shown in the ESI† (point 1.1) and the results show that the adsorbed amounts of 4-MPBA on the Au@Ag-GO surface are 25 times more than on pure Au@AgNP, which indicates that the ability to capture bacteria can be greatly increased with an Au@Ag-GO nanocomposite.
Fabrication and characterization of antimicrobial peptide modified Fe3O4NPs
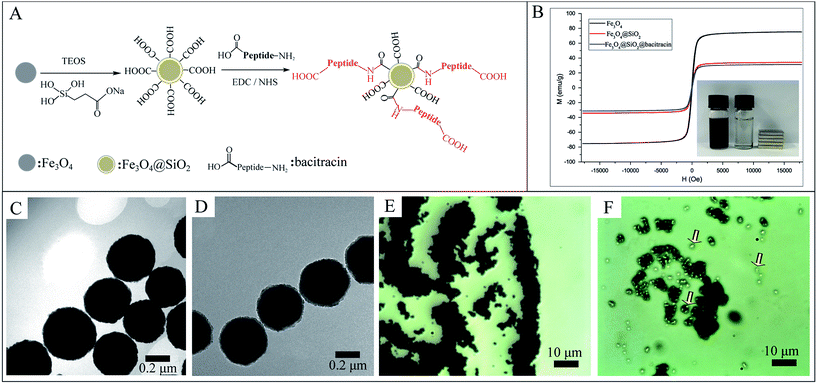
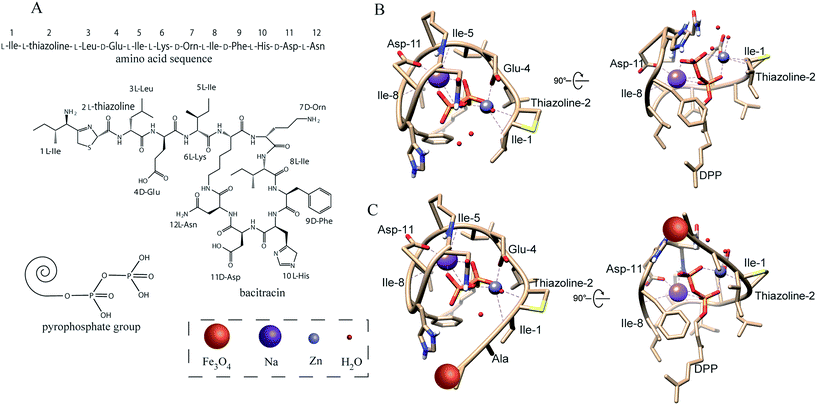
The design and fabrication of an AMP modified Fe3O4 magnetic capture substrate are depicted in Fig. 4A, and mainly include the coating of SiO2, functionalization of the carboxyl group, and further modification of AMP. The AMP used here is bacitracin A, with an amino sequence of L-Ile-L-thiazoline-L-Leu-D-Glu-L-Ile-L-Lys-D-Orn-L-Ile-D-Phe-L-His-D-Asp-L-Asn.35The magnetic hysteresis loops of the as-prepared Fe3O4, Fe3O4@SiO2 and Fe3O4@SiO2@AMP are shown in Fig. 4B. All show good ferromagnetic behavior at room temperature. The magnetic saturation (Ms) value of the Fe3O4 has decreased from 75.3 emu g−1 to 34.2 emu g−1 and the further modification of AMP does not apparently decrease the Ms value (31.4 emu g−1). The decrease in the Ms value of Fe3O4@SiO2 compared with Fe3O4 may be due to the coating with a layer of amorphous SiO2. The further slight decrease in the Ms value of Fe3O4@SiO2@AMP compared with Fe3O4@SiO2 may be due to the coating with a layer of thin carboxyethyl silanetriol and AMP. Such an excellent magnetic property means that all of the as-prepared materials have a strong magnetic response before and after the modification and can easily separate the analytes from the mixture under an external magnetic field. Besides, the inset of Fig. 4B shows that Fe3O4@SiO2@AMP can be concentrated on the side of the vials within 30 s upon the placement of an external magnet next to the vials, which further confirmed the strong magnetic response of Fe3O4@SiO2@AMP. For the morphology measurement, Fig. 4C shows the TEM images of the as-synthesized spherical Fe3O4NPs, with average diameters of 440 ± 30 nm. The TEM image (Fig. 4D) also revealed the core–shell nanostructures of Fe3O4@SiO2NPs with ∼20 nm of shell thickness. The FTIR (Fig. S6, point 1.4 in the ESI†) also showed the successful synthesis of AMP modified Fe3O4NPs.
The bacteria capture ability of the as-prepared Fe3O4@SiO2@AMP has also been preliminarily evaluated through a microscope. Fig. 4E is the microscope photograph of the unmodified Fe3O4NPs mixed with E. coli after magnetic separation, which shows that only a black substance (Fe3O4 aggregates) can be found in the images. In contrast, when we mixed the Fe3O4@SiO2@AMP with E. coli and separated them under the magnetic field, some white points appeared, as indicated with arrows in Fig. 4F. Thus, the AMP is not only successfully modified onto the surface of the Fe3O4NPs, but also has good bacteria capture ability.
Molecular details of target recognition by antimicrobial peptide
In general, the antimicrobial peptide (bacitracin A, Fig. 5A) will recognize the pyrophosphate group of the lipid target on the bacteria due to the AMP–lipid interactions and indirect interactions mediated by the zinc ion and sodium ion.36,37Fig. 5B shows that the interactions are augmented by interactions between the pyrophosphate and two metal ions. The zinc ion adopts an octahedral coordination geometry. Two of the interactions are with oxygen atoms from the lipid (1, 2 of the phosphate groups). The other interactions are the zinc ion interacting with the side chain of D-Glu-4, L-thiazoline-2 and L-Ile-1 from the AMP. A sixth ligand is the interaction between zinc ion and a water molecule. The sodium ion interacts with three oxygen atoms from the lipid (1, 2, 3 of the phosphate groups) and the side chain of L-Ile-5, D-Asp-11, L-Ile-8 from the AMP. Fig. S7A† also shows that AMP was wrapped tightly around the lipid pyrophosphate. As a result, the AMP forms a compact structure that completely envelopes the ligand's pyrophosphate group, which with the co-existence of zinc and sodium ions, thus results in a strong interaction between bacteria and AMP.Now, we turn our attention to the interaction between peptide modified Fe3O4NPs and pyrophosphate. The published ternary complex of bacitracin A was used as the initial model (PDB entry: 4K7T).38 Then the predicted structure was adjusted in the software COOT with minor changes at its amino-terminus.39 In the peptide modification, the N-terminal amino of AMP (1 L-Ile) and the side chain of 7 D-Orn reacts with the carboxyl group on the Fe3O4NPs through a condensation reaction. However, the above simulation results showed that the N-terminal amino contributes to the interaction between bacteria and AMP, which may be different after modification. Thus, we simulated the interaction between the pyrophosphate group and the peptide in which the N-terminal amino has been reacted with the carboxyl group to form an amide group. As shown in Fig. 5C, the amidation of the N-terminal amino will not change the interaction between AMP and bacteria. Fig. S7B† also shows that AMP is still wrapped tightly around the lipid pyrophosphate even after modification onto the Fe3O4NPs. Thus, the modification of the peptide onto the Fe3O4NPs will not change its recognition site. Though some AMP may react with Fe3O4NPs by the side chain of 7 D-Orn (less AMP reacts with the side chain of 7 D-Orn than with the N-terminal amino due to a higher steric-hinderance effect21,40), this will not influence the interaction between AMP and bacteria, as the side chain of ornithine does not contribute to the interaction, according to the simulation results above.
Detection of bacteria by SERS
To prove the practicality and great potential of the sandwich strategy in application, the 4-MPBA modified Au@Ag-GO nanocomposites and AMP based Fe3O4NPs were all applied in bacterial detection. As illustrated in Fig. 1, the SERS sandwich structure based on AMP recognition included the following processes. First of all, the AMP based Fe3O4 capture element was mixed with bacteria under shaking conditions, and then the bacteria were bound to the AMP based Fe3O4NPs. After magnetic separation, the bacteria–AMP–Fe3O4 complexes were easily separated from the complex samples and washed with PBS to remove the excess free bacteria. Subsequently, the 4-MPBA modified Au@Ag-GO nanocomposites (SERS tags) were mixed with the above complexes to bind together with the bacteria, which was mainly caused by the interaction between the boronic acid and the diol group from the saccharides of the bacteria wall to form cyclic boronate esters.41 Hence, the SERS tags/bacteria/Fe3O4NPs sandwich structure was formed, creating many “hotspots” for the following SERS detection.Fig. 6B, which corresponds to Fig. 6A, shows the substance acquired from the SERS detection process under the Raman microscope. In this situation, no bacteria were added to form the sandwich structure and only the AMP based Fe3O4NPs were acquired. Hence, the dark substance distributed in this image can be identified as Fe3O4NPs aggregates, and no Raman signals were collected from it (Fig. 7A(e)). Fig. 6D, which corresponds to Fig. 6C, shows the sandwich structures formed under the Raman microscope. In this situation, bacteria were added to form the sandwich structure, and thus the SERS tags could be collected. Compared with Fig. 6B, some bright areas which were caused by the SERS tags appeared, as indicated by the arrows. When we focus the laser point onto the bright area, strong Raman signals will be collected (Fig. 7A(b–d)).
Fig. 6F shows the TEM image of the SERS tags/E. coli/Fe3O4 sandwich structure, in which the Fe3O4NPs are surrounding the bacteria, and the Au@Ag-GO nanocomposites can be identified on the bacterial surface, as indicated by arrows. Fig. 6E also shows the TEM image of pure E. coli, which shows great differences from the sandwich structure.
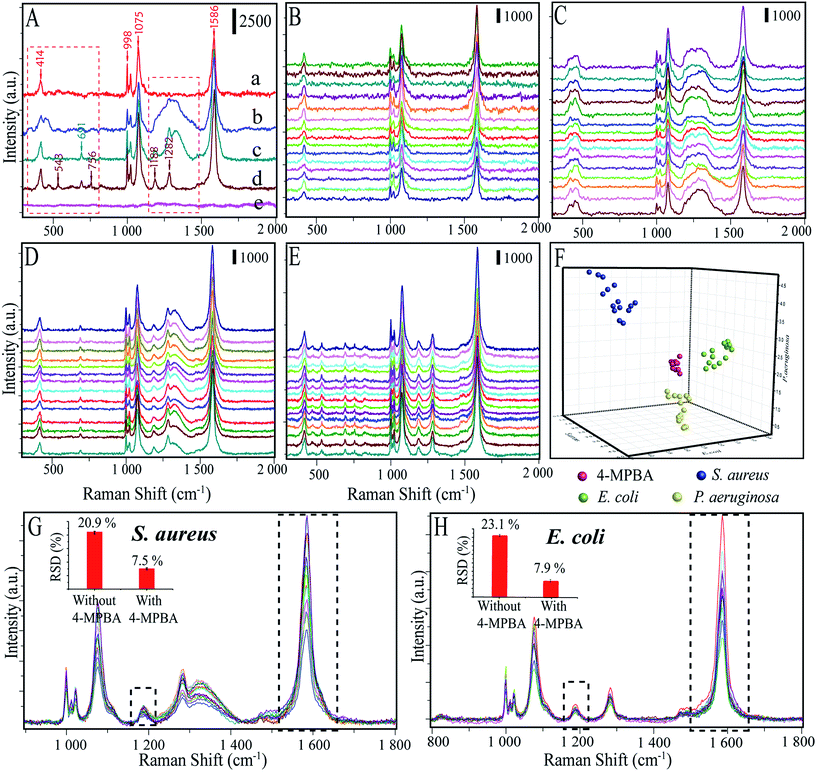
We further investigated the applicability of this biosensor in distinguishing among different kinds of bacteria by comparing the SERS spectra of E. coli, S. aureus and P. aeruginosa. The Raman spectra of pure 4-MPBA–Au@Ag-GO without bacteria were measured and the results showed that five apparent peaks (i.e. at 414, 998, 1022, 1075, and 1586 cm−1) can be identified in the SERS spectrum of 4-MPBA (Fig. 7A(a)). Such Raman bands from 4-MPBA do not interfere with the representative bacterial fingerprint bands in the range between 1100 cm−1 and 1400 cm−1, or the peaks between 400 cm−1 and 800 cm−1. Thus, different kinds of bacteria can be discriminated from their fingerprint recognition bands. Specifically, two sharp peaks at 1188 cm−1 (amide III) and 1282 cm−1 (δ(CH2) amide III) were observed in the SERS spectrum of E. coli (Fig. 7A(d)). For S. aureus (Fig. 7A(c)), peaks at 1188 cm−1 (amide III) and 1282 cm−1 (δ(CH2) amide III) still exist, and a new broad peak at 1333 cm−1 (υ(COO–) and δ(C–H)) appeared. For the P. aeruginosa, only one broad peak at 1333 cm−1 (υ(COO–) and δ(C–H)) between 1100 cm−1 and 1400 cm−1 is observed (Fig. 7A(b)). Additionally, peaks between 400 and 800 cm−1 also show some differences between these three bacteria. A sharp peak appeared at 691 cm−1 in the detection of S. aureus while three sharp peaks appeared at 543 cm−1, 691 cm−1 and 756 cm−1 in the detection of E. coli. Additionally, blank samples have also been investigated and the results show that no Raman signal can be acquired due to no bacteria being added to form the sandwich structure (Fig. 7A(e)). To make the discrimination results more reliable, Raman spectra of 4-MPBA modified Au@Ag-GO, P. aeruginosa, S. aureus and E. coli from 45 batches (each kind of bacteria was represented by 15 batches) were recorded and are shown in Fig. 7B–E. Discriminant analysis (DA) was used to maximize the spectral differences resulting from the data arrays in the discrimination of the bacteria. Fig. 7F shows the 3D map of the DA results, where red balls represent pure 4-MPBA modified Au@Ag-GO, green balls represent E. coli, blue balls represent S. aureus and yellow balls represent P. aeruginosa. It shows that these three different kinds of bacteria and pure 4-MPBA can be completely separated from each other in space, which means that this method combined with DA could be used to discriminate among different kinds of bacteria.
Furthermore, we compared our method (labeling) to the label-free detection of bacteria. In this experiment, AgNPs were simply mixed with the bacteria. As depicted in Fig. S8,† Raman signals in the label-free detection mode (without 4-MPBA) are apparently weaker than in the label detection mode (with 4-MPBA), though the bacteria were in high concentration (1 × 108 CFU mL−1). Importantly, the label-free detection showed no characteristic peaks from the “fingerprints” of these three kinds of bacteria, which can hardly be discriminated with the naked eye. All these results imply that combined with 4-MPBA, our sandwich strategy can enhance detection sensitivity, as well as featuring good capability for discriminating among different kinds of bacteria.
The 4-MPBA utilized here can also act as an IS to eliminate the influence of electromagnetic heterogeneity in enhancing the substrate. As shown in Fig. 7G and H, we extracted the Raman data (800–1800 cm−1) from the above raw spectra of the DA analysis (Fig. 7D and E). The band at 1188 cm−1 and the ratio of I1586/I1188 were used for relative standard deviation (RSD, %) analysis. Fig. S9A† (E. coli) and C (S. aureus) showed that the Raman intensities were very unstable without the 4-MPBA. In contrast (Fig. S9B and D†), the ratio of I1586/I1188 apparently showed higher stability with 4-MPBA. The results are shown in the inset photographs of Fig. 7G and H. RSDs were significantly reduced from 20.9% to 7.5% (S. aureus), and 23.1% to 7.9% (E. coli) by using the 4-MPBA IS to eliminate Raman intensity variation.
This novel sandwich strategy also displays excellent enrichment and separation abilities in bacterial detection, which gives it a potential ability to detect very low concentrations of pathogenic bacteria. The performance of the biosensor in the detection of different concentrations of bacteria was further measured through recording the changes in Raman intensity of the peaks at 1586 cm−1 from the SERS mapping. As depicted in Fig. S10A–F,† the red squares in the SERS mapping results are the SERS signals acquired from different concentrations of E. coli (101 to 106 CFU mL−1). As can be seen, the increase in numbers of red squares corresponded to an increase in bacterial concentration, and a low limit of detection (LOD) of 101 CFU mL−1 was achieved.
Inactivation and detection of bacteria in blood
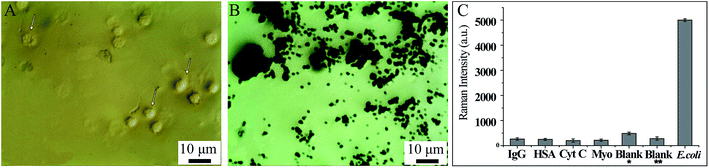
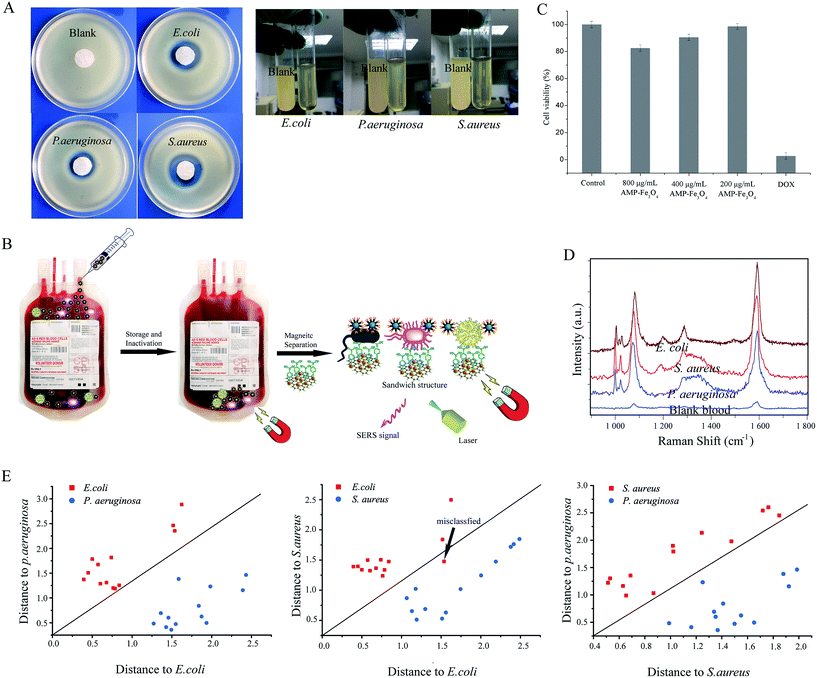
Firstly, the specificity of our SERS biosensors has been evaluated in the presence of IgG, HAS, Cyt C, Myo at the same concentrations of 4 ng mL−1, as well as with blank whole blood with and without dilution. Fig. 8A shows the blood cells under a microscope: we can see that the blood cells can be easily identified (indicated by arrows). When AMP modified Fe3O4NPs were mixed with the blank blood and magnetically separated, we can see that no blood cells can be identified under the microscope (Fig. 8B). Fig. 8C also shows that the signal intensity increased significantly in the presence of bacteria, while there were no obvious changes in SERS intensities for IgG, HAS, Cyt C, Myo or blank whole blood. This demonstrates that our SERS biosensor has good selectivity for the detection of bacteria in blood samples.The AMP modified Fe3O4NPs also feature good antibacterial ability. The antibacterial activity of carboxyl modified Fe3O4NPs and AMP modified Fe3O4NPs in a solid medium were evaluated by the disc-diffusion test, in which antibacterial ability is determined by measuring the zones of inhibition (ZoI).42Fig. 9A(left) shows the ZoI of carboxyl modified Fe3O4NPs and AMP modified Fe3O4NPs towards S. aureus, E. coli and P. aeruginosa. After 24 h of incubation, carboxyl modified Fe3O4NPs showed negligible inhibitory action against the bacteria, while AMP modified Fe3O4NPs showed significant efficacy against the bacteria.43 For the antibacterial ability in whole blood (right), we spiked the three different kinds of bacteria into the whole blood with AMP modified Fe3O4NPs or pure Fe3O4NPs and cultivated them. After that, a small amount of whole blood was added to the LB medium and cultivated again. Finally, the antibacterial ability was compared by the turbidity of the LB medium by the naked eye (Fig. 9A(right)). Compared with the blank, the LB medium spiked with whole blood (cultivated with bacteria and AMP modified Fe3O4NPs) is much higher in clarity. Hence, the AMP modified Fe3O4NPs can act as a good antibacterial agent in whole blood.
Nowadays, blood transfusion safety is a serious problem that is causing general public concern. After the long-term storage of blood, bacteria will grow to some extent and the patient could suffer morbidity or mortality from a transfusion-transmitted infection. Thus, the detection of bacteria in whole blood before the blood transfusion would greatly reduce the infection. Additionally, bacterial inactivation is also an option to reduce infection risks. Hence, the development of multi-functional nanomaterials with both antibacterial and detection abilities is of great significance.44 Our SERS tags/bacteria/Fe3O4NPs sandwich structure can well fulfil both the above functions. As shown in Fig. 9B, AMP modified Fe3O4NPs can first be added to the blood to act as an antibacterial agent in the storage of blood. Before a blood transfusion, the AMP modified Fe3O4NPs were magnetically separated from the blood, and mixed with 4-MPBA modified Au@Ag-GO nanocomposites. If bacteria exist in the blood, the sandwich structure will be formed, and will show corresponding SERS signals to indicate that the blood is not safe for transfusion. Additionally, the in vitro cytotoxicity of the AMP modified Fe3O4NPs has been approved to be of low cytotoxicity through an evaluation of murine macrophage RAW264.7 (RAW) cells. Fig. S11† shows the cell morphological changes after exposure to AMP modified Fe3O4NPs: we can observe no obvious difference between the control cells and the AMP modified Fe3O4NPs-treated cells. Fig. 9C shows the cellular toxicology evaluation results: different concentrations of AMP modified Fe3O4NPs (800, 400, 200 μg mL−1) showed no obvious cytotoxicity, while the positive control group, doxorubicin (DOX), showed high cytotoxicity. For real applications, infected blood samples (3 times dilution) from 39 patients (provided by First Affiliated Hospital of Jinan University, Guangzhou, China; bacteria were identified by the VITEK 2 System) were immediately processed using our universal sample preparation process and detected by SERS. Fig. 9D shows one of the SERS spectra acquired from infected blood samples of patients (all the SERS spectra of infected blood samples are shown in Fig. S12†). We then analyzed the SERS results using algorithm-based DA over the whole range from 800 cm−1 to 1800 cm−1. The DA based Mahalanobis distance plots of every sample to the centre of gravity of two classes (‘E. coli and P. aeruginosa group’, ‘E. coli and S. aureus group’, and ‘S. aureus and P. aeruginosa group’) are shown in Fig. 9E. A diagonal line was used to identify the boundary of two classes. The spectra were divided into three clusters according to the species. In the validation analysis, 97.3% of the real blood sample can be classified effectively (only one patient infected with E. coli was misclassified). Additionally, blood samples (3 times dilution) spiked with bacteria down to 1 × 104 CFU mL−1 were also tested. Fig. S13† shows that different kinds of bacteria are obviously detectable in the SERS. Comparatively, signals were rarely collected in the pure blood sample. Importantly, the SERS spectra of blood spiked with bacteria are similar to those of a real infected blood sample, indicating that this method is suitable for the sensitive and specific discrimination of bacteria in real applications.
Conclusions
In summary, we herein present a sandwich strategy for the rapid, sensitive detection and discrimination of three different kinds of bacteria from a matrix sample. This novel approach involves combining a sandwich strategy including modified magnetic Fe3O4NPs for the capture and enrichment of bacteria, and a SERS tag to provide and enhance the Raman signals. For the magnetic Fe3O4NPs, AMP was first modified as a capture element in the SERS detection of bacteria. Compared with previously reported capture elements in the SERS detection of pathogenic bacteria, as shown in Table 1, this proposed method possesses the advantages of high sensitivity,9,18 high stability,13–15 low cost13–17 and high specificity.9 What is more, it is more efficient in bacteria capture due to the long chain of the peptide.17 For the SERS tags, Au@Ag-GO nanocomposites with high SERS activity and stability have been fabricated, and further modified with 4-MPBA to act as SERS tags. In SERS detection, the Raman spectrum of the bacteria will show changes in its “fingerprints” corresponding to different kinds of bacteria. In this way, E. coli, S. aureus and P. aeruginosa were discriminated using DA analysis with an LOD of 101 CFU mL−1. In applications, this novel method can be applied in the detection of bacteria from clinical patients who are infected with bacteria and the results showed that 97.3% of the real blood samples (39 patients) can be classified effectively (only one patient infected with E. coli was misclassified). More importantly, the AMP modified Fe3O4NPs also showed good antibacterial activities, which means this biosensor can act as a multifunctional platform in the simultaneous capture, discrimination and inactivation of bacteria.| Method/capture element | Pathogenic bacteria | LOD | Substrate | Characteristic | Ref | |
|---|---|---|---|---|---|---|
| Capture substrate | SERS substrate | |||||
| SERS/antibody | S. aureus | 101 CFU mL−1 | MnFe2O4@Au | AuNR | High specificity but with low stability and high cost | 13 |
| SERS/antibody | E. coli | 101 CFU mL−1 | Fe3O4 | AuNPs | High specificity but with low stability and high cost | 14 |
| SERS/antibody | MRSA, E. coli, S. typhimurium | 101 CFU mL−1 | Fe3O4@Ag | AgNPs | High specificity, simultaneous detection but with low stability, high cost and tedious modification process | 15 |
| SERS/aptamer | S. aureus | 101 CFU mL−1 | MnFe2O4@Au | AuNR | High specificity, high stability but aptamers available for specific capture of bacteria are limited | 16 |
| SERS/aptamer | S. typhimurium, S. aureus | ∼101 CFU mL−1 | Fe3O4@Au | AuNPs | High specificity, high stability, simultaneous detection but with tedious modification process and aptamers available for specific capture of bacteria are limited | 17 |
| SERS/boric acid | E. coli, S. aureus | 102 CFU mL−1 | Silicon wafer decorated with AgNPs | High stability, low cost, simultaneous detection but with poor specificity (not specific for bacteria) | 9 | |
| SERS/antibiotic | E. coli, S. aureus, MRSA | 102 CFU mL−1 | Fe3O4@Ag | High stability, low cost, specific for bacteria but are limited in recognition sites as a small molecule | 18 | |
| Electrical detection/AMP | E. coli | 1 bacterium/μL | Microelectrode array | High stability, low cost, high specificity for bacteria and more recognition sites due to the long chain of the peptide | 45 | |
| Infrared-integrated sensors/AMP | L. monocytogenes | 1 bacterium/μL | Microfluidic channel on a biomaterial cantilever | High stability, low cost, high specificity for bacteria and more recognition sites due to the long chain of the peptide | 46 | |
| SERS/AMP | E. coli, S. aureus, P. aeruginosa | 101 CFU mL−1 | Fe3O4 | Au@Ag-GO | High stability, low cost, high specificity for bacteria and more recognition sites due to the long chain of the peptide | This work |
Experimental section
Chemicals, biochemicals, and instruments
Antimicrobial peptide (bacitracin A), rhodamine 6G (R6G), 4-mercaptophenylboronic acid (4-MPBA), 2-aminoethanethiol (AET), iron chloride hexahydrate (FeCl3·6H2O), ethylene glycol, polyethylene glycol 6000, sodium acetate trihydrate (NaAc·3H2O), ammonia solution, tetraethyl orthosilicate (TEOS) were all supplied by Macklin (Shanghai, China). 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were purchased from Energy Chemical (Shanghai, China). Carboxyethylsilanetriol sodium salt (carboxyl-silane) was purchased from J&K Scientific Ltd. Luria–Bertani (LB) medium was purchased from Qingdao Hope Bio-Technology Co., Ltd. (Qingdao, China). Fetal bovine serum (FBS), Dulbecco's Modified Media (DMEM), and penicillin-streptomycin were all obtained from Saiguo Bio-Technology Inc (Guangzhou, China). All chemicals were of the analytical grade. E. coli (ATCC8739), S. aureus (ATCC6538), and P. aeruginosa (PAO1) shock-frozen strains were purchased from Guangdong Microbial Culture Center (Guangdong, China).The morphologies and microstructures of the Fe3O4NPs, Au@Ag-GO nanocomposites and other related nanomaterials were all investigated by field-emission transmission electron microscopy (JEM-2100F). The FTIR spectra were conducted with a Gilson 306 FT-IR Spectrometer (France). Magnetic characterization was conducted with a superconducting quantum interference device (VSM KakeShore 7404, Lakeshore, America). Powder XRD patterns of the products were investigated on a Bruker diffractometer D8 Advance (Germany).
All SERS measurements were detected with a Raman microscope (LabRAM HR, HORIBA Scientific, Japan). A 638 nm laser was used as the excitation source. A 50× microscope objective was used to focus the light from the laser and for collection of the Raman signals. The power density of the laser is 16.0 mW μm−2. The Raman spectra were acquired within the range 400–2000 cm−1 with an exposure time of 4 s and 50% of maximum laser power. For comparison purposes, the acquired Raman spectra were presented after adjusting the baselines.
Fabrication of AMP modified Fe3O4NPs
Fe3O4NPs were synthesised according to a previous report with minor modification.47 First, 2.7 g of FeCl3·6H2O was added into 80 mL of ethylene glycol and stirred until completely dissolved. Subsequently, polyethylene glycol 6000 (2.0 g) and NaAc·3H2O (7.2 g) were added and kept under stirring until fully dissolved. Then 75 mL of the mixture was transferred into a Teflon-lined autoclave with a capacity of 100 mL and heated at 200 °C for 8 h. With magnetic separation, the as-prepared Fe3O4NPs were then collected and washed with deionized water and ethanol three times each.For the synthesis of Fe3O4@SiO2, 22.5 mg of Fe3O4NPs was added into 3 mL of ammonia solution (1.2%) and ultrasonicated for 5 min. Then 200 μL of TEOS was added and kept under ultrasonication for 90 min. After that, 10 μL of TEOS was added and kept under ultrasonication for another 90 min. The resulting products were magnetically separated and washed with deionized water for further use.
For the synthesis of carboxyl modified Fe3O4NPs, 80 μL of carboxyl-silane as a sodium salt was added to 1 mL of Fe3O4@SiO2 suspension (10 mg mL−1) in 20 mM phosphate-buffered saline (PBS, pH 7.4) and mixed for 8 h. Then the particles were magnetically separated and washed three times with 10 mM PBS. Finally, the Fe3O4NPs were diluted in 20 mM phosphate-buffered saline (PBS, pH 7.4).
For the synthesis of AMP modified Fe3O4NPs, 10 mg of carboxyl modified Fe3O4NPs were dispersed in 25 mL of PBS, then 16.5 mg of EDC and 10 mg of NHS were added, followed by a mixing for 4 h. Subsequently, 20 mg of bacitracin A were added and mixed for another 3 h. The as-prepared AMP modified Fe3O4NPs were collected by magnetic separation, washed 3 times with BPS to remove impurities, and diluted with 400 mM phosphate-buffered saline containing 150 mM zinc acetate to form a solution with a final concentration of 1 mg mL−1, and stored at 4 °C for further use.48
Fabrication of 4-MPBA modified Au@Ag-GO nanocomposites
GO was synthesized using a modified Hummer method from natural graphite,49 while AuNPs, AgNPs, and Au@AgNPs were synthesized in the same way as in our previous report.34For the preparation of thiol functionalized graphene (HS-GO), 100 mg of GO were added to 50 mL of ethanol to produce a 2 mg mL−1 solution. The mixture was then sonicated to form a homogeneous suspension. Subsequently, 1.9 g of EDC was added to the above GO suspension and mixed for 12 h to ensure the surface activation of residual carboxylated groups on the GO surface. After that, 50 mL of AET (1 mM) was added and kept under stirring for 4 h. The AET modified GO were then collected by centrifugation at 9000 rpm, and washed twice with deionized water to remove excess ATE. Thus, HS-GO were acquired.
For the preparation of Au@Ag-GO nanocomposites, 10 mL of as-prepared Au@AgNPs were added to 2 mL of HS-GO (0.1 mg mL−1) and stirred for 2 h. The Au@Ag-GO nanocomposites were collected by centrifugation at 9000 rpm and washed twice with deionized water to further remove excess Au@AgNPs.
For the modification of 4-MPBA onto the Au@Ag-GO nanocomposites, 6 mL of 4-MPBA (0.01 mg mL−1) were mixed with Au@Ag-GO nanocomposites for 4 h. The as-prepared 4-MPBA modified Au@Ag-GO nanocomposites were collected by centrifugation at 9000 rpm and washed twice with deionized water to remove excess 4-MPBA.
Bacteria culture
Shock-frozen E. coli, P. aeruginosa, and S. aureus were used as models in our experiment. Bacterial cells were cultivated in Luria–Bertani (LB) medium in a gyratory shaker at 100 rpm and 37 °C for 16 h. Afterwards, the bacterial cells at 5 mL of LB were collected through centrifugation at 4000 rpm and 4 °C, then washed twice with PBS. Finally, the bacterial cells were diluted to the desired concentrations with PBS buffer, which were measured by the optical density (OD) of media at 600 nm (OD600).Bacterial detection from PBS media and human blood
Prior to SERS analysis, the bacterial cells were diluted with PBS or human blood from healthy volunteers (3 times dilution) to the desired concentrations. In a typical experiment, 50 μL of AMP modified Fe3O4NPs and 1 mL of bacterial suspension (the concentrations differ with different experimental aims) were added to a 1.5 mL centrifuge tube. The mixture was then incubated under shaking for 1 h. After that, bacteria@Fe3O4NPs complexes were separated under a magnetic field and washed with PBS to remove unbound bacteria. Subsequently, 100 μL of SERS tag (4-MPBA modified Au@Ag-GO nanocomposites) and 100 μL of PBS were added to the above bacteria@Fe3O4NPs complexes. The resulting SERS tags/bacteria/Fe3O4NPs sandwich structures were magnetically separated, washed with PBS buffer 3 times, and dispersed in 50 μL of deionized water for further SERS detection. Real infected blood samples (3 times dilution) were directly added to AMP modified Fe3O4NPs, and treated by the same procedure described above.Strain identification of real infected blood samples
The bacteria were directly identified after blood samples were cultured in blood culture bottles for 24 h. Firstly, fluids in the bottles were centrifuged (1000 rpm) to remove the blood cells. Then bacterial cells were collected by centrifugation (3000 rpm) and diluted with 0.45% saline to the equivalent of 0.5 McFarland turbidity standard. The bacterial suspensions were used for identification with the VITEK 2 system (bioMerieux, Inc). Suspensions for the comparative identification method were made according to the manufacturer's instructions, using a GN Test Kit and a GP Test Kit for the identification of Gram-negative and Gram-negative bacteria. The cards were read by kinetic fluorescence measurement and the final results were obtained automatically.Antibacterial ability and cellular toxicity evaluation
The antibacterial ability of AMP modified Fe3O4NPs on a solid medium was evaluated by a disc-diffusion test. A filter paper disk of 6 mm in diameter was dropped with 20 μL of AMP modified Fe3O4NPs (400 μg mL−1) and incubated for 36 h at 37 °C. The diameter of the clear zone around the filter disk was measured.The antibacterial ability of AMP modified Fe3O4NPs on whole blood was evaluated by the following process. Firstly, whole blood treated with AMP modified Fe3O4NPs (200 μg mL−1) or pure Fe3O4NPs (200 μg mL−1) was spiked with E. coli, S. aureus, or P. auruginosa, and then cultivated in a gyratory shaker at 100 rpm and 37 °C for 16 h. After cultivation, 10 μL of whole blood with AMP modified Fe3O4NPs or pure Fe3O4NPs were added to LB medium, and then cultivated under the same conditions as the whole blood. After that, the turbidities of the LB medium were compared with the naked eye.
The growth inhibitory effect of AMP modified Fe3O4NPs toward murine macrophage RAW264.7 cells was assessed with a classical MTT assay.50 The cells were plated in 96-well microplates with a density of 5 × 103 per well and incubated in 100 μL of medium (DMEM, 10% FBS, 1% penicillin-streptomycin solution) at 37 °C in a tissue culture incubator (5% CO2) for 12 h. After that, the culture medium was replaced by fresh medium (DMEM, 10% FBS, 1% penicillin-streptomycin solution, 0.2% DMSO) containing AMP modified Fe3O4NPs with concentrations of 800, 400 and 200 μg mL−1, and cultured for another 72 h. Then 20 μL of MTT (5 mg mL−1) were added into each well and incubated under the same conditions for 4 h. Finally, the culture medium was removed, 120 μL of DMSO was added, and the 96 well microplates were shaken for 10 min. The adsorption at 570 nm was recorded using a microplate-reader (Bio-Tek).
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81773684, 21505053), Guangdong Natural Science Funds for Distinguished Young Scholars (2018B030306033), Pearl River S&T Nova Program of Guangzhou (201806010060), Science and Technology Planning Project of Guangdong Province, China (2016A030310089), Medical Scientific Research Foundation of Guangdong Province of China (A2018133), and the Fundamental Research Funds for the Central Universities (21618407). We thank Prof. Xiangjiang Liu from Zhejiang University for his help and advice.Notes and references
- R. Wu, Y. Ma, J. Pan, S.-H. Lee, J. Liu, H. Zhu, R. Gu, K.-J. Shea and G. Pan, Biosens. Bioelectron., 2018, 101, 52–59 CrossRef CAS PubMed.
- Q. Mei, H. Jing, Y. Li, W. Yisibashaer, J. Chen, B.-N. Li and Y. Zhang, Biosens. Bioelectron., 2016, 75, 427–432 CrossRef CAS PubMed.
- H. Zhou, D. Yang, N.-P. Ivleva, N.-E. Mircescu, S. Schubert, R. Niessner, A. Wieser and C. Haisch, Anal. Chem., 2015, 87, 6553–6561 CrossRef CAS PubMed.
- N. Reta, C.-P. Saint, A. Michelmore, B. Prieto-Simon and N.-H. Voelcker, ACS Appl. Mater. Interfaces, 2018, 10, 6055–6072 CrossRef CAS PubMed.
- Z. Farka, T. Juřík, D. Kovář, L. Trnková and P. Skládal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- H. Zhou, D. Yang, N. P. Ivleva, N. E. Mircescu, R. Niessner and C. Haisch, Anal. Chem., 2014, 86, 1525–1533 CrossRef CAS PubMed.
- H. Liu, X. Du, Y. Zang, P. Li and S. Wang, J. Agric. Food Chem., 2017, 65, 10290–10299 CrossRef CAS PubMed.
- W. Gao, B. Li, R. Yao, Z. Li, X. Wang, X. Dong, H. Qu, Q. Li, N. Li, H. Chi, B. Zhou and Z. Xia, Anal. Chem., 2017, 89, 9836–9842 CrossRef CAS PubMed.
- H. Wang, Y. Zhou, X. Jiang, B. Sun, Y. Zhu, H. Wang, Y. Su and Y. He, Angew. Chem., Int. Ed., 2015, 54, 5132–5136 CrossRef CAS PubMed.
- J. H. Chen, S. M. Andler, J. M. Goddard, S. R. Nugen and V. M. Rotello, Chem. Soc. Rev., 2017, 46, 1272–1283 RSC.
- X. Cao, M. Qin, P. Li, B. Zhou, X. H. Tang, M. H. Ge, L. B. Yang and J. H. Liu, Sens. Actuators, B, 2018, 268, 350–358 CrossRef CAS.
- P. Li, B. B. Zhou, X. M. Gao, X. H. Tang, L. B. Yang, L. Hu and J. H. Liu, Chem.–Eur. J., 2017, 23, 14278–14285 CrossRef CAS PubMed.
- J. Wang, X. Wu, C. Wang, Z. Rong, H. Ding, H. Li, S. Li, N. Shao, P. Dong, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 19958–19967 CrossRef CAS PubMed.
- I. H. Cho, P. Bhandari, P. Patel and J. Irudayaraj, Biosens. Bioelectron., 2015, 64, 171–176 CrossRef CAS PubMed.
- H. Kearns, R. Goodacre, L.-E. Jamieson, D. Graham and K. Faulds, Anal. Chem., 2017, 89, 12666–12673 CrossRef CAS PubMed.
- J. Wang, X. Wu, C. Wang, N. Shao, P. Dong, X. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 20919–20929 CrossRef CAS PubMed.
- H. Zhang, X. Ma, Y. Liu, N. Duan, S. Wu, Z. Wang and B. Xu, Biosens. Bioelectron., 2015, 74, 872–877 CrossRef CAS PubMed.
- C. Wang, B. Gu, Q. Liu, Y. Pang, R. Xiao and S. Wang, Int. J. Nanomed., 2018, 13, 1159–1178 CrossRef CAS PubMed.
- S. Deshwal and E.-B. Mallon, Dev. Comp. Immunol., 2014, 42, 240–243 CrossRef CAS PubMed.
- R. Kanchanapally, B.-P.-V. Nellore, S.-S. Sinha, F. Pedraza, S.-J. Jones, A. Pramanik, S.-R. Chavva, C. Tchounwou, Y. Shi, A. Vangara, D. Sardar and P.-C. Ray, RSC Adv., 2015, 24, 18881–18887 RSC.
- H. Etayash, L. Norman, T. Thundat, M. Stiles and K. Kaur, ACS Appl. Mater. Interfaces, 2014, 6, 1131–1138 CrossRef CAS PubMed.
- H. Etayash, K. Jiang, T. Thundat and K. Kaur, Anal. Chem., 2014, 86, 1693–1700 CrossRef CAS PubMed.
- M. Hoyos-Nogués, S. Brosel-Oliu, N. Abramova, F.-X. Muñoz, A. Bratov, C. Mas-Moruno and F.-J. Gil, Biosens. Bioelectron., 2016, 86, 377–385 CrossRef PubMed.
- Z. Li, H. Yang, L. Sun, H. Qi, Q. Gao and C. Zhang, Sens. Actuators, B, 2015, 210, 468–474 CrossRef CAS.
- E. Babich, A. Redkov, I. Reduto and A. Lipovskii, Phys. Status Solidi RRL, 2018, 12, 1700226 CrossRef.
- S. Saha, M. Ghosh, B. Dutta and J. Chowdhury, Appl. Surf. Sci., 2016, 362, 364–373 CrossRef CAS.
- M. S. Hizir, N. M. Robertson, M. Balcioglu, E. Alp, M. Rana and M. V. Yigit, Chem. Sci., 2017, 8, 5735–5745 RSC.
- X. Li, J. Li, X. Zhou, Y. Ma, Z. Zheng, X. Duan and Y. Qu, Carbon, 2014, 66, 713–719 CrossRef CAS.
- Y. X. Zou, S. Q. Huang, Y. X. Liao, X. P. Zhu, Y. Q. Chen, L. Chen, F. Liu, X. X. Hu, H. J. Tu, L. Zhang, Z. K. Liu, Z. Chen and W. H. Tan, Chem. Sci., 2018, 9, 2742–2849 Search PubMed.
- W. Shen, X. Lin, C. Jiang, C. Li, H. Lin, J. Huang, S. Wang, G. Liu, X. Yan, Q. Zhong and B. Ren, Angew. Chem., Int. Ed., 2015, 54, 7308–7312 CrossRef CAS PubMed.
- Y. Zou, L. Chen, Z. Song, D. Ding, Y. Chen, Y. Xu, S. Wang, X. Lai, Y. Zhang, Y. Sun, Z. Chen and W. Tan, Nano Res., 2016, 9, 1418–1425 CrossRef CAS.
- S. Saito, T. L. Massie, T. Maeda, H. Nakazumi and C. L. Colyer, Anal. Chem., 2012, 84, 2452–2458 CrossRef CAS PubMed.
- M. Kale and M. S. Shaikh, Int. J. Pharm. Sci., 2014, 6, 27–35 CAS.
- K. S. Yuan, J. X. Zheng, D. T. Yang, B. J. Sanchez, X. J. Liu, X. J. Guo, C. S. Liu, N. E. Dina, J. Y. Jian, Z. J. Bao, Z. W. Hu, Z. H. Liang, H. B. Zhou and Z. J. Jiang, ACS Omega, 2018, 3, 2855–2864 CrossRef CAS PubMed.
- B. Wagner, D. Schumann, U. Linne, U. Koert and M. A. Marahiel, J. Am. Chem. Soc., 2006, 128, 10513–10520 CrossRef CAS PubMed.
- A. C. Pawlowski, W. L. Wang, K. Koteva, H. A. Barton, A. G. McArthur and G. D. Wright, Nat. Commun., 2016, 7, 13803 CrossRef CAS PubMed.
- K. J. Stone and J. L. Strominger, Proc. Natl. Acad. Sci. U. S. A., 1971, 68, 3223–3227 CrossRef CAS.
- N. J. Economou, S. Cocklin and P. J. Loll, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14207–14212 CrossRef CAS PubMed.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- F. Costa, I. F. Carvalho, R. C. Montelaro, P. Gomes and M. C. L. Martins, Acta Biomater., 2011, 7, 1431–1440 CrossRef CAS PubMed.
- X. Bi, X. Du, J. Jiang and X. Huang, Anal. Chem., 2015, 87, 2016–2021 CrossRef CAS PubMed.
- A. Melaiye, R. S. Simons, A. Milsted, F. Pingitore, C. Wesdemiotis, C. A. Tessier and W. J. Youngs, J. Med. Chem., 2004, 47, 973–977 CrossRef CAS PubMed.
- W. Zhang, X. Shi, J. Huang, Y. Zhang and Z. Wu, ChemPhysChem, 2012, 13, 3388–3396 CrossRef CAS PubMed.
- L. Wang, J. Luo, S. Shan, E. Crew and J. Yin, Anal. Chem., 2011, 83, 8688–8695 CrossRef CAS PubMed.
- M. S. Mannoor, S. Y. Zhang, A. J. Link and M. C. McAlpine, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 19207–19212 CrossRef CAS PubMed.
- H. Etayash, M. F. Khan, K. Kaur and T. Thundat, Nat. Commun., 2016, 7, 12947 CrossRef CAS PubMed.
- H. Hu, Z. Wang, L. Pan, S. Zhao and S. Zhu, J. Phys. Chem. C, 2010, 114, 7738–7742 CrossRef CAS.
- J. E. Smith, K. E. Sapsford, W. Tan and F. S. Ligler, Anal. Biochem., 2011, 410, 124–132 CrossRef CAS PubMed.
- P. Chettri, V. S. Vendamani, A. Tripathi, A. P. Pathak and A. Tiwari, Appl. Surf. Sci., 2016, 362, 221–229 CrossRef CAS.
- M. S. Zheng, Y. K. Lee, Y. Li, K. Hwangbo, C. S. Lee, J. R. Kim, S. K. S. Lee, H. W. Chang and J. K. Son, Arch. Pharmacal Res., 2010, 33, 1307–1315 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional data and 13 supplementary figures. See DOI: 10.1039/c8sc04637a |
| ‡ K. Y. and Q. M. contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |