Transcriptomic and microRNAomic profiling reveals molecular mechanisms to cope with silver nanoparticle exposure in the ciliate Euplotes vannus†
Yongbo
Pan
a,
Wenjing
Zhang
 *a and
Senjie
Lin
ab
*a and
Senjie
Lin
ab
aState Key Laboratory of Marine Environmental Science, Marine Biodiversity and Global Change Research Center, College of Ocean and Earth Sciences, Xiamen University, Xiamen 361005, China. E-mail: zhangwenjing@xmu.edu.cn
bDepartment of Marine Sciences, University of Connecticut, Groton, CT 06340, USA
First published on 24th October 2018
Abstract
In spite of many reports on the toxicity of silver nanoparticles (AgNPs), the mechanisms underlying the toxicity are far from clear. The present study conducted transcriptome and microRNAome sequencing for Euplotes vannus to understand the molecular mechanisms by which this protist responds to and copes with AgNP exposure. By transcriptomic profiling, 1884 and 5834 differentially expressed genes (DEGs) were identified after one hour and 12 hours of exposure to 15 mg L−1 AgNPs, respectively. The DEGs were significantly enriched in macropinocytosis and phagocytic vesicles, suggesting that endocytic pathways may mediate the uptake of AgNPs, while the differential expression of ABC transporters and copper-transporting ATPase implicates active efflux transport of Ag. Several DNA repair pathways were also significantly enriched with differentially expressed cell cycle control genes, implying that exposure to AgNPs might have caused DNA damage and G2/M cell cycle arrest. The damage might have resulted from increased ROS production, as evidenced by elevated expression of several antioxidant genes to combat oxidative stress. From microRNAomic profiling, a total of 16 differentially expressed microRNAs were identified under AgNP stress. Integrated analysis of the microRNA and mRNA expression profiles indicated that the differentially expressed microRNAs target a series of genes involved in many important biological processes, such as the cell cycle, TCA cycle and mismatch repair, suggesting that AgNP exposure elicited a broad post-transcriptional regulatory mechanism in E. vannus to cope with the toxicity.
Environmental significanceThe broad applications of AgNPs have high potential to cause environmental pollution and toxicity in organisms, particularly benthic organisms which are likely to be exposed to higher accumulated concentrations than their planktonic counterparts. However, information about how AgNPs impact organisms and how organisms direct the cell to a detoxifying reaction and repair under AgNP stress at the molecular level is limited. This study provides a comprehensive insight into how a benthic ciliate copes with AgNPs, with multiple lines of evidence for novel mechanisms revealed by integrated analysis of transcriptomic, microRNAomic and biochemical approaches. This information is critical for assessing the toxicological impact of ENMs on living cells. |
1. Introduction
AgNPs are one of the most highly commercialized inorganic metallic nanoparticles due to their unique physico-chemical properties and antimicrobial potential.1,2 They are important in many industries, such as pharmaceuticals, cosmetics, textiles, surface coatings, electronic components, and food packaging.3,4 Because AgNPs could be released into the environment during the production, transport, erosion, washing, and disposal of AgNP products,5 the broad industrial applications have high potential to increase environmental exposure, cause toxicity in organisms and bring detrimental effects to ecosystems.In recent years, the toxicity of AgNPs has been demonstrated in many organisms, including zebrafish,6 arthropods (Daphnia magna),7 fish (medaka),8 mammalian cells9 and the bacterial model Escherichia coli.10 Various adverse effects have been identified in organisms under AgNP exposure. For example, Asmonaite et al.6 found that AgNPs caused adverse effects on the development, inhibited the hatchability and altered the locomotion of zebrafish. Shi et al.11 demonstrated that exposure to AgNPs could inhibit proliferation, damage the cell membrane and seriously induce apoptosis in human umbilical vein endothelial cells. Although several toxicity mechanisms of AgNPs have been suggested, such as 1) the release of Ag+ from AgNPs;12 2) the generation of reactive oxygen species (ROS);13,14 and 3) cell membrane damage caused by direct interaction with AgNPs,15 the molecular basis behind these complex toxicity mechanisms is largely unknown, especially regarding what key enzymes and signaling pathways may be responsible for the toxicity.13 Conventional toxicity assays cannot fully capture the complexities of cellular responses toward NPs. Thus, new studies using more integrated approaches are needed.
Using next-generation sequencing technologies to characterize genome-wide transcriptional regulation (mRNA) and post-transcriptional regulation (for example, via microRNA) can help address this need. RNA sequencing (RNA-Seq) provides a useful tool to identify the difference in the expression level of genes under different conditions.16 MicroRNAs, a class of endogenous small noncoding RNAs (∼22 nt in length), can bind to the 3′ untranslated region of their target mRNAs, leading to degradation of these mRNAs or inhibition of their translation, and thus, to the regulation of multiple cellular functions.17–19 The use of these analyses should aid in revealing the molecular basis underlying an organism's stress response to AgNP exposure. Indeed, studies on gene expression have uncovered multi-faceted mechanisms to cope with exposure to Ag salts or different forms of AgNPs in different test models (e.g. in E. coli,10D. magna,20Enchytraeus crypticus,21Eisenia fetida,22Chlamydomonas reinhardtii23 and human lung epithelial cells24), which would not be possible via standard tests (i.e. survival and reproduction endpoints). However, compared with the extensively studied mammalian cells or other model organisms, the transcriptional and post-transcriptional regulation in response to AgNPs has remained unexplored for protists. Additionally, information on temporal resolution of AgNP bioaccumulation, ROS production, DNA damage and other biochemical responses in protists is limited. This study aimed to address this gap in research by investigating genome-wide responses to AgNP stress in a marine ciliate.
Ciliates are single-celled protozoa and are abundant in freshwater, marine environments and terrestrial ecosystems. They play an important role in the food chain as transmitters of energy from primary producers to consumers, and as components of the planktonic and benthic microecosystems in aquatic environments. With their extraordinary advantages of high diversity, short life cycles, cosmopolitan distribution, simplicity, high degree of reproducibility, and quick responses to environmental disturbances, ciliates have been recognized as ideal biological indicators for environmental risk assessment.25,26 Additionally, as they share characteristics with eukaryotic cells, their biological response to various stressors can be used to predict the response of higher organisms.27 Furthermore, since sediments form a reservoir of nanoparticles, marine benthic organisms that inhabit these sediments, such as benthic ciliates, can be ideal organisms for researching AgNP ecotoxicology.
We conducted transcriptome and microRNAome sequencing for E. vannus, a marine benthic ciliate, grown under exposure to AgNPs. Our data provide a comprehensive insight into how a ciliate copes with AgNPs, revealing some novel mechanisms. This work also demonstrates the power of combined physiological response, transcriptomic and microRNAomic analysis in gaining a panoramic view of protist responses to AgNP stress.
2. Materials and methods
2.1 Chemicals
The uncoated AgNP powder (99% pure, cat. #484059-5G) and AgNO3 (99% pure, cat. #204390-1G) were purchased from Sigma-Aldrich. Other chemicals were purchased from Shanghai Macklin Biochemical Co. Ltd. (Shanghai, China), and all of them were of analytical grade. Ultrapure water (18.2 MΩ) was provided by a Millipore Advantage System (Merck Millipore, Darmstadt, Germany). Oshima's artificial seawater (Oshima's ASW, salinity 30 ‰) was prepared by dissolving NaCl (28 g), KCl (0.8 g), MgCl2·6H2O (5 g) and CaCl2·H2O (1.2 g) in 1 L ultrapure water and subsequently adjusting the pH to 8.2 with 0.5 M NaOH,28 then filtered through a 0.22 μm-pore size, 47 mm-diameter polycarbonate membrane (Millipore, Billerica, MA, USA) to remove undissolved salt particles.Twenty-five mg of Ag nanopowder was mixed with 50 ml of ultrapure water to yield a 500 mg L−1 stock suspension, which was vortexed and sonicated (40 W, Branson probe sonicator, USA) for 60 minutes before use. AgNO3 powder dissolved in ultrapure water (stock concentration 50 mg L−1 of Ag+) was used as an Ag+ control to distinguish between particle specific effects and effects caused by Ag+. AgNP and Ag+ stock solutions were prepared and stored in the dark at 4 °C.
2.2 Nanoparticle (NP) characterization
Characterization of NP dispersions in the exposure medium was performed at 15 mg L−1. Scanning electron microscopy (SEM, LEO 1530, Germany) and transmission electron microscopy (TEM, Tecnai G2 Spirit BioTWIN TEM, USA) analyses were used to characterize the size and shape of the AgNPs. The samples were prepared by placing a droplet of the AgNPs on a carbon-coated copper grid followed by obtaining images. The characteristic surface plasmon absorption band of AgNPs was recorded with a UV-vis spectrophotometer (Shimadzu UV 2450, Shimadzu Corporation, Japan). The hydrodynamic size and zeta potential of the AgNPs were measured using a Malvern Zetasizer Nano-ZS (Malvern Instruments, Malvern, UK). At each time point, three replicates were prepared, and five consecutive measurements were made per replicate. Dissolution of the AgNPs was determined by separating dissolved silver from the AgNPs using centrifugal ultrafiltration with a 3 kDa filter (Millipore Amicon Ultra). The dispersions were centrifuged at 6000g for 10 min (Centrifuge 5424, Eppendorf, Germany), and the dissolved silver present in the filtrate was acidified and analyzed in triplicate at 0, 1, 3, 6, 12 and 24 h by inductively coupled plasma mass spectrometry (ICP-MS; Agilent 7700X, USA).2.3 Culture of E. vannus
E. vannus was isolated from the coast of Xiamen, situated in the southeast of China. Identification of the species was performed using morphological characteristics and a molecular marker (18S rRNA, sequence has been deposited to NCBI, MG999513). Clonal cultures were established and maintained in Oshima's ASW at 25 °C under cool white fluorescent light (16 h light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 h dark) at a light intensity of ∼40 μmol photons m2 s−1 and fed with E. coli. The ciliates were cultured to the stationary growth phase before they were transferred to a new culture medium for the following toxicity tests.
8 h dark) at a light intensity of ∼40 μmol photons m2 s−1 and fed with E. coli. The ciliates were cultured to the stationary growth phase before they were transferred to a new culture medium for the following toxicity tests.
2.4 Acute toxicity tests for determining 12 h-LC50
The toxicity tests were conducted in 24 well flat-bottom culture plates (height: 1 mm; diameter: 16 mm) at 25 °C. E. vannus cells were inoculated into a series of AgNP solutions of different concentrations (0, 5, 10, 15, 20, 25, 35, 45 and 55 mg L−1) for 12 hours. Each treatment was randomly assigned to triplicate wells, and each well was stocked with 24 E. vannus cells in a 1 ml solution. To minimize contamination through food (mainly bacteria), E. vannus were starved for 12 h before use and not fed throughout the entire experimental period. Those E. vannus unable to swim or creep on the bottom of the well, together with disappearing cells, were regarded as dead.29 Mortality was measured under a dissecting microscope (10–40×). Results were analyzed and the median lethal dose (LC50) was determined using the probit method.30 The 12 h-LC50 of Ag+ was also determined.2.5 Bioaccumulation, lipid peroxidation, ROS levels, ATP content and enzyme activity
For the exposure experiments, E. vannus in the stationary growth phase were added to a final cell density of 2.5 × 103 cells per mL to the exposure medium, which was pre-equilibrated with 15 mg L−1 (1/2 12 h-LC50) AgNPs. After treatment for 0, 1, 3, 6, 12 and 24 hours, E. vannus was harvested by filtration with a 20 μm pore-size sieve and washed with Oshima's ASW five times in order to mechanically remove nanoparticles and bacteria. The harvested samples were used for detecting various physiological parameters and enzyme activity, including silver bioaccumulation, ROS content, lipid peroxidation (MDA content), intracellular ATP levels, and activities of glutathione peroxidase (GPx) and glutathione reductase (Gr). The total protein level of each sample was measured using a BCA Protein assay kit (Beyotime Co., China) according to the manufacturer's instructions, and the measured total protein level was used for normalizing the MDA, ATP, GPx and Gr content. All exposure experiments were carried out in three biological replicates. The assays were performed as described in the ESI.†2.6 RNA isolation and sequencing
We conducted transcriptomic and microRNAomic analyses after 0, 1 and 12 hours of AgNP treatment (denoted as T0, T1 and T12, respectively), because our measured parameters at T1 and T12 exhibited the strongest responses to AgNPs, and T0 was a time control for comparison. Each treatment was conducted in triplicate cultures. About 5 × 104 cells were collected on each culture and homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and RNA was extracted according to the manufacturer's protocol. Ethanol precipitation instead of an RNA binding column was used in this isolation, which enables us to analyze both mRNA and microRNA from one sample.31 Agarose gel electrophoresis (1.5%), Nanodrop 2000 (Thermo Scientific, Wilmington, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) were used to check the integrity, concentration and quality of RNA. The RNA integrity number of the samples was all above 8.0. The same RNA samples were used for mRNA and microRNA sequencing as well as real-time quantitative PCR (RT-qPCR) analysis.A total of 3 μg RNA per sample was used to generate a paired-end RNA-Seq library using a NEBNext® Ultra™ RNA Library Prep Kit (NEB, USA), following the manufacturer's instructions. Small RNA libraries were generated using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, USA) from 1.5 μg total RNA. The libraries were purified (AMPure XP system) and quantified using the Agilent Bioanalyzer 2100 system and then sequenced on an Illumina HiSeq X Ten platform. Sequence files of mRNA and microRNA have been deposited into the GEO databank under the accession number GSE118418.
2.7 Bioinformatic analyses
Raw reads for RNA and microRNA sequencing were cleaned up by trimming adapter sequences, removing poly-N containing reads (≥5% of nucleotides in each read) and filtering low quality reads (Q-score ≤20 for ≥50% of nucleotides in each read) using a proprietary program.32 All the downstream analyses were based on clean data with high quality.Transcriptome assembly was performed using Trinity (min_kmer_cov = 2, and all other parameters set as default). The assemblies were blasted against the NCBI non-redundant protein database (NR), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) to predict the protein function with a cutoff e-value of 1 × 10−5. Differentially expressed genes (DEGs) were estimated using edgeR (V3.6)33 in the Bioconductor package. Genes showing |log2 (fold change)| > 1 and FDR < 0.01 (adjusted p-value, determined by the Benjamini and Hochberg multiple-testing correction implemented in the ‘p.adjust’ method of R) at any of the three comparison groups (T1 vs. T0, T12 vs. T0 and T12 vs. T1) were defined as DEGs.
For the microRNA sequencing data set, reads smaller than 18 nt or longer than 30 nt in length were removed. Sequence tags originating from non-coding RNAs (rRNA, scRNA, snoRNA, snRNA and tRNA) deposited in the Rfam 10.1 database were also excluded. The remaining reads were used to detect known microRNAs and novel microRNAs predicted by comparison with known microRNAs from miRBase 21.0. Randfold tools software34 was used for novel microRNA secondary structure prediction, which allowed us to distinguish real and background precursors by examining the stability of the secondary structure. The expression level of all microRNAs were normalized by dividing the absolute read count with the total read count of the sample and multiplying the result by 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. After normalization, the microRNA expression profiles between each pair of microRNA libraries were compared. The differentially expressed microRNAs were defined with the criteria of |log2 (fold change)| > 1 and p-value < 0.05 shown at any of the three comparison groups (T1 vs. T0, T12 vs. T0 and T12 vs. T1), while microRNAs with a stricter criterion, FDR < 0.05, were also identified.
000. After normalization, the microRNA expression profiles between each pair of microRNA libraries were compared. The differentially expressed microRNAs were defined with the criteria of |log2 (fold change)| > 1 and p-value < 0.05 shown at any of the three comparison groups (T1 vs. T0, T12 vs. T0 and T12 vs. T1), while microRNAs with a stricter criterion, FDR < 0.05, were also identified.
Previous studies have suggested that when the expression levels of microRNAs and mRNAs are simultaneously profiled, a microRNA and the mRNAs that it targets for degradation should exhibit inverse relative expressions.35–37 To determine reliable target mRNAs of the differentially expressed microRNAs, an integrated analysis of the microRNA and mRNA expression profiles was conducted in this study. Firstly, the 3′ UTR sequence of each unigene was used to predict animal type targets using miRanda38 with stringent criteria (miRanda-3.3a, -sc 140 -en -15 -scale 4 -strict). Secondly, Spearman's rank correlation analyses between the microRNA and mRNA expression data in all samples were conducted via the cor.test () function in the R statistical package. Only microRNA–mRNA pairs with a correlation coefficient Rho < = −0.4 were retained as potential target genes. Thirdly, only genes passing both miRanda and Spearman's rank correlation coefficient criteria were recognized as the final target genes, and their associated functions were analyzed.
GO enrichment was analyzed for the DEGs and the predicted target genes of the differentially expressed microRNAs using DAVID39 with UNIPROT ACCESSION terms as previously reported.40 All the transcripts of E. vannus with Swiss-Prot annotation were used as a background gene list for the enrichment analyses. KEGG pathways were analyzed using TBtools.41 All the transcripts of E. vannus with KO annotation were used as a background gene list. A corrected Benjamini p-value < 0.05 as cutoff was used to yield statistically significant gene enrichment for particular pathways or GO categories.
2.8 Gene expression validation
From the RNA samples, 500 ng RNA of each sample was used for reverse transcription to cDNA using a PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa Biotech Co., Ltd., Dalian, China). gDNA Eraser degrades any genomic DNA that might remain in the RNA extract. From these cDNA samples, 1 μL (50 ng) from each sample was used in qPCR with a Faststart Universal SYBR Green Master (Roche, Germany) on LightCycler 96 (Roche, Germany). All reactions were performed in triplicate. All quantifications were performed with 18S rRNA as a reference for normalization, and the relative amount of mRNA was calculated using the 2−ΔΔCt method. Primer specificity was confirmed through assessment of the product melting curves. For microRNA RT-qPCR, the TransScript Green miRNA Two-Step qRT-PCR SuperMix (TransGen Biotech, China) was used to facilitate cDNA synthesis, then amplified using a specific primer. The microRNA expression levels were normalized to U6 snRNA. The primer sets are listed in ESI† Table S8.2.9 Statistical analyses
Data are presented as the mean ± SD derived from the biological triplicates. The statistical comparisons of means were performed using one-way analysis of variance (ANOVA) unless otherwise indicated. A value of p < 0.05 was accepted as significantly different. The statistical analysis was performed using the Statistical Package for the Social Sciences software (SPSS 20.0).3. Results
3.1 Characterization of AgNPs
The initial SEM and TEM measurement showed that AgNPs were spherical particles with an average size of 73.82 nm (Fig. S1A and B†). The hydrodynamic size of AgNPs in the E. vannus exposure medium was found to change from 160 ± 6.5 nm at 0 h to 294.4 ± 18.2 nm at 12 h (Fig. S1F†), indicating slight aggregation. These results were also confirmed by the ζ potentials. Values indicate strong electrostatic repulsion and good stability at 0 h (−25.2 ± 2.7 mV) but a loss of electrostatic repulsion, and likely agglomeration, at 12 h (−16.6 ± 2.1 mV, Fig. S1E†). AgNPs showed an absorption band at 401 nm, which is the typical absorption band of AgNP surface plasmons, but the absorbance decreased slightly after 12 h of exposure indicating AgNP sedimentation (Fig. S1C†). The Ag particle dissolution rate was determined, and the results demonstrated a slow increase in Ag shedding after 1, 3, 6, 12, and 24 h of incubation in the exposure medium, with the mean dissolution rate ranging from 1.13% at 0 h to 7.55% at 24 h (Fig. S1D†).3.2 Toxicity test
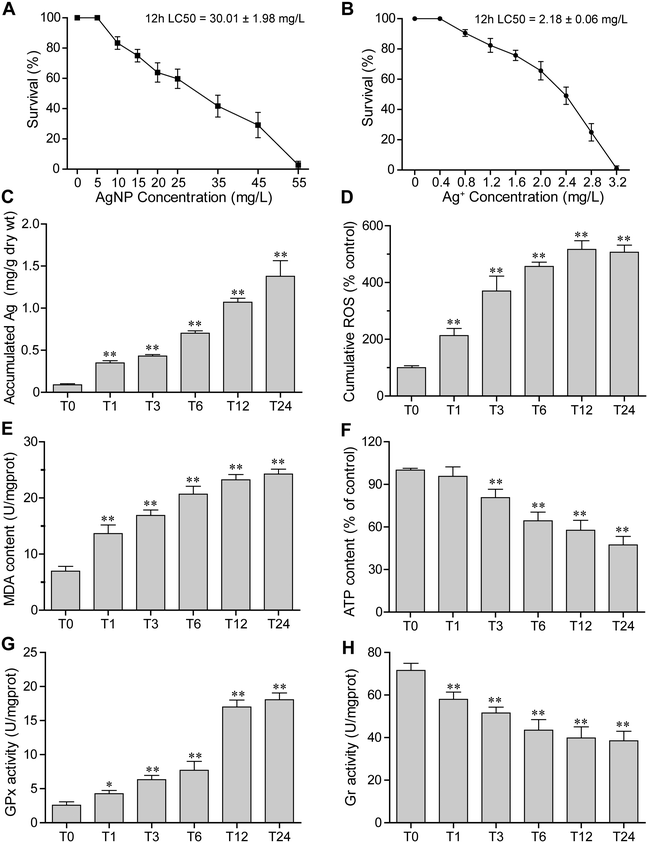
The survival rates of E. vannus after 12 h of exposure to different concentrations of AgNPs are shown in Fig. 1A. The toxicity of AgNPs increased in a concentration-dependent manner. When the concentration of AgNPs reached 55 mg L−1, E. vannus could hardly survive. Based on the survival rate, the 12 h LC50 was calculated to be 30.01 ± 1.98 mg L−1. Meanwhile, the toxicity of AgNO3 to E. vannus was determined, and the result showed that the 12 h LC50 of Ag+ was 2.18 ± 0.06 mg L−1 (Fig. 1B). Based on these results, we choose 15 mg L−1 (close to 1/2 LC50) AgNPs as the exposure concentration in the subsequent experiments.3.3 Bioaccumulation of Ag
Intracellular concentrations of silver were measured by ICP-MS. When E. vannus was exposed to 15 mg L−1 of AgNPs, the intracellular concentration of silver significantly increased over time. Compared with the Ag content of 0.09 ± 0.01 mg g−1 dry weight (dry wt) of E. vannus in the time control group (0 hour), the Ag content in E. vannus after 24 hours of exposure reached 1.38 ± 0.15 mg g−1 dry wt (Fig. 1C), suggesting that E. vannus was taking up Ag fast and the bioconcentration factor was high.3.4 ROS production
The production of ROS in E. vannus induced by AgNPs was measured through DCF fluorescence. We found that the ROS production as measured by a flow cytometer significantly increased after AgNP exposure in a time-dependent manner (Fig. 1D). Fluorescence micrographs also show that the ROS-induced fluorescence intensity after 12 h of exposure (T12) was significant higher than that at 0 h of exposure (T0) (Fig. S2†). These results were consistent with the increase in intracellular concentrations of Ag, indicating that ROS production was intracellular AgNP dose-dependent.3.5 Lipid peroxidation
The malondialdehyde (MDA) content, a measure of the degree of lipid peroxidation, was examined after AgNP exposure. Results showed that AgNPs caused significant lipid peroxidation of the cell membrane (Fig. 1E), and the MDA content showed a similar increasing trend to ROS generation, clearly suggesting oxidative stress-induced lipid peroxidation.3.6 ATP levels
Intracellular ATP levels could be affected by the dysfunction of mitochondria. We detected the intracellular ATP levels, and found that they significantly decreased over time under exposure to 15 mg L−1 of AgNPs (Fig. 1F), indicating that exposure to high-concentration AgNPs has likely caused the mitochondria dysfunction of E. vannus.3.7 Redox enzyme activities
The enzyme activities of glutathione peroxidase (GPx) and glutathione reductase (Gr) were detected at different exposure durations. We found that the GPx activity increased while the Gr activity decreased as the exposure prolonged (Fig. 1G and H). These results indicated that exposure to AgNPs caused an alteration of activities of enzymes potentially involved in detoxification.3.8 Transcriptomic and microRNAomic responses
Approximately 21.41–33.81 million of 150 bp pair-end reads were generated for the nine samples through RNA sequencing (Table S1†). After sequence trimming, the retained high-quality reads of all the samples were pooled and assembled de novo into 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 343 unigenes (practically defined as the longest sequence of the Trinity transcript cluster) with an average length of 797 bp, a maximum length of 15
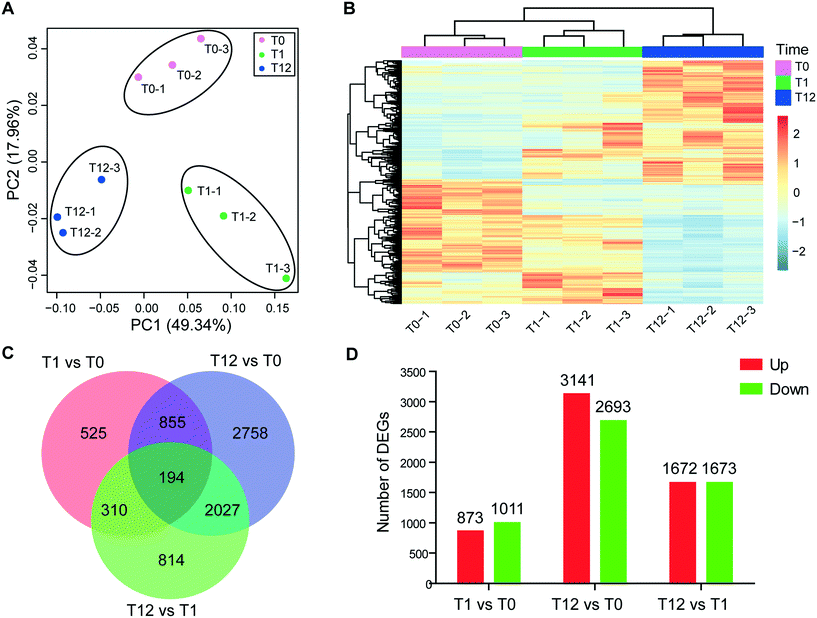
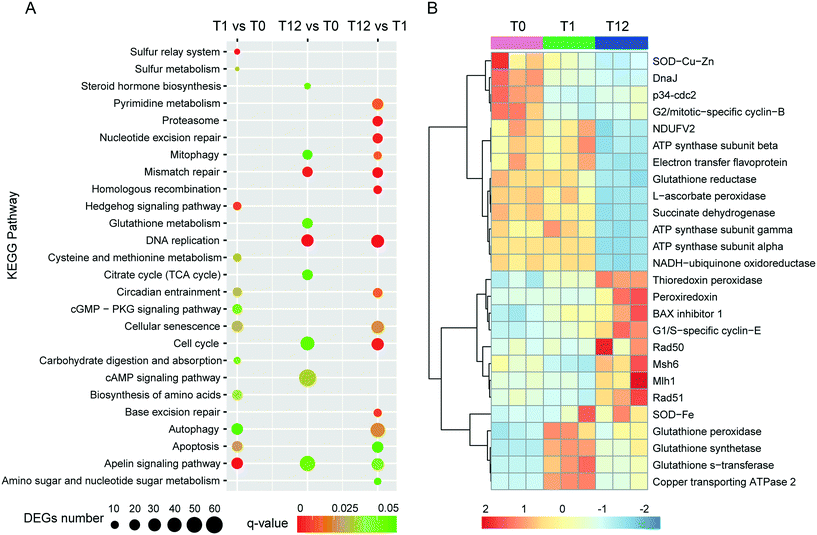
343 unigenes (practically defined as the longest sequence of the Trinity transcript cluster) with an average length of 797 bp, a maximum length of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 756 bp, and an N50 of 1304 bp, indicating a good assembly quality of the transcriptome. About 51% of these unigenes had significant matches to documented genes in public databases (Table S2†). All unigenes were used for principal component analysis (PCA), and the result showed that the biological replicates of each time point were close to each other and far from the other time points, verifying the reproducibility and reliability of the RNA-Seq data (Fig. 2A). Using a cutoff |log2 (fold change)| > 1 and FDR < 0.01 as the criteria, a total of 7483 DEGs were detected, among which 1884, 5834 and 3345 DEGs were detected for T1 vs. T0, T12 vs. T0 and T12 vs. T1 comparisons, respectively (Fig. 2B and D). The number of up-regulated genes was similar to the number of down-regulated genes at each time point, but the number of DEGs at 12 h of exposure was higher than that at one h of exposure (Fig. 2D). The KEGG enrichment of these DEGs indicated their involvement in various pathways at different comparison groups. For instance, apoptosis and autophagy were significantly enriched at T1 vs. T0 and T12 vs. T1, and the cell cycle and mitophagy were significantly enriched at T12 vs. T0 and T12 vs. T1, while the TCA cycle and glutathione metabolism were significantly enriched T12 vs. T0 (Fig. 3A, q-value < 0.05). GO analysis showed that macropinocytosis was significantly enriched at T12 vs. T0 and T12 vs. T1, and the phagocytic vesicles were significantly enriched at T12 vs. T0 (Table S5†).
756 bp, and an N50 of 1304 bp, indicating a good assembly quality of the transcriptome. About 51% of these unigenes had significant matches to documented genes in public databases (Table S2†). All unigenes were used for principal component analysis (PCA), and the result showed that the biological replicates of each time point were close to each other and far from the other time points, verifying the reproducibility and reliability of the RNA-Seq data (Fig. 2A). Using a cutoff |log2 (fold change)| > 1 and FDR < 0.01 as the criteria, a total of 7483 DEGs were detected, among which 1884, 5834 and 3345 DEGs were detected for T1 vs. T0, T12 vs. T0 and T12 vs. T1 comparisons, respectively (Fig. 2B and D). The number of up-regulated genes was similar to the number of down-regulated genes at each time point, but the number of DEGs at 12 h of exposure was higher than that at one h of exposure (Fig. 2D). The KEGG enrichment of these DEGs indicated their involvement in various pathways at different comparison groups. For instance, apoptosis and autophagy were significantly enriched at T1 vs. T0 and T12 vs. T1, and the cell cycle and mitophagy were significantly enriched at T12 vs. T0 and T12 vs. T1, while the TCA cycle and glutathione metabolism were significantly enriched T12 vs. T0 (Fig. 3A, q-value < 0.05). GO analysis showed that macropinocytosis was significantly enriched at T12 vs. T0 and T12 vs. T1, and the phagocytic vesicles were significantly enriched at T12 vs. T0 (Table S5†).
For microRNA analysis, nine samples were sequenced. After sequence quality control, approximately 11.99–19.27 million clean reads for each sample were obtained (Table S3†). A total of 190 mature microRNAs were identified, and all of these were novel microRNAs (Dataset S1†). The lengths of these microRNAs ranged from 18 to 25 nt, with a peak at 23 nt (Fig. S4†). Using a cutoff of |log2 (fold change)| > 1 and p < 0.05, we identified 16 differentially expressed microRNAs, among which only five had FDR < 0.05 (Table S4†). The 16 differentially expressed microRNAs showed varied expression patterns in the three comparison groups, with 8 (8 up-regulated, 0 down-regulated), 10 (8 up-regulated, 2 down-regulated) and 10 (6 up-regulated, 4 down-regulated) differentially expressed in T1 vs. T0, T12 vs. T0 and T12 vs. T1, respectively (Table 1).
| Total | Up | Down | Total target gene | Up microRNA target gene | Down microRNA target gene | |
|---|---|---|---|---|---|---|
| MicroRNA | 190 | — | — | — | — | — |
| DE microRNA | 16 | — | — | 4640 | — | — |
| T1 vs. T0 | 8 | 8 | 0 | 2986 | 2986 | — |
| T12 vs. T0 | 10 | 8 | 2 | 3436 | 2633 | 809 |
| T12 vs. T1 | 10 | 6 | 4 | 1931 | 879 | 1090 |
3.9 Integrated analysis of mRNA and microRNA
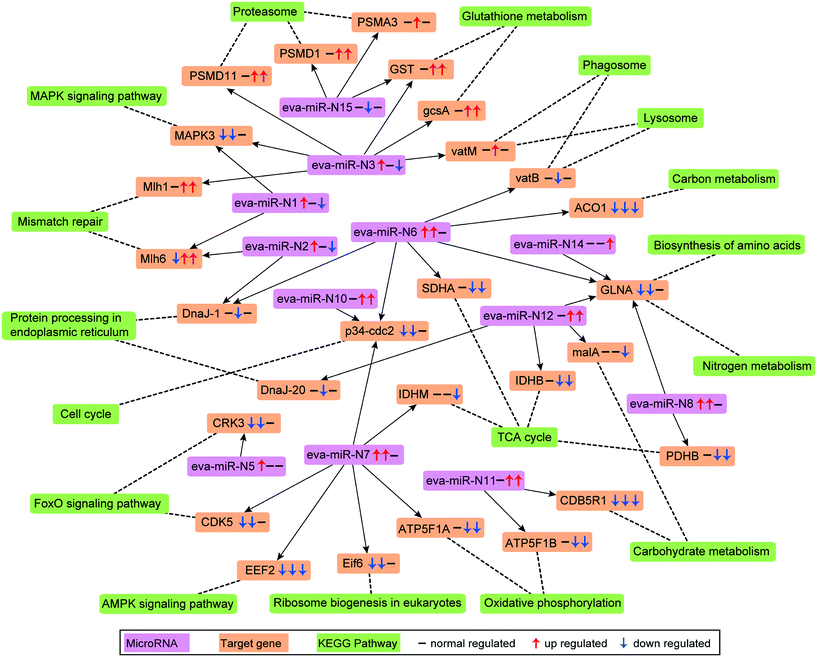
Combing the structural and expression profile based analysis results, a total of 4640 protein-coding genes were found as targets of the 16 differentially expressed microRNAs. Among these predicted target genes, 2986, 3436 and 1931 were regulated by 8, 10 and 10 microRNAs that were differentially expressed in T1 vs. T0, T12 vs. T0 and T12 vs. T1 comparisons, respectively (Tables 1 and S4†). KEGG pathway analysis indicated that the targets of these microRNAs might be involved in a series of biological processes including the FoxO signaling pathway, lysosome, proteasome, cell cycle, mismatch repair and TCA cycle in AgNP-exposed E. vannus (Fig. 4 and Table S6†). To visually demonstrate the relationship between the differentially expressed microRNAs and their target genes and associated biological functions, a simplified microRNA–target gene-biological function regulatory network (Fig. 4) was constructed using Cytoscape.42 We found that one microRNA could regulate several biological processes by targeting different mRNAs, and one biological process could be associated with multiple microRNAs. For example, three up-regulated microRNAs (eva-miR-N6, eva-miR-N7 and eva-miR-N10) co-regulated p34cdc2, which was a key gene that controls cell cycle progression from the G2 phase to M phase. Consistent with the general negative regulator function of microRNAs, the p34cdc2 mRNA level decreased after AgNP exposure for one and 12 hours (Fig. 4), indicating that these microRNAs may be involved in post-transcriptional regulation of the cell cycle.3.10 RNA sequencing validation by qPCR
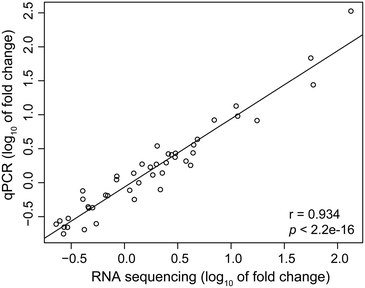
To validate the expression data obtained from RNA-Seq, 12 mRNAs and 3 microRNAs that showed different expression patterns or known to be involved in important biological functions were selected for further analysis using RT-qPCR. The results showed a strong spearman correlation with the data of RNA sequencing (r = 0.934, p < 2.2 × 10−16, Fig. 5). For each gene, the expression count values of transcriptome and microRNA data exhibited a similar expression profile in comparison with the results of RT-qPCR at all the three time stages (Fig. S5†).4. Discussion
The present study demonstrates the toxicity of AgNPs to E. vannus, and the molecular responses of this ciliate at the exposure concentration of 15 mg L−1. This concentration of AgNPs was markedly higher than that measured (1–5 μg L−1) in sewage treatment plants43 or predicted (<1 μg L−1) in surface water,44 but was selected: 1) because E. vannus is a benthic ciliate which would likely be in contact with higher concentrations of AgNPs than that in surface water as NPs tend to homo- and hetero-aggregate and accumulate on the sediment; 2) to ensure that E. vannus would trigger a fast response at the molecular level; and 3) to allow for comparison of our results with the existing toxicological literature based on lethality.45 Consequently, caution is needed when interpreting our results presented in this paper in regard to possible responses of E. vannus to silver nanoparticles in the natural environment where the concentrations of silver nanoparticles are substantially lower.4.1 Uptake and efflux of silver in E. vannus
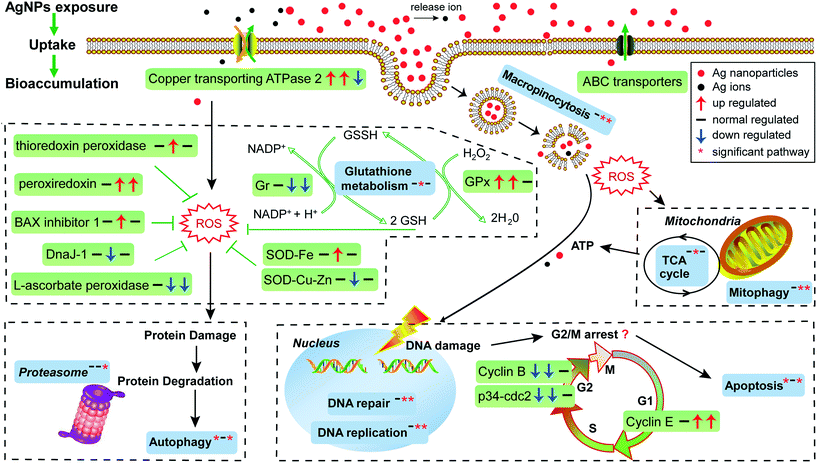
One factor determining the outcome of exposure to AgNPs is the intracellular concentration. By comparing the Ag content in our experimental ciliates with that in the time control group, our study demonstrates that Ag in E. vannus is taken up fast and the bioconcentration factor is high (Fig. 1C). We speculated that both AgNPs and the ions released from the particles contributed to the intracellular bioaccumulation of Ag in E. vannus. Previous studies have used various techniques to provide experimental evidence for the internalization of Ag nanoparticles in different organisms, although the uptake mechanisms and routes have remained unclear.15,46,47 Endocytic pathways had been considered a potential mechanism of nanomaterial uptake for animal cells, such as clathrin-mediated endocytosis, caveolae-mediated endocytosis, macropinocytosis and phagocytosis.48,49 In our study, GO enrichment analysis showed that the GO terms of macropinocytosis and phagocytic vesicles were significantly enriched after 12 hours of exposure to AgNPs (q < 0.01, Table S5†). A previous study found that the human lung cells treated with pharmacologic inhibitors of different endocytic pathways decreased the uptake of AgNPs, thus suggesting that the uptake of AgNPs was a combination of active mechanisms.50 These results suggest that endocytic pathways may be involved in the uptake of AgNPs in E. vannus. For the uptake of Ag+, earlier studies have hypothesized that it occurred via Cu+ transport systems.23,46,51 Indeed, our transcriptomic and qPCR analysis indicated that ATP7B (copper-transporting ATPase 2 protein gene) was significantly up-regulated both at T1 vs. T0 and T12 vs. T0, suggesting that Ag+ transport by E. vannus might also be mediated via the Cu+ transport system, but this transport pathway might become less active as the exposure prolonged because this gene was down-regulated at T12 compared to T1 (Fig. 3B and S5†).The high intracellular concentrations of silver are expected to trigger an efflux of silver response. At the transcriptome level, we observed that several of ATP-binding cassette (ABC) transporters, including ABC transporter A, B, C, F and G subfamilies, were differentially expressed (Table S9†). These transporters are members of one of the largest protein superfamily, existing widely from bacteria to humans.52 They are transmembrane proteins and the subfamilies of ABC transporter B (ABC-B) and C (ABC-C) are known to contribute to detoxification through transporting endogenous metabolites and exogenous xenobiotics out of cells,53–55 including heavy metal ions.56 Thus, we speculate that the differentially expressed ABC transporters may play a key role in the efflux of AgNPs although Ag transport by these transporters has not been reported. In addition, the copper-transporting ATPase as mentioned above is a bidirectional transporter and has also been suggested as a pump for the extrusion of monovalent copper and silver ions in Enterococcus hirae.51 However, E. vannus seems to be unable to effectively eliminate the metal at high exposure concentrations (1/2 12 h-LC50) because the cells still contained detectable Ag+ (Fig. 1C), casting a question whether the Cu+ transport system in effect serves more as a Ag+ uptake than an export mechanism.
4.2 The effects of AgNPs in E. vannus
At the submicroscopic level, a very recent study using differential interference, TEM and SEM methods demonstrated that AgNPs could cause a series of adverse effects in Euplotes aediculatus, including mitochondrial damage, chromatin condensation and cilium shedding, but the mechanisms at the molecular level have remained unclear.57 ROS generation and oxidative stress are generally accepted as the likely mechanisms of nanoparticle-induced toxicity.13,14 We measured the presence of ROS in E. vannus after exposure to AgNPs, and found a time-dependent increase (Fig. 1D). It is postulated that ROS generation occurs on the surface of AgNPs as a result of particle surface reactions.58,59 In addition, the inherent catalytic activity of nanoparticles and mitochondrial damage induced by nanoparticles may be sources of oxidative stress and ROS production.60 At the transcriptomic level, the pathway of mitophagy was significantly enriched after 12 hours of exposure (Fig. 3A), which is suggestive of serious mitochondria damage and explains the high content of ROS. We detected a significant decrease in ATP content, confirming the compromised function of mitochondria (Fig. 1F), although detoxifying mechanisms, such as active efflux of AgNPs and Ag+, also require ATP, impacting the total content within the cell. Our transcriptomic data showed a potential decrease in several proteins involved in energy metabolism as a result of AgNP exposure. NADH-ubiquinone oxidoreductase, succinate dehydrogenase (SDHA), electron transfer flavoprotein (ETFA) and NADH dehydrogenase [ubiquinone] flavoprotein 2 (NDUFV2), all of which are involved in transport of electrons, were significantly down-regulated after 12 h of exposure (Fig. 3B). ATP-synthase subunits alpha (ATP5F1A), subunits beta (ATP5F1B) and subunits gamma (ATP5F1C) were also down-regulated after 12 h of exposure (Fig. 3B). Interestingly, several up-regulated microRNAs were predicted to regulate ATP metabolism (Fig. 4). For instance, eva-miR-N6, eva-miR-N7, eva-miR-N8 and eva-miR-N12 presumably regulate the SDHA, IDHM (isocitrate dehydrogenase [NAD]), PDHB (pyruvate dehydrogenase E1 component subunit beta) and IDHB (isocitrate dehydrogenase [NADP]) gene, respectively, all of which are key components in the TCA cycle. Besides, eva-miR-N7 and eva-miR-N11 presumably regulate the ATP5F1A and ATP5F1B gene, respectively, both of which are involved in oxidative phosphorylation. Up-regulation of these microRNAs under AgNPs is consistent with the depression of ATP synthesis.High ROS content is detrimental to cells. One of the cellular targets of ROS is lipids. The resulting lipid peroxides decompose rapidly and produce malondialdehyde (MDA) as the major product. We observed a significant time-dependent increase in MDA in E. vannus after 24 h of exposure to AgNPs (Fig. 1E), suggesting membrane damage caused by overload of ROS, as has been reported in bacteria61 and human liver cells62 previously.
In cell culture studies, DNA damage has been reported upon exposure to AgNPs through both direct measurements63 and through the induction of genes or proteins involved in DNA damage repair.64,65 KEGG enrichment analysis in the present study showed that several pathways exhibited significant enrichment upon AgNP exposure in DNA repair, including mismatch repair (T12 vs. T0 and T12 vs. T1), homologous recombination (T12 vs. T1), base excision repair (T12 vs. T1) and nucleotide excision repair (T12 vs. T1) (Fig. 3A). Furthermore, several DNA repair genes were up-regulated after 12 hours of exposure, including Mlh1, Msh6, Rad 50 and Rad 51 (Fig. 3B), among which Mlh1 was regulated by eva-miR-N3, and Msh6 was co-regulated by eva-miR-N1 and eva-miR-N2 (Fig. 4). These results suggest that exposure to AgNPs has caused DNA damage in E. vannus and triggered response to repair the damage via transcriptional and post-transcriptional regulation. DNA damage is usually accompanied by cell cycle arrest. Eom and Choi66 reported that after 12 and 24 hours of exposure, AgNPs lead to serious decreases in cells in the G1 phase and increases in cells in the G2/M and S phases of Jurkat T cells. In our transcriptome analysis, G1/S specific cyclin E (CycE), which is a key gene whose encoded protein controls G1 to S transition, was up-regulated at T12 vs. T0 and T12 vs. T1, while the genes G2/mitotic specific cyclin B (CycB) and p34cdc2 that control the G2 to M progression were down-regulated at T1 vs. T0 and T12 vs. T0 comparisons, suggesting that exposure to AgNPs might have caused G2/M arrest, which can lead to apoptosis (Fig. 3B and 6). This is speculative, but nevertheless consistent with previous studies showing that G2/M arrest can result from a decrease in cyclin B expression.67 Additionally, three up-regulated microRNAs (eva-miR-N6, eva-miR-N7 and eva-miR-N10) were predicted to co-regulate the p34cdc2 gene (Fig. 4), suggesting that these microRNAs may be involved in post-transcriptional regulation of the cell cycle (leading to slowdown of cell division).
Our transcriptomic and microRNAomic data also show signs of protein damage as a response to exposure to AgNPs. The oxidative damage caused by AgNPs can produce a large amount of protein fragments or cell debris that needs to be removed, which would involve proteasome and lysosome. The significant enrichment of the proteasome pathway in the AgNP-treated E. vannus observed in this study is evidence of protein damage (Fig. 3A). Indeed, we found 32 DEGs involved in proteasome and all of them showed up-regulation under AgNP exposure; 35 DEGs were involved in lysosome, 27 of which showed up-regulation (Dataset S2†). Additionally, three important genes involved in proteasome were regulated by microRNAs, PSMA3 (proteasome subunit alpha type-3) and PSMD1 (26S proteasome non-ATPase regulatory subunit 1) were regulated by eva-miR-N15, while PSMD11 was regulated by eva-miR-N3. The vatM (vacuolar proton translocating ATPase) and vatB (V-type proton ATPase subunit B) genes that are involved in lysosome were presumably targeted by eva-miR-N3 and eva-miR-N6, respectively (Fig. 4). These results illustrate that exposure to high concentration of AgNPs could cause protein damage in E. vannus (Fig. 6).
Although the toxic effect of AgNPs has been explicitly proposed as mentioned above, due to the lack of Ag+ control in our study, we were unable to distinguish the mechanism between nanoparticles and ions in E. vannus. Some information can be obtained from what has been done in other model organisms, however. For example, Bicho et al.68 showed that Ag NM300K and AgNO3 toxicity occurs in distinct life stages of E. crypticus, indicating multiple routes of toxicity. Our study is in support of this view because the release of Ag+ of AgNPs at 15 mg L−1 (1/2 LC50) was estimated to be 0.792 ± 0.024 mg L−1 (15 × 5.28%), which was significantly lower than the 1/2 LC50 of Ag+ (1.09 ± 0.03 mg L−1, p < 0.01, Fig. 1B), indicating that the ions released from nanoparticles cannot totally explain the toxicity. Hence, we believe that the toxicity of AgNPs is multifactorial, where size, shape, surface functionalization and the released ions could all be important. To determine how much each of these parameters contributes to AgNP toxicity, more experimental work is needed.
4.3 The defense response against the adverse effects of AgNPs in E. vannus
In addition to the active efflux of silver, our transcriptomic and microRNAomic data indicated that exposure of E. vannus to AgNPs elicited a temporally dynamic defense response against ROS, directing the cell to a detoxifying reaction and repair. Genes of antioxidant molecules and enzymes such as glutathione synthetase, thioredoxin peroxidase, peroxiredoxin, Gr, GPx, GST, BAX inhibitor 1, SOD, DnaJ and L-ascorbate peroxidase were significantly regulated at different time-point comparisons (Fig. 3B and 6). Among of them, glutathione synthetase, GPx and GST were up-regulated at T1 vs. T0 and T12 vs. T0, and thioredoxin peroxidase, SOD-Fe and BAX inhibitor 1 were up-regulated at T12 vs. T0, while peroxiredoxin was up-regulated at T12 vs. T0 and T12 vs. T1. In contrast, SOD-Cu–Zn and DnaJ-1 were down-regulated at T12 vs. T0, while Gr and L-ascorbate peroxidase were down-regulated at T12 vs. T0 and T12 vs. T1. Interestingly, the GST gene, which was involved in glutathione metabolism, was co-regulated by eva-miR-N3 and eva-miR-N15 (Fig. 4). Usually, antioxidant enzymes are produced to protect the cells from damage by the excess production of ROS. GPx uses glutathione to reduce hydrogen peroxide into water and lipid hydroperoxides into their corresponding alcohols,69 while Gr catalyzes the reduction of glutathione disulfide (GSSG) to the sulfhydryl form glutathione (GSH), which is a critical molecule in resisting oxidative stress and maintaining the reducing environment of the cell.70 We measured the enzyme activity of GPx and Gr and found that the regulation of these enzymes was consistent with the transcriptome response, confirming that these enzymes contributed to the cell defense oxidative stress. The transcriptomic and microRNAomic analysis therefore indicated that AgNPs induce an oxidative stress response, likely triggering detoxification and repair. However, when the induced oxidative stress exceeds the scavenging ability of the antioxidant enzymes, cellular damage would occur. Chen et al.71 described the stress responses of Phanerochaete chrysosporium under cadmium exposure and considered the increase of exposure time and concentration part as an exhaustion stage in which the defense systems were overloaded, resulting in chronic damage and cell death. In our results, only a few DEGs were shared by the three compartment groups, while most of the DEGs were unique at each time point (Fig. 2C), suggesting that E. vannus regulated its genes to cope with AgNP stress with a temporal dynamics. Further evidence is that the number of DEGs in E. vannus at 12 h of exposure was significantly greater than that at one h of exposure. We should note that because our study did not include a time-matched control, the difference observed here might be due to natural temporal dynamics of gene expression or a response to the change of experimental conditions over time. However, many of the regulated genes are related to stress responses, making it more likely that the differential expression of the genes resulted from AgNP stress. Under this scenario, it is probable that the early response (one hour) at the transcriptome level is not sufficient to allow E. vannus to cope with the toxicity, and more genes responded to the toxicity at prolonged exposure (12 hour). In fact, all the parameters detected in this study showed a temporally progressive manner, consistent with a time-dependent toxicity aggravation of AgNPs.Additionally, the expression of microRNA also showed temporal dynamics (Tables 1 and S4†). As the general function of a microRNA is binding to mRNA to yield mRNA degradation or translation repression,19 the up-regulation of these microRNAs would presumably lead to inhibition of their target genes' expression and encoded biological functions. This suggests that E. vannus possesses a broad post-transcriptional regulation through a microRNA–mRNA regulatory network to cope with AgNPs' toxicity. Previous reports had provided solid evidence that protists possess a microRNA regulatory network comparable with multicellular organisms.72 To our knowledge, the present study is the first to investigate the global microRNA expression patterns in protists under AgNP stress. In total, we have identified 190 mature microRNAs that meet the criteria of microRNA recognition and all of them are novel microRNAs that have not been reported, suggesting that more experimental work is needed to verify the target genes and to understand the biological processes these microRNAs are involved in.
5. Conclusion
The physiological, transcriptomic and microRNAomic profiles obtained in this study provide insights into the mechanisms of toxicity, detoxification, and repair at different biological levels in E. vannus on exposure to AgNPs. From the detailed analysis of the perturbations of the cell's functional networks, we were able to characterize AgNPs' uptake and toxicity pathways for AgNPs in E. vannus. The initiating event is E. vannus uptake of AgNPs through endocytic pathways and uptake of Ag+ by copper transporters, as our data suggest. The internalization of sliver elicits several effects, including disruption of the function of mitochondria, damage of proteins and DNA, and possibly cell cycle arrest. The damage is likely to have mainly resulted from increased ROS production. As a defense mechanism against oxidative stress, the protist elicited an antioxidant response. Another defense mechanism is active efflux of AgNPs and Ag+ possibly via ABC transporters and copper transporters, respectively. Our data reveal a broad microRNA–mRNA regulatory network in E. vannus to cope with AgNPs' toxicity. Finally, this study underscores the importance of time serial observations and analysis (including time-match sampling, which is unfortunately lacking in this study) to capture the dynamics of progressive response to and detoxification from AgNPs' toxicity in this protist.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank Xinguo Shi, Xiu Gao, and Jing Fu from the State Key Laboratory of Marine Environmental Science, Xiamen University, China for experimental help. This study was supported by the Fundamental Research Funds for the Central Universities (20720160115) and the National Marine Public Welfare Project of China (201505034).References
- C. Marambio-Jones and E. M. V. Hoek, A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment, J. Nanopart. Res., 2010, 12, 1531–1551 CrossRef CAS.
- N. Durán, M. Durán, M. B. de Jesus, A. B. Seabra, W. J. Fávaro and G. Nakazato, Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity, J. Nanomed. Nanotechnol., 2016, 12, 789–799 CrossRef PubMed.
- M. N. Nadagouda, T. F. Speth and R. S. Varma, Microwave-assisted green synthesis of silver nanostructures, Acc. Chem. Res., 2011, 44, 469–478 CrossRef CAS PubMed.
- T. Benn, B. Cavanagh, K. Hristovski, J. D. Posner and P. Westerhoff, The release of nanosilver from consumer products used in the home, J. Environ. Qual., 2010, 39, 1875–1882 CrossRef CAS PubMed.
- N. C. Mueller and B. Nowack, Exposure modeling of engineered nanoparticles in the environment, Environ. Sci. Technol., 2008, 42, 4447–4453 CrossRef CAS PubMed.
- G. Asmonaite, S. Boyer, K. B. Souza, B. Wassmur and J. Sturve, Behavioural toxicity assessment of silver ions and nanoparticles on zebrafish using a locomotion profiling approach, Aquat. Toxicol., 2016, 173, 143–153 CrossRef CAS PubMed.
- S. M. Hoheisel, S. Diamond and D. Mount, Comparison of nanosilver and ionic silver toxicity in Daphnia magna and Pimephales promelas, Environ. Toxicol. Chem., 2012, 31, 2557–2563 CrossRef CAS PubMed.
- Y. Wu and Q. Zhou, Dose- and time-related changes in aerobic metabolism, chorionic disruption, and oxidative stress in embryonic medaka (Oryzias latipes): Underlying mechanisms for silver nanoparticle developmental toxicity, Aquat. Toxicol., 2012, 124–125, 238–246 CrossRef CAS PubMed.
- Y. Li, T. Qin, T. Ingle, J. Yan, W. He, J. J. Yin and T. Chen, Differential genotoxicity mechanisms of silver nanoparticles and silver ions, Arch. Toxicol., 2017, 91, 509–519 CrossRef CAS PubMed.
- A. Ivask, A. Elbadawy, C. Kaweeteerawat, D. Boren, H. Fischer, Z. Ji, C. H. Chang, R. Liu, T. Tolaymat and D. Telesca, Toxicity mechanisms in Escherichia coli vary for silver nanoparticles and differ from ionic silver, ACS Nano, 2014, 8, 374–386 CrossRef CAS PubMed.
- J. Shi, X. Sun, Y. Lin, X. Zou, Z. Li, Y. Liao, M. Du and H. Zhang, Endothelial cell injury and dysfunction induced by silver nanoparticles through oxidative stress via IKK/NF-κB pathways, Biomaterials, 2014, 35, 6657–6666 CrossRef CAS PubMed.
- Z. M. Xiu, Q. B. Zhang, H. L. Puppala, V. L. Colvin and P. J. Alvarez, Negligible particle-specific antibacterial activity of silver nanoparticles, Nano Lett., 2012, 12, 4271–4275 CrossRef CAS PubMed.
- D. McShan, P. C. Ray and H. Yu, Molecular toxicity mechanism of nanosilver, J. Food Drug Anal., 2014, 22, 116–127 CrossRef CAS PubMed.
- A. Nel, T. Xia, L. Mädler and N. Li, Toxic potential of materials at the nanolevel, Science, 2006, 311, 622–627 CrossRef CAS PubMed.
- R. Sekine, K. L. Moore, M. Matzke, P. Vallotton, H. Jiang, G. M. Hughes, J. K. Kirby, E. Donner, C. R. M. Grovenor, C. Svendsen and E. Lombi, Complementary imaging of silver nanoparticle interactions with green algae: dark-field microscopy, electron microscopy, and nanoscale secondary ion mass spectrometry, ACS Nano, 2017, 11, 10894–10902 CrossRef CAS PubMed.
- J. C. Marioni, C. E. Mason, S. M. Mane, M. Stephens and Y. Gilad, RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays, Genome Res., 2008, 18, 1509–1517 CrossRef CAS PubMed.
- D. P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 2004, 116, 281–297 CrossRef CAS PubMed.
- B. P. Lewis, C. B. Burge and D. P. Bartel, Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets, Cell, 2005, 120, 15–20 CrossRef CAS PubMed.
- D. P. Bartel, MicroRNAs: target recognition and regulatory functions, Cell, 2009, 136, 215–233 CrossRef CAS PubMed.
- H. C. Poynton, J. M. Lazorchak, C. A. Impellitteri, B. J. Blalock, K. Rogers, H. J. Allen, A. Loguinov, J. L. Heckman and S. Govindasmawy, Toxicogenomic responses of nanotoxicity in Daphnia magna exposed to silver nitrate and coated silver nanoparticles, Environ. Sci. Technol., 2012, 46, 6288–6296 CrossRef CAS PubMed.
- S. I. L. Gomes, C. P. Roca, J. J. Scott-Fordsmand and M. J. B. Amorim, High-throughput transcriptomics reveals uniquely affected pathways: AgNPs, PVP-coated AgNPs and Ag NM300K case studies, Environ. Sci.: Nano, 2017, 4, 929–937 RSC.
- M. Novo, E. Lahive, M. Díez-Ortiz, M. Matzke, A. J. Morgan, D. J. Spurgeon, C. Svendsen and P. Kille, Different routes, same pathways: Molecular mechanisms under silver ion and nanoparticle exposures in the soil sentinel Eisenia fetida, Environ. Pollut., 2015, 205, 385–393 CrossRef CAS PubMed.
- S. Pillai, R. Behra, H. Nestler, M. J. Suter, L. Sigg and K. Schirmer, Linking toxicity and adaptive responses across the transcriptome, proteome, and phenotype of Chlamydomonas reinhardtii exposed to silver, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3490–3495 CrossRef CAS PubMed.
- R. Foldbjerg, E. S. Irving, Y. Hayashi, D. S. Sutherland, K. Thorsen, H. Autrup and C. Beer, Global gene expression profiling of human lung epithelial cells after exposure to nanosilver, Toxicol. Sci., 2012, 130, 145–157 CrossRef CAS PubMed.
- J. Li, L. Zhou, X. Lin, Z. Yi and K. A. Al-Rasheid, Characterizing dose–responses of catalase to nitrofurazone exposure in model ciliated protozoan Euplotes vannus for ecotoxicity assessment: Enzyme activity and mRNA expression, Ecotoxicol. Environ. Saf., 2014, 100, 294–302 CrossRef CAS PubMed.
- A. Gomiero, A. Dagnino, C. Nasci and A. Viarengo, The use of protozoa in ecotoxicology: application of multiple endpoint tests of the ciliate E. crassus for the evaluation of sediment quality in coastal marine ecosystems, Sci. Total Environ., 2013, 442, 534–544 CrossRef CAS PubMed.
- F. Trielli, A. Amaroli, F. Sifredi, B. Marchi, C. Falugi and M. U. D. Corrado, Effects of xenobiotic compounds on the cell activities of Euplotes crassus, a single-cell eukaryotic test organism for the study of the pollution of marine sediments, Aquat. Toxicol., 2007, 83, 272–283 CrossRef CAS PubMed.
- H. Xu, M. Zhu, Y. Jiang, S. Gao, G.-S. Min and K. A. S. Al-Rasheid, Population dynamics of marine ciliate Euplotes vannus (Protozoa, Ciliophora) in different artificial seawaters, Chin. J. Oceanol. Limnol., 2011, 29, 109–117 CrossRef.
- P. Madoni and M. G. Romeo, Acute toxicity of heavy metals towards freshwater ciliated protists, Environ. Pollut., 2006, 141, 1–7 CrossRef CAS PubMed.
- D. J. Finney, Probit analysis, Cambridge Univ. Press, Cambridge, 3rd edn, 1971, p. 333 Search PubMed.
- M. Mraz, K. Malinova, J. Mayer and S. Pospisilova, MicroRNA isolation and stability in stored RNA samples, Biochem. Biophys. Res. Commun., 2009, 390, 1–4 CrossRef CAS PubMed.
- S. L. Tian, G. Y. Zhou and R. Q. Li, Method and device for processing sequencing data, CN102831330A, 2012.
- M. D. Robinson, D. J. McCarthy and G. K. Smyth, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 2010, 26, 139–140 CrossRef CAS PubMed.
- E. Bonnet, J. Wuyts, P. Rouzé and d. P. Y. Van, Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences, Bioinformatics, 2004, 20, 2911–2917 CrossRef CAS PubMed.
- L. D. Aicher, S. L. Lederer, E. R. Rosenzweig, W. Kathie-Anne, Y. Li, X. Peng, P. Sean and M. G. Katze, Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers, BMC Genomics, 2009, 10, 373 CrossRef PubMed.
- J. C. Huang, T. Babak, T. W. Corson, G. Chua, S. Khan, B. L. Gallie, T. R. Hughes, B. J. Blencowe, B. J. Frey and Q. D. Morris, Using expression profiling data to identify human microRNA targets, Nat. Methods, 2009, 4, 1045 CrossRef PubMed.
- Y. Yadong, L. Xiaoying and D. Fei, microRNA regulatory mechanism by which PLLA aligned nanofibers influence PC12 cell differentiation, J. Neural. Eng., 2015, 12, 046010 CrossRef PubMed.
- A. J. Enright, B. John, U. Gaul, T. Tuschl, C. Sander and D. S. Marks, MicroRNA targets in Drosophila, Genome Biol., 2003, 5, R1 CrossRef PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 2009, 4, 44 CrossRef CAS PubMed.
- N. Rosic, E. Y. S. Ling, C.-K. K. Chan, H. C. Lee, P. Kaniewska, D. Edwards, S. Dove and O. Hoegh-Guldberg, Unfolding the secrets of coral–algal symbiosis, ISME J., 2014, 9, 844–856 CrossRef PubMed.
- C. Chen, R. Xia, H. Chen and Y. He, TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface, bioRxiv, 2018 DOI:10.1101/289660.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res., 2003, 13, 2498–2504 CrossRef CAS PubMed.
- P. S. Bäuerlein, E. Emke, P. Tromp, J. A. M. H. Hofman, A. Carboni, F. Schooneman, P. de Voogt and A. P. van Wezel, Is there evidence for man-made nanoparticles in the Dutch environment?, Sci. Total Environ., 2017, 576, 273–283 CrossRef PubMed.
- F. Gottschalk, T. Sun and B. Nowack, Environmental concentrations of engineered nanomaterials: review of modeling and analytical studies, Environ. Pollut., 2013, 181, 287–300 CrossRef CAS PubMed.
- O. Bondarenko, K. Juganson, A. Ivask, K. Kasemets, M. Mortimer and A. Kahru, Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review, Arch. Toxicol., 2013, 87, 1181–1200 CrossRef CAS PubMed.
- S. Leclerc and K. J. Wilkinson, Bioaccumulation of nanosilver by Chlamydomonas reinhardtii—nanoparticle or the free ion?, Environ. Sci. Technol., 2014, 48, 358–364 CrossRef CAS PubMed.
- M. Mortimer, A. Kahru and V. I. Slaveykova, Uptake, localization and clearance of quantum dots in ciliated protozoa Tetrahymena thermophila, Environ. Pollut., 2014, 190, 58–64 CrossRef CAS PubMed.
- S. T. Stern, P. P. Adiseshaiah and R. M. Crist, Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity, Part. Fibre Toxicol., 2012, 9, 20 CrossRef CAS PubMed.
- R. Behra, L. Sigg, M. J. Clift, F. Herzog, M. Minghetti, B. Johnston, A. Petrifink and B. Rothenrutishauser, Bioavailability of silver nanoparticles and ions: from a chemical and biochemical perspective, J. R. Soc., Interface, 2013, 10, 20130396 CrossRef PubMed.
- A. R. Gliga, S. Skoglund, I. Odnevall Wallinder, B. Fadeel and H. L. Karlsson, Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- M. Solioz and A. Odermatt, Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae, J. Biol. Chem., 1995, 270, 9217–9221 CrossRef CAS PubMed.
- I. B. Holland and M. A. Blight, ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans, J. Mol. Biol., 1999, 293, 381–399 CrossRef CAS PubMed.
- H. Kim, B. Yim, J. Kim, H. Kim and Y.-M. Lee, Molecular characterization of ABC transporters in marine ciliate, Euplotes crassus: Identification and response to cadmium and benzo[a]pyrene, Mar. Pollut. Bull., 2017, 124, 725–735 CrossRef CAS PubMed.
- A. C. Hart, M. S. Schwartz, J. L. Benci, D. S. Selote, A. K. Sharma, A. G. Y. Chen, H. Dang, H. Fares and O. K. Vatamaniuk, Detoxification of multiple heavy metals by a half-molecule ABC transporter, HMT-1, and coelomocytes of Caenorhabditis elegans, PLoS One, 2010, 5, e9564 CrossRef PubMed.
- Y. M. Mamnun, C. Schüller and K. Kuchler, Expression regulation of the yeast PDR5 ATP-binding cassette (ABC) transporter suggests a role in cellular detoxification during the exponential growth phase, FEBS Lett., 2004, 559, 111–117 CrossRef CAS PubMed.
- I. T. Paulsen and S. M. Jr, A novel family of ubiquitous heavy metal ion transport proteins, J. Membr. Biol., 1997, 156, 99–103 CrossRef CAS PubMed.
- Y. Ren, B. Ni, X. Fan and F. Gu, a study on silver nanoparticles cytotoxicity of Euplotes aediculatus (Ciliophora, Hypotrichida), Zhongguo Haiyang Daxue Xuebao, Ziran Kexueban, 2016, 46, 57–64 Search PubMed , (in Chinese).
- X. Yang, A. P. Gondikas, S. M. Marinakos, M. Auffan, J. Liu, H. Hsu-Kim and J. N. Meyer, Mechanism of silver nanoparticle toxicity Is dependent on dissolved silver and surface coating in Caenorhabditis elegans, Environ. Sci. Technol., 2012, 46, 1119–1127 CrossRef CAS PubMed.
- J. Liu and R. H. Hurt, Ion release kinetics and particle persistence in aqueous nano-silver colloids, Environ. Sci. Technol., 2010, 44, 2169–2175 CrossRef CAS PubMed.
- E. J. Petersen and B. C. Nelson, Mechanisms and measurements of nanomaterial-induced oxidative damage to DNA, Anal. Bioanal. Chem., 2010, 398, 613–650 CrossRef CAS PubMed.
- B. Das, S. K. Dash, D. Mandal, T. Ghosh, S. Chattopadhyay, S. Tripathy, S. Das, S. K. Dey, D. Das and S. Roy, Green synthesized silver nanoparticles destroy multidrug resistant bacteria via reactive oxygen species mediated membrane damage, Arabian J. Chem., 2017, 10, 862–876 CrossRef CAS.
- M. J. Piao, K. A. Kang, I. K. Lee, H. S. Kim, S. Kim, J. Y. Choi, J. Choi and J. W. Hyun, Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis, Toxicol. Lett., 2011, 201, 92–100 CrossRef CAS PubMed.
- R. Foldbjerg, P. Olesen, M. Hougaard, D. A. Dang, H. J. Hoffmann and H. Autrup, PVP-coated silver nanoparticles and silver ions induce reactive oxygen species, apoptosis and necrosis in THP-1 monocytes, Toxicol. Lett., 2009, 190, 156–162 CrossRef CAS PubMed.
- J. Ma, X. Lü and Y. Huang, Genomic analysis of cytotoxicity response to nanosilver in human dermal fibroblasts, J. Biomed. Nanotechnol., 2011, 7, 263 CrossRef CAS PubMed.
- M. Ahamed, M. Karns, M. Goodson, J. Rowe, S. M. Hussain, J. J. Schlager and Y. Hong, DNA damage response to different surface chemistry of silver nanoparticles in mammalian cells, Toxicol. Appl. Pharmacol., 2008, 233, 404–410 CrossRef CAS PubMed.
- H. J. Eom and J. Choi, p38 MAPK activation, DNA damage, cell cycle arrest and apoptosis as mechanisms of toxicity of silver nanoparticles in Jurkat T cells, Environ. Sci. Technol., 2010, 44, 8337–8342 CrossRef CAS PubMed.
- Y. H. Choi, L. Zhang, W. H. Lee and K. Y. Park, Genistein-induced G2/M arrest is associated with the inhibition of cyclin B1 and the induction of p21 in human breast carcinoma cells, Int. J. Oncol., 1998, 13, 391–396 CAS.
- R. C. Bicho, T. Ribeiro, N. P. Rodrigues, J. J. Scott-Fordsmand and M. J. B. Amorim, Effects of Ag nanomaterials (NM300K) and Ag salt (AgNO3) can be discriminated in a full life cycle long term test with Enchytraeus crypticus, J. Hazard. Mater., 2016, 318, 608–614 CrossRef CAS PubMed.
- S. S. Gill and N. Tuteja, Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants, Plant Physiol. Biochem., 2010, 48, 909–930 CrossRef CAS PubMed.
- M. Deponte, Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 3217–3266 CrossRef CAS PubMed.
- A. Chen, G. Zeng, G. Chen, L. Liu, C. Shang, X. Hu, L. Lu, M. Chen, Y. Zhou and Q. Zhang, Plasma membrane behavior, oxidative damage, and defense mechanism in Phanerochaete chrysosporium under cadmium stress, Process Biochem., 2014, 49, 589–598 CrossRef CAS.
- W. C. Lin, S. C. Li, W. C. Lin, J. W. Shin, S. N. Hu, X. M. Yu, T. Y. Huang, S. C. Chen, H. C. Chen, S. J. Chen, P. J. Huang, R. R. C. Gan, C. H. Chiu and P. Tang, Identification of microRNA in the protist Trichomonas vaginalis, Genomics, 2009, 93, 487–493 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8en00924d |
| This journal is © The Royal Society of Chemistry 2018 |