Open Access Article
Open Access ArticleDual polarity MALDI imaging mass spectrometry on the same pixel points reveals spatial lipid localizations at high-spatial resolutions in rat small intestine†
Ibrahim
Kaya
 ab,
Eva
Jennische
c,
Stefan
Lange
c and
Per
Malmberg
ab,
Eva
Jennische
c,
Stefan
Lange
c and
Per
Malmberg
 *bd
*bd
aDepartment of Chemistry and Molecular Biology, University of Gothenburg, 412 96, Sweden
bThe Gothenburg Imaging Mass Spectrometry (Go:IMS) Platform, University of Gothenburg and Chalmers University of Technology, Gothenburg 412 96, Sweden. E-mail: malmper@chalmers.se
cInstitute of Biomedicine, Gothenburg 413 90, Sweden
dDepartment of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg 412 96, Sweden
First published on 3rd May 2018
Abstract
Sensitive laser desorption/ionization obtained via a sublimation-coated 1,5-diaminonaphthalene (1,5-DAN) matrix allowed dual polarity MALDI-IMS analysis on the same pixel points across the jejunal mucosal region in rat small intestine which yielded high-spatial-resolution (10 μm) ion images of several lipid species correlated with the same histological features.
Introduction
The ongoing epidemic of obesity and metabolic diseases has increased the importance of intestinal lipidomics research. The small intestine has important roles in the integrated regulation of lipid homeostasis including glucose metabolism, energy utilization, and inflammation.1 It has been widely reported that lipids are taken up by enterocytes (intestinal absorptive cells) and packaged as chylomicrons (lipoprotein particles) for export into the lymphatic system through exocytosis which is regulated by several biochemical and genetic regulatory pathways along with the role of nuclear hormone receptors and circadian rhythms in continuation of ordinary lipid homeostasis.2 Several lipid species including glycerides, fatty acids, glycerophospholipids, cardiolipins and bile acids were shown to play important roles in intestinal lipid absorption, digestion and the regulation of homeostatic processes.1,2 Therefore, knowledge of the spatial localization of individual lipid species in the small intestine would provide a basis to construe their physiological and/or pathophysiological roles in situ.Chemical imaging techniques, such as immunohistochemistry and fluorescent dye staining, are typically used for the detection and localization of lipids. Staining with Oil Red O, 1,6-diphenyl-1,3,5-hexatriene (DPH) or BODIPY 493/503 reveals the localization of only the alkyl chains of phospholipids and/or neutral lipids.3 Moreover, the ability to image individual lipid species in tissues has been limited as there is no specific antibody for every single lipid species. Therefore, there is a need for an effective technique to reveal spatial distribution profiles for specific lipid molecular species in tissue.
Imaging mass spectrometry (IMS) is a powerful technique to probe spatial molecular distributions within tissues in a single experiment.4 Matrix-assisted laser desorption/ionization (MALDI) imaging MS has been demonstrated to be an effective method for probing lipids in different tissue sections.5,6 For MALDI-IMS, a tissue slice coated with a desorption/ionization enhancing “matrix” molecule is raster scanned by energetic laser pulses and the resulting spectra from each pixel point are used to generate ion distribution maps of biomolecules. The sublimation matrix-coating strategy was previously demonstrated to be a good approach for high-spatial resolution MALDI-IMS analysis of lipids.7 The sublimation-coated 1,5-diaminonaphthalene (1,5-DAN) matrix enabled ionization of lipid molecules via laser desorption/ionization in dual polarity on different tissue sections.8 This approach was particularly efficient in negative ionization polarity using relatively lower laser pulse energies compared to commonly used acidic matrix molecules.8,9 On the other hand, multiple laser desorption/ionization analyses can be made on the same tissue sections using matrix molecules assisting the ionization in dual polarity which enhances the coverage of the lipid molecular information correlated with the same histological or histopathological features.8,10–12
In this communication, we report high-spatial resolution (10 μm), dual polarity MALDI-IMS via 1,5-DAN sublimation-coating not only on the same tissue sections but also on the same pixel points to reveal spatial distributions of lipids across the rat small intestinal mucosa. Lipid species ionized in dual polarity including fatty acids, lysophospholipids, bile acids, phospholipids, sphingolipids, cardiolipins and cholesterol sulfate were detected on the same pixel points and the resulting ion images were correlated with the intestinal histological features including enterocytes, the lamina propria, the lumen and even the mucous goblet cell layer. MS/MS using laser-induced fragmentation was subsequently employed on the same tissue section after MALDI-IMS analysis which provided tandem MS information useful for the structural elucidation of several lipid species. This methodology can be a powerful approach for probing local roles of lipids in small intestinal lipid absorption, digestion and the lipid pathology of metabolic and infectious diseases.
Experimental section
Chemicals and reagents
All chemicals for matrix and solvent preparation were of pro-analysis grade and obtained from Sigma-Aldrich (Stockholm, Sweden), unless otherwise specified. The TissueTek optimal cutting temperature (OCT) compound was purchased from Sakura Finetek (AJ Alphen aan den Rijn, The Netherlands). Deionized H2O was obtained from a MilliQ purification system (Merck Millipore, Darmstadt, Germany).Animals
Male Sprague Dawley rats weighing 220–260 g were used. The rats were housed with a 12 h light cycle at standardized humidity and temperature, and had access to pelleted feed and water ad libitum. All animal procedures were approved by the animal experimental ethical committee in Gothenburg, and performed in accordance with local and federal guidelines (EU86/609/EEC).Tissue sampling and sectioning
The animals (n = 3) were deeply anesthetized with isoflurane and sacrificed by opening of the thorax and removal of the heart. The proximal part of the jejunum was quickly, within 3 min, dissected out and frozen in liquid nitrogen. The samples were kept at −80 °C until sectioning. Frozen tissue sections (10 μm thick) were cut in a cryostat microtome (Leica CM 1520, Leica Biosystems, Nussloch, Germany) at −20 °C, and collected on indium tin-oxide (ITO) coated glass slides (Bruker Daltonics, Bremen, Germany) and stored at −80 °C.Sublimation for matrix deposition
Prior to matrix deposition by sublimation, tissue sections were thawed in a desiccator for ∼30 minutes under reduced pressure (SpeedVac, Eppendorf, Hamburg, Germany). The sections were scanned in a slide scanner (PathScan Enabler IV, Electron Microscopy Sciences, Hatfield, PA, USA) to correlate the brightfield images with Flex Imaging software v3.0 (Bruker Daltonics). Matrix deposition was carried out, as previously described,9,13 using a vacuum sublimation apparatus (Ace Glass, New York, USA) comprising an inner flat top and an outer bottom attached to each other by an O-ring-sealed flange. The chamber was connected to a diaphragm pump attached to a digital vacuum gauge controller and placed in a heated sand bath (SiO2, 50–70 mesh particle size, Sigma-Aldrich) on a hot plate (C-MAG HP 4, IKA Werke GmbH & Co. KG, Staufen, Germany) connected to an electronic contact thermometer (ETS-D5, IKA Werke GmbH & Co. KG, Staufen, Germany). Sublimation was performed by the following steps. First, vacuum-dried (∼20 minutes) ITO-glass slides with the thaw-mounted mouse brain tissues were attached to the flat top of the chamber using double-sided heat conductive copper tape (12 mm × 16.5 m copper, 3 M, United States). Then, ∼300 mg of 1,5-diaminonaphthalene (1,5-DAN) matrix powder was spread evenly on the outer bottom of the sublimation chamber, which was then attached to the top using the O-ring seal. A vacuum of 0.8 mbar was provided by a membrane pump and the system was allowed to equilibrate under vacuum for a period of ∼20 minutes. The cooler was filled with ice slush (≥0 °C) for condensation of the matrix on the sample slides. The process was halted by removing the sublimation apparatus from the sand bath and closing the vacuum valve. The sublimation apparatus was allowed to slowly re-equilibrate to atmospheric pressure before removal of the sample plate. The amount of deposited matrix on ITO glass was determined using a high-precision scale (AX224, Sartorius, Göttingen, Germany) weighing the amount before and after the sublimation experiments. The sublimation protocol was optimized with respect to the temperature, deposition time and total amount of deposited matrix to obtain the best detection efficiency for lipids in rat small intestine tissue sections. With this setup, the optimum matrix layer was found to be ∼120 μg cm−2 for optimum lipid ionization and high signals. For that, we used optimized sublimation conditions: 20 minutes at a temperature of ∼130 °C under a stable vacuum of ∼0.8 mbar.MALDI-IMS and MALDI-MS/MS analysis
Imaging MS analysis of tissue sections was performed using a MALDI TOF/TOF UltrafleXtreme mass spectrometer equipped with a SmartBeam II Nd:YAG/355 nm laser operating at 1 kHz providing a laser spot diameter down to ∼10 μm for the ‘minimum’ focus setting (Bruker Daltonics). Since the laser pulse energy was previously demonstrated to be crucial for enhanced MALDI-IMS analysis using the 1,5-DAN matrix compound,9 we provide detailed information about the laser pulse energy settings as previously indicated for lipid analysis in our previous communication;9 the global laser attenuator setting was kept stable at 10% throughout all the experiments and the laser focus was set to “minimum”. Attenuator offset and attenuator range settings were 40% and 10%, respectively for the minimum laser focus. For dual polarity lipid analysis, imaging data acquisitions were performed in reflective ion mode under optimized delayed extraction conditions over a mass range of 0–2000 Da. First, IMS data were acquired with 10 laser shots per pixel point with a source accelerating voltage of 20 kV in negative polarity which was followed by 30 laser shots per pixel point in positive polarity with a source accelerating voltage of 25 kV. The detector gain value was kept stable at 2926 V for both ionization modes. A mass resolution of M/ΔM—20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 was achieved in the mass window of lipids (i.e., 650–1000 Da). Calibration was carried out using peptide calibration standard I (Bruker Daltonics spotted next to the intestinal tissue sections and analyzed with MALDI-MS using the same experimental parameters used for MALDI-IMS. For sensitive lipid ionization using relatively low laser-pulse energies, laser mirrors, laser lenses, mirror plates and potential plates of the instrument should be cleaned according to the manufacturer's maintenance manual (Bruker Daltonics, Bremen, Germany).
000 was achieved in the mass window of lipids (i.e., 650–1000 Da). Calibration was carried out using peptide calibration standard I (Bruker Daltonics spotted next to the intestinal tissue sections and analyzed with MALDI-MS using the same experimental parameters used for MALDI-IMS. For sensitive lipid ionization using relatively low laser-pulse energies, laser mirrors, laser lenses, mirror plates and potential plates of the instrument should be cleaned according to the manufacturer's maintenance manual (Bruker Daltonics, Bremen, Germany).
Image data were reconstructed and visualized using Flex Imaging v3.0 (Bruker Daltonics). Lipid classifications were performed by examining MS/MS spectra obtained in LID-LIFT-TOF/TOF mode14 and mass accuracy data from the LIPID MAPS database (Nature Lipidomics Gateway, www.lipidmaps.org) and previous results obtained on intestinal tissue sections.15 Briefly, the LIFT TOF/TOF mass spectrometer (Ultraflex TOF/TOF, Bruker) consists of a gridless MALDI ion source with delayed extraction (DE) electronics, a high-resolution timed ion selector (TIS), a “LIFT” device for raising the potential energy of the ions, a further velocity focusing stage with subsequent post-acceleration, a post lift metastable suppressor (PLMS), a gridless space-angle and energy focusing reflector, and fast ion detectors for the linear and reflector mode. After MALDI-IMS, acquisition conditions in LIFT-TOF/TOF are modified to generate high fragment yields of lipid molecules at their spatial locations in the tissue sections. The laser power was increased to provide a high number of precursor ions and a low initial accelerating voltage of 8 kV is used to provide a long flight time (10–20 μs) during which fragmentation occurs. The selected precursor and fragment ions are post-accelerated by an additional 19 kV and mass analyzed in one spectrum. Then, the selected precursor ion and its fragments were selected with the timed ion selector (TIS) which deflects all ion families except the one under investigation by switching the gate voltage off while the selected ions pass through.
Post-MALDI-IMS H&E staining
The sections were immersed in methanol for 1 min in order to remove remnants of the 1,5-DAN matrix. After air drying, the sections were immersed in Harris hematoxylin (Histolab Products AB, Gothenburg, Sweden) for 20 seconds, rinsed in water and immersed in 0.2% eosin in ethanol (Histolab Products AB, Gothenburg, Sweden) for 30 seconds. The stained sections were rapidly dehydrated in ethanol and mounted in DPX mounting solution (BDH chemicals). The sections were scanned using a slide scanner (NanoZoomer SQ, Hamamatsu Photonics, Japan) and the areas corresponding to the area used for MALDI imaging MS were captured.Results and discussion
High-spatial resolution MALDI-IMS reveals high-throughput small intestinal lipid molecular information
For a successful MALDI-IMS analysis, a sensitive matrix molecule for the targeted analytes applied with a compatible matrix application strategy is needed to achieve enhanced laser desorption/ionization and high-spatial resolution.8,9It was previously demonstrated that 1,5-diaminonaphthalene (1,5-DAN) can assist sensitive laser desorption/ionization of a wide range of lipid molecules in dual polarity including sphingolipids and phospholipids8,13 along with relatively small molecules such as lysophospholipids16 and certain metabolites.17 Regarding MALDI-IMS, sublimation-based matrix coating provides uniform, purified, small crystals of matrix molecules on the tissue sections7 which hinders heat dissipation on a laser irradiated raster spot18 and favors sensitive desorption/ionization and high spatial resolution analysis.7,9,19 For optimum lipid ionization using 1,5-DAN via the sublimation approach, the thickness of the matrix coating and the laser pulse energy were shown to be crucial.9 Compared to commonly used acidic matrix molecules, relatively lower laser pulse energies on a thin-coated 1,5-DAN layer (120 μg cm−2)9,16 provided an efficient ionization of lipids and suppression of matrix clusters resulting in enhanced lipid signals in a wide spectral range at high spatial resolutions.8,9
The low ionization energy and strong UV absorption of 1,5-DAN can rationalize its relatively higher sensitivity for lipids. In detail, the low ionization energy of 1,5-DAN likely favors the formation of odd-electron molecular ions such as M+˙ and M−˙ contrary to what is usually observed by laser irradiation of other MALDI matrices (with the sole formation of the MH+ and MH− ions of the matrix).20 The radical nature of odd-molecular ions of 1,5-DAN formed by laser irradiation might favor quick reactions with the analyte molecules in the laser desorption plumes and provide sensitive lipid ionization. However, these radicals can also react with each other and neutral 1,5-DAN molecules in the gas phase resulting in a variety of oligomeric matrix ions20,21 which can interfere with small-molecule analyte signals in the spectra and hinder lipid ionization.9 Previously, it was demonstrated that the initial few low-energy laser shots provide an efficient ionization of lipid molecules and minimize the matrix-derived signals (particularly in negative ion mode) on a 1,5-DAN coated mice brain tissue section.9 This is possibly due to the optimum matrix/analyte ratio and the desorption/ionization process in the initial laser pulses which can provide effective lipid ionization and optimum matrix ion suppression.
We applied dual polarity MALDI-IMS in the mucosal area of a small intestinal tissue section to reveal the lipid molecular composition. Intense and diverse lipid signals were observed in negative ion mode (Fig. 1a) compared to positive ion mode (Fig. 1b) which is in line with the previous results on brain tissue sections.8,9 This can be due to the basic nature of the 1,5-DAN molecule and its reductive features.20 Several lipid species including fatty acids, lysophospholipids, bile acids, sphingolipids, phospholipids and cholesterol sulfate were detected in negative ion mode whereas lysophosphatidylcholines and phosphatidylcholines were observed in positive ion mode in a mass range of 200–1000 Da (Fig. 1). Further, cardiolipins and lipid dimers were observed in a high mass range (1300–1700 Da) in negative and positive ion modes, respectively (see ESI†). Matrix-derived oligomeric ion signals were few and they didn't interfere with the signals from certain fatty acids or the low molecular weight lysophospholipids, bile acids and cholesterol sulfate in negative ion mode (Fig. 1a).
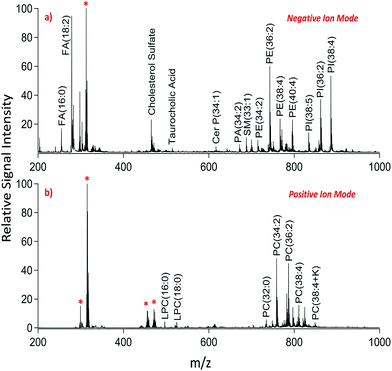 | ||
| Fig. 1 Dual polarity MALDI-MS spectra obtained from jejunal mucosal regions of the rat small intestinal tissue section. Total ion spectra of (a) negatively charged lipids including fatty acids (FA), cholesterol sulfate, taurocholic acid, ceramide phosphates (CerP), sphingomyelins (SM), phosphatidic acids (PA), phosphatidylethanolamines (PE), and phosphatidylinositols (PI) and (b) positively charged lipids including lysophosphatidylcholines (LPC) and phosphatidylcholines (PC) obtained on the same pixel points. * indicates matrix-derived oligomeric structural ion signals as previously described.20,21 Ions are [M ± H]± unless indicated. | ||
In positive ion mode, more matrix clusters were observed compared to negative ion mode (Fig. 1). These were dimeric and trimeric structures formed by the interaction of laser-induced radicals either with themselves or with the neutral 1,5-DAN molecules. Lysophosphatidylcholines (LPC) were clearly observed without any interference from matrix derived signals in positive ion mode (Fig. 1b).
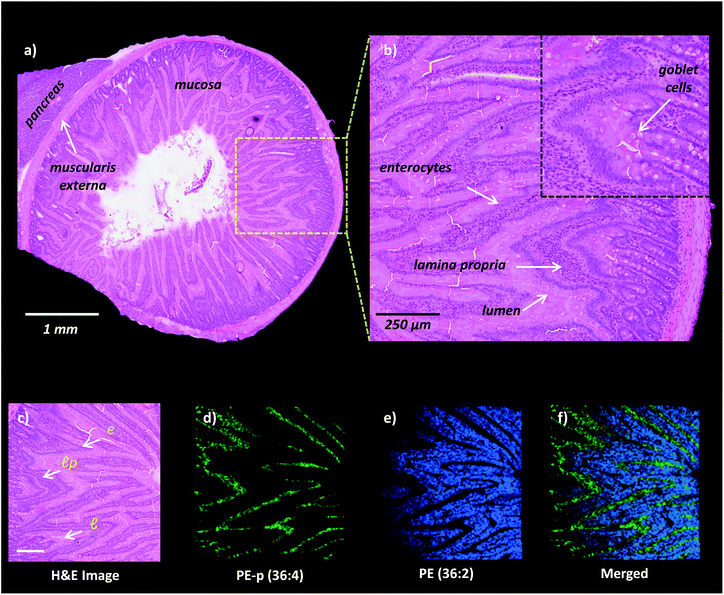
High-spatial resolution MALDI-IMS is a strong approach to determine the localization of individual lipid molecules across the spatially confined histological features.8,9,16 We applied high-resolution (10 μm) MALDI-IMS using 1,5-DAN as the matrix via the sublimation approach to reveal spatial distributions of lipid species in the mucosa of a rat small intestinal tissue section (Fig. 2). The H&E staining on the same tissue section revealed mucosal histological features in jejunum including absorptive enterocytes which are the predominant cells in the intestinal epithelium and mucous goblet cells interspersed between the enterocytes and lamina propria which consists of loose connective tissue and blood vessels (Fig. 2a and b). For example, ion images of PE (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and PE-p (36
2) and PE-p (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) obtained in negative polarity were correlated with the lamina propria and enterocytes & lumen, respectively (Fig. 2c, d, e, f).
4) obtained in negative polarity were correlated with the lamina propria and enterocytes & lumen, respectively (Fig. 2c, d, e, f).
High-spatial resolution dual polarity MALDI-IMS on the same pixel points reveals high-throughput lipid molecular information at high-spatial resolutions
While lipids are highly abundant in tissue sections and efficiently ionized in MALDI, the structural diversity of lipids and the complexity of the matrix-assisted laser desorption/ionization process limit comprehensive lipid profiling within a single IMS analysis.22 Therefore, since histological features can vary from one tissue section to another, correlation of the lipid ion images obtained in different polarities with the same histological features is restricted due to the need for multiple laser desorption/ionization analyses on consecutive tissue sections. Dual polarity MALDI-IMS analysis on the same tissue sections can increase the coverage of the lipid molecular information correlated with the same histological features.8,11 However, use of a high number of laser shots and/or high laser pulse energies required for enhanced MALDI of lipids can result in laser ablation of the matrix coating on tissue sections which hinders the subsequent laser desorption/ionization analysis on the same pixel points. Therefore, a stage offset between the grid arrays of the negative and positive ion mode analysis is required for a dual polarity MALDI-IMS analysis on the same tissue section.8,11 However, certain histological features can be smaller than the stage offset values. This is of relevance for tissues which have small and morphologically varying histological features in adjacent sections such as the small intestine. Therefore, there is a need for a technique to perform dual polarity MALDI-IMS on the same pixel points without sacrificing spatial resolution.The 1,5-DAN matrix compound requires relatively lower laser pulse energies for the optimum laser desorption/ionization of lipids in tissue sections in negative ion mode.8,9 This limits the laser ablation damage of the tissue surface morphology9 and minimizes laser ablation on the matrix coating while still conserving the spectral quality. Therefore, a subsequent laser desorption/ionization analysis can be performed on the same pixel points without significant loss of signals which enhances the lipid molecular information obtained in different polarities at high spatial resolutions.16 We applied high-spatial resolution, dual polarity MALDI-IMS on the same pixel points to reveal small intestinal lipid molecular information obtained in dual polarity across the jejunal mucosa (Fig. 3).
Firstly, we analyzed a mucosal region of interest in negative polarity due to the need of lower laser pulse energies for optimum lipid ionization.9 Then, we switched the ionization polarity to positive and analyzed the same imaging sequence by raster scanning the same pixel points. The quality of the spectra obtained in positive ion mode on laser-ablated spots analyzed in negative ion mode was comparable with the quality of the spectra obtained in positive ion mode in a non-laser irradiated histologically equivalent region (data not shown).
On the other hand, dual polarity acquisitions take at least twice amount of time as long as a single polarity acquisition acquired with the same resolution. This can be a limitation when large areas of the tissue sections and/or large tissue cohorts are needed to be analysed. However, if there is a preliminary knowledge about the histopathology of a disease in an adjacent tissue section, the targeted areas for dual polarity MALDI-IMS can be chosen which can be compatible with the time limit of this technique.16 Previously, Thomas et al. demonstrated the consistent performance of the 1,5-DAN matrix during a total acquisition time of ∼16 h which proves the high-vacuum stability of 1,5-DAN.8 We recently demonstrated that the need of a relatively lower number of laser shots for optimum lipid ionization on a 1,5-DAN-coated tissue section can decrease the acquisition time per pixel point, particularly in negative ion mode.9 Recently, it was also demonstrated that the use of new, high-speed MALDI-MSI instrumentation using a self-scanning laser beam facilitates the extremely high-throughput acquisition of dual-polarity lipid MSI data at high spatial resolutions over 20–50 times faster than other MALDI-MSI systems.10
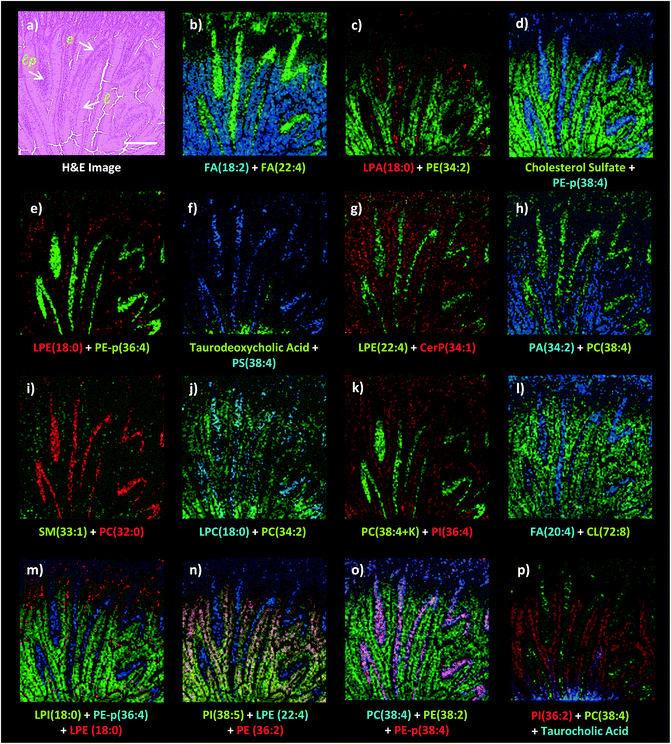
Dual polarity MALDI-IMS on the same pixel points revealed that ion images of fatty acids (FA), sphingomyelins (SM), ceramides (Cer), phosphatidylethanolamines (PE), phosphatidic acids (PA), phosphatidylinositols (PI), phosphatidylserines (PS), lysophosphatidylethanolamines (LPE), lysophosphatidylinositols (LPI), lysophosphatidic acids (LPA), cardiolipins (CL), bile acids and cholesterol sulfate obtained in negative polarity can be correlated with the ion images of phosphatidylcholines (PC) and lysophosphatidylcholines (LPC) obtained in positive polarity at 10 μm spatial resolution (Fig. 3). For example, FA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), LPE (22
2), LPE (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), LPA (18
4), LPA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PE-p (38
0), PE-p (38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PE-p (36
4), PE-p (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PS (38
4), PS (38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), and FA (20
4), and FA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) obtained in negative ion mode and PC (38
4) obtained in negative ion mode and PC (38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), LPC (18
4), LPC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PC (32
0), PC (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), and PC (34
0), and PC (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) obtained in positive ion mode can be simultaneously correlated with the lamina propria in the intestinal mucosa (Fig. 3a–l). This is significant as lipid species ionized in opposite polarities can have interrelated physiological or pathophysiological roles in the same histological regions at high-spatial resolutions. For example, LPA is generated through several enzymatic pathways such as their generation from membrane phospholipids through the action of phospholipases as well as through the enzymatic action of autotaxin (ATX) which metabolizes LPC to LPA and which can be altered in disease states. Our method enables their simultaneous probing in small histological areas in the same tissue sections (Fig. 3c and j).
2) obtained in positive ion mode can be simultaneously correlated with the lamina propria in the intestinal mucosa (Fig. 3a–l). This is significant as lipid species ionized in opposite polarities can have interrelated physiological or pathophysiological roles in the same histological regions at high-spatial resolutions. For example, LPA is generated through several enzymatic pathways such as their generation from membrane phospholipids through the action of phospholipases as well as through the enzymatic action of autotaxin (ATX) which metabolizes LPC to LPA and which can be altered in disease states. Our method enables their simultaneous probing in small histological areas in the same tissue sections (Fig. 3c and j).
On the other hand, cholesterol sulfate, FA (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PE (34
2), PE (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), CerP (34
2), CerP (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PA (22
1), PA (22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), SM (33
4), SM (33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC (34
1), PC (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PI (36
2), PI (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), CL (72
4), CL (72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8), LPI (18
8), LPI (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), and PI (36
0), and PI (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were correlated with the enterocytes and lumen in the intestinal mucosa (Fig. 3). Interestingly, an intense accumulation of LPE (18
2) were correlated with the enterocytes and lumen in the intestinal mucosa (Fig. 3). Interestingly, an intense accumulation of LPE (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) species was observed in the mucous goblet cell layer while it also exists in the lumen and enterocytes (Fig. 3e and m). This is a similar phenomenon to the accumulation of LPC (18
0) species was observed in the mucous goblet cell layer while it also exists in the lumen and enterocytes (Fig. 3e and m). This is a similar phenomenon to the accumulation of LPC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) and the depletion of PC (36
0) and the depletion of PC (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) in the Purkinje cell layer in the cerebellum region of the mice brain tissue section as previously demonstrated using high-spatial resolution MALDI-IMS.9 Moreover, bile acids, taurodeoxycholic and taurocholic acids were found to be intensely accumulated in the direction of the duodenal lumen but less in the mucosa (Fig. 3f and p).
4) in the Purkinje cell layer in the cerebellum region of the mice brain tissue section as previously demonstrated using high-spatial resolution MALDI-IMS.9 Moreover, bile acids, taurodeoxycholic and taurocholic acids were found to be intensely accumulated in the direction of the duodenal lumen but less in the mucosa (Fig. 3f and p).
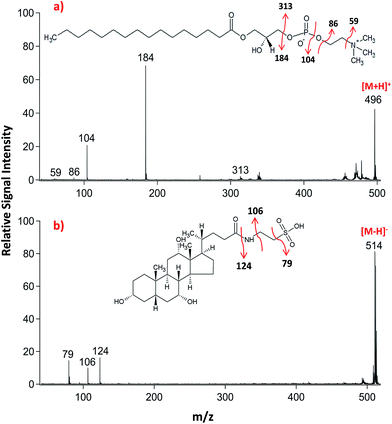
Lipid classifications were performed by examining MS/MS spectra obtained in LID-LIFT-TOF/TOF mode from the corresponding regions of the ion images after MALDI-IMS. For example, specific fragments of LPC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, m/z 496.3) and TCA (m/z, 514.3) were observed in positive and negative polarities, respectively (Fig. 4).
0, m/z 496.3) and TCA (m/z, 514.3) were observed in positive and negative polarities, respectively (Fig. 4).
Conclusions
Dual polarity MALDI-IMS on the same pixel points was successfully applied on rat small intestinal tissue sections. Several lipid species including fatty acids, lysophospholipids, bile acids, phospholipids, sphingolipids, cardiolipins and cholesterol sulfate were detected in dual polarity, identified and correlated with histological features at high-spatial resolutions. It was demonstrated that fatty acids along with low-molecular-weight lysophopholipids can be detected without any interference from the matrix clusters when the laser power is optimized for efficient desorption/ionization on 1,5-DAN sublimation-coated intestinal tissue sections. LID fragmentation analyzed in LIFT-TOF/TOF mode provided tandem MS information useful for the structural elucidation of lipid species in dual polarity. Spatial distribution profiles of several rat small intestinal lipid species including cardiolipins, sphingolipids and phospholipids were for the first time demonstrated in this communication.Statement
This manuscript and its contents in some other form have not been published previously by any of the authors and are not under consideration for publication in another journal at the time of submission.Affirmation
All authors have seen and approved the submission of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MALDI-IMS experiments were performed at the go:IMS imaging MS platform at the University of Gothenburg/Chalmers University of Technology (www.go-ims.gu.se) headed by Prof. Andrew Ewing. This work was financially supported by the Knut and Alice Wallenberg Foundation, the USA National Institutes of Health, European Research Council (ERC), and the Swedish Research Council. The authors thank Professor Andrew G. Ewing and Associate Professor John S. Fletcher for their constructive comments on the manuscript.References
- N. A. Abumrad and N. O. Davidson, Physiol. Rev., 2012, 92, 1061–1085 CrossRef PubMed.
- J. Iqbal and M. M. Hussain, Am. J. Physiol.: Endocrinol. Metab., 2009, 296, E1183–E1194 CrossRef PubMed.
- K. A. Z. Berry, B. Li, S. D. Reynolds, R. M. Barkley, M. A. Gijón, J. A. Hankin, P. M. Henson and R. C. Murphy, J. Lipid Res., 2011, 52, 1551–1560 CrossRef PubMed.
- L. A. McDonnell and R. Heeren, Mass Spectrom. Rev., 2007, 26, 606–643 CrossRef PubMed.
- D. S. Cornett, M. L. Reyzer, P. Chaurand and R. M. Caprioli, Nat. Methods, 2007, 4, 828–833 CrossRef PubMed.
- R. C. Murphy, J. A. Hankin and R. M. Barkley, J. Lipid Res., 2009, 50, S317–S322 CrossRef PubMed.
- J. A. Hankin, R. M. Barkley and R. C. Murphy, J. Am. Soc. Mass Spectrom., 2007, 18, 1646–1652 CrossRef PubMed.
- A. l. Thomas, J. L. Charbonneau, E. Fournaise and P. Chaurand, Anal. Chem., 2012, 84, 2048–2054 CrossRef PubMed.
- I. Kaya, W. Michno, D. Brinet, Y. Iacone, G. Zanni, K. Blennow, H. Zetterberg and J. r. Hanrieder, Anal. Chem., 2017, 89, 4685–4694 CrossRef PubMed.
- N. Ogrinc Potočnik, T. Porta, M. Becker, R. Heeren and S. R. Ellis, Rapid Commun. Mass Spectrom., 2015, 29, 2195–2203 CrossRef PubMed.
- S. R. Ellis, J. Cappell, N. O. Potočnik, B. Balluff, J. Hamaide, A. Van der Linden and R. M. Heeren, Analyst, 2016, 141, 3832–3841 RSC.
- A. R. Korte and Y. J. Lee, J. Am. Soc. Mass Spectrom., 2013, 24, 949–955 CrossRef PubMed.
- I. Kaya, D. Brinet, W. Michno, S. Syvänen, D. Sehlin, H. Zetterberg, K. Blennow and J. r. Hanrieder, ACS Chem. Neurosci., 2017, 8, 347–355 CrossRef PubMed.
- D. Suckau, A. Resemann, M. Schuerenberg, P. Hufnagel, J. Franzen and A. Holle, Anal. Bioanal. Chem., 2003, 376, 952–965 CrossRef PubMed.
- A. Seyer, M. Cantiello, J. Bertrand-Michel, V. Roques, M. Nauze, V. Bézirard, X. Collet, D. Touboul, A. Brunelle and C. Coméra, PLoS One, 2013, 8, e58224 Search PubMed.
- I. Kaya, D. Brinet, W. Michno, M. Başkurt, H. Zetterberg, K. Blenow and J. r. Hanrieder, ACS Chem. Neurosci., 2017, 8, 2778–2790 CrossRef PubMed.
- A. R. Korte and Y. J. Lee, J. Mass Spectrom., 2014, 49, 737–741 CrossRef PubMed.
- T. W. Jaskolla, M. Karas, U. Roth, K. Steinert, C. Menzel and K. Reihs, J. Am. Soc. Mass Spectrom., 2009, 20, 1104–1114 CrossRef PubMed.
- J. Yang and R. M. Caprioli, Anal. Chem., 2011, 83, 5728–5734 CrossRef PubMed.
- L. Molin, R. Seraglia, F. R. Dani, G. Moneti and P. Traldi, Rapid Commun. Mass Spectrom., 2011, 25, 3091–3096 CrossRef PubMed.
- K. Scheffler and K. Strupat, Appl. Notes, 2010, 30218, 1–8 Search PubMed.
- K. A. Zemski Berry, J. A. Hankin, R. M. Barkley, J. M. Spraggins, R. M. Caprioli and R. C. Murphy, Chem. Rev., 2011, 111, 6491–6512 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ay00645h |
| This journal is © The Royal Society of Chemistry 2018 |