Investigation and intervention of autophagy to guide cancer treatment with nanogels†
Xudong
Zhang‡
abc,
Xin
Liang‡
d,
Jianjun
Gu‡
e,
Danfeng
Chang
ab,
Jinxie
Zhang
ab,
Zhaowei
Chen
 c,
Yanqi
Ye
c,
Chao
Wang
c,
Wei
Tao
ab,
Xiaowei
Zeng
ab,
Gan
Liu
ab,
Yongjun
Zhang
c,
Yanqi
Ye
c,
Chao
Wang
c,
Wei
Tao
ab,
Xiaowei
Zeng
ab,
Gan
Liu
ab,
Yongjun
Zhang
 *e,
Lin
Mei
*ab and
Zhen
Gu
*c
*e,
Lin
Mei
*ab and
Zhen
Gu
*c
aSchool of Life Sciences, Tsinghua University, Beijing 100084, P.R. China
bThe Shenzhen Key Lab of Gene and Antibody Therapy, Division of Life and Health Sciences, Graduate School at Shenzhen, Tsinghua University, Shenzhen 518055, P.R. China. E-mail: mei.lin@sz.tsinghua.edu.cn
cJoint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA. E-mail: zgu@email.unc.edu
dShenzhen Key Laboratory of two dimensional materials and devices/Shenzhen Engineering Laboratory of Phosphorene and Optoelectronics, SZUNUS Collaborative Innovation Center for Optoelectronic Science and Technology, and Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, Shenzhen University, Shenzhen 518060, China
eKey Laboratory of Functional Polymer Materials, Institute of Polymer Chemistry, College of Chemistry, Nankai University, and Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300071, China. E-mail: yongjunzhang@nankai.edu.cn
First published on 14th November 2016
Abstract
Cancer cells use autophagy to resist poor survival environmental conditions such as low PH, poor nutrients as well as chemical therapy. Nanogels have been used as efficient chemical drug carriers for cancer treatment. However, the effect of nanogels on autophagy is still unknown. Here, we used Rab proteins as the marker of multiple trafficking vesicles in endocytosis and LC3 as the marker of autophagy to investigate the intracellular trafficking network of Rhodamine B (Rho)-labeled nanogels. The nanogels were internalized by the cells through multiple protein dependent endocytosis and micropinocytosis. After inception by the cells, the nanogels were transported into multiple Rab positive vesicles including early endosomes (EEs), late endosomes (LEs), recycling endosomes (REs) and lipid droplets. Finally, these Rab positive vesicles were transported to lysosome. In addition, GLUT4 exocytosis vesicles could transport the nanogels out of the cells. Moreover, nanogels could induce autophagy and be sequestered in autophagosomes. The crosstalk between autophagosomes and Rab positive vesicles were investigated, we found that autophagosomes may receive nanogels through multiple Rab positive vesicles. Co-delivery of autophagy inhibitors such as chloroquine (CQ) and the chemotherapeutic drug doxorubicin (DOX) by nanogels blocked the autophagy induced by DOX greatly decreasing both of the volume and weight of the tumors in mice tumor models. Investigation and intervention of the autophagy pathway could provide a new method to improve the therapeutic effect of anticancer nanogels.
Introduction
Biodegradable materials such as synthetic polymers, lipids and nanogels have received considerable interest as drug delivery vehicles.1–3 Nanogels have been considered to be one of the most promising nanoparticles for drug delivery and pathological imaging.4–6 The nanogels are mainly formulated based on natural polymers such as chitosan and synthetic polymers such as poly(vinyl alcohol), poly(ethylene oxide), poly(ethyleneimine), poly(vinyl pyrrolidone) and poly-N-isopropylacrylamide.7 The hydrophilic groups present on the surface of hydrogels – including OH, –CONH–, –CONH2–, and –SO3H make the nanogels hydrophilic.5 Therefore, nanogels provide a tip-top vehicle for both hydrophilic chemical drugs and hydrophilic biomolecular drugs such as proteins and peptides.8 Nanogels with hydrate properties can be formulated with various sizes from 20 nm to 300 nm.9 Thus, nanogels represent an optimal choice for loading and delivery of the hydrophilic drugs. Of particular interest, studying their interactions with bio-systems would provide important information for optimizing the parameters for maximizing therapy efficiency.Nano-bio interactions, especially the in vivo tissue uptake of nanoparticles, are important for determining the therapeutic effect of nanomedicine.10,11 Nanoparticles have to reach the tumor tissue and then be taken up by the tumor cells through passive targeting or active targeting.12,13 When the nanoparticles are incepted by the cells, the intracellular trafficking fate determines its therapeutic effect. The inner membrane vesicle system is a complex transport system mainly including endocytosis, exocytosis and autophagy, all of which are responsible for the transportation of various macromolecules between different organelles in the cells.14 Endocytosis pathways mainly include clathrin dependent and clathrin independent pathways.15 The clathrin independent pathways include micropinocytosis, and caveolin dependent and caveolin independent (Arf-6, flotillin, Cdc42 and RhoA-dependent) pathways.16–19 Nanoparticles are transported to the early endosomes (EEs), then to late endosomes (LEs), and finally delivered to the lysosome for degradation.19 Besides the EE-LE-lysosome pathway, other endocytosis pathways also exist. For instance, a recycling endosome system is responsible for the delivery of the protein receptors back to the cell membrane.19 The exocytosis pathway is mainly responsible for the transportation of the mature proteins from the Golgi body to the cell membrane or secretion of the proteins out of the cells.20 Autophagy is a cellular stress response process, which could engulf cellular components such as cytoplasm, aggregated proteins, damaged mitochondria and invaded pathogens to make the cells under go stress condition.20,21 However, most of the studies focus on the cell trafficking pathway on the classic pathway: the EE-LE-lysosome pathway. Other pathways such as the recycling endosome system, exocytosis and autophagy of the nanoparticles in the cells remain rarely explored.
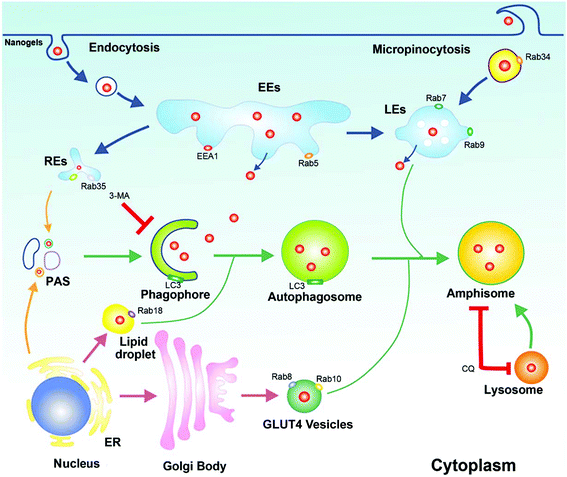
In this work, by employing the Rab family proteins as the markers of intracellular vesicle networks, we investigated the intracellular trafficking pathways of nanogels. The Rab protein family, a type of GTPase/GTP-binding protein, has various cellular functions including protein trafficking, transmembrane signal transduction and fusion of vesicles between different organelles.22 By using this unique advantage, we identified two novel transport pathways: the micropinocytosis (Rab34 positive)-LE (Rab7 positive)-lysosome pathway and the EE-liposome (Rab18 positive)-lysosome pathway, besides the classic pathway – early endosomes (EEs)/late endosomes (LEs) to lysosome. Moreover, we found that the cells used the slow recycling pathway (Rab35 positive vesicles) and the GLUT4 exocytosis pathway (Rab8 and Rab10 positive vesicles) to transport the nanogels out of the cells. Most importantly, we found that autophagosomes could act as crossing sites, sequestering nanogels by receiving them from different vesicles. To further prove our investigation, on inhibiting the autophagy pathways with 3-MA or CQ (autophagy inhibitor), greatly enhanced retention of the drug-loaded nanogels was observed, which significantly improved the therapeutic efficiency of the nanomedicine both in vitro and in vivo.
Experimental
Materials
N-Isopropylacrylamide (NIPAM), N,N′-methylenebis(acrylamide) (BIS), acrylic acid (AAc), potassium persulfate (KPS), sodium dodecyl sulfate (SDS), chloroquine hydrochloride (HCQ) and 3-methyladenine (3-MA) were purchased from Sigma-Aldrich (St Louis, MO). Doxorubicin (DOX) was purchased from Huafenglianbo Co. Methacryloxyethyl thiocarbamoyl rhodamine B (PolyFluor™ 570) was purchased from Polysciences. NIPAM was purified by recrystallization from a hexane/acetone mixture and dried in a vacuum. AAc was distilled under reduced pressure. Acetonitrile and methanol were purchased from EM Science (HPLC grade, Mallinckrodt Baker, USA). All other chemicals used here were of the highest quality and were commercially available. Antibodies against EEA1, Rab7, LC3B and LAMP1 were from Cell Signaling Technology, Inc. (Danvers, MA). The antibody against β-actin was obtained from Abmart, Inc. (Shanghai, China).Plasmid and transfection
The pEGFP-LC3 and pDsRed-LC3 plasmids were constructed in our lab. The KSHV Flag-vBcl-2 plasmid was kindly provided by Professor Beth Levine, University of Texas, Department of Medicine. DsRed-Rab5, DsRed-Rab7 and Flag-Rubicon plasmids were from Addgene. Rab1–37 genes in T vector were from Professor Jiahuai Han's Lab, and then sub-cloned to pEGFP-C1 and pDsRed-C1, respectively. All plasmids were confirmed by automated DNA sequencing. Cells were transiently transfected with the plasmids using lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.Synthesis of fluorescence-labeled PNIPAM nanogels (PN1–3) and P(NIPAM-Aac) nanogels (PNA) for drug loading
The nanogels were synthesized by free-radical precipitation polymerization. Briefly, a predetermined amount of monomers, cross-linker (BIS) and SDS (see Table 2) were dissolved in 95 mL of water. The mixture was transferred to a 250 mL, three-necked flask, purged with N2 and heated to 70 °C. One hour later, 0.081 g of KPS (dissolved in 5 mL of water) was added to initiate the polymerization. The reaction was allowed to proceed for 5 h. After purification by 2-week dialysis against water (cut off 8000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), the resultant nanogels were lyophilized. This Rho B contains a vinyl group which can be copolymerized with the monomers. To label the nanogels with Rho-B, (commercial name: PolyFluorTM 570) was added to the reaction mixture and reacted with monomers before polymerization.
000), the resultant nanogels were lyophilized. This Rho B contains a vinyl group which can be copolymerized with the monomers. To label the nanogels with Rho-B, (commercial name: PolyFluorTM 570) was added to the reaction mixture and reacted with monomers before polymerization.
| Protein markers | Co-localization with nanogels | Subcellular localization | Function |
|---|---|---|---|
| Membrane internalization | |||
| Clathrin | Yes | Cell membrane | Clathrin dependent endocytosis |
| Caveolin | Yes | Cell membrane | Caveolin dependent endocytosis |
| RhoA | Yes | Cell membrane | RhoA dependent endocytosis |
| Cdc42 | Yes | Cell membrane | Cdc42 dependent endocytosis |
| Flotillin | Yes | Cell membrane | Flotillin dependent endocytosis |
| Arf6 | Yes | Cell membrane | Arf6 dependent endocytosis |
| Rab proteins in trafficking | |||
| Rab1 | Yes | ER–Golgi intermediate | ER–Golgi trafficking |
| Rab2 | No | ER–Golgi intermediate | ER–Golgi trafficking |
| Rab3 | No | TGN-apico-lateral membranes | Exocytosis of secretory granules and vesicles from TGN to apico-lateral |
| Rab4 | No | Early and recycling endosomes | Endocytic recycling to plasma membrane |
| Rab5 | Yes | Early endosome | Endocytic internalization and early endosome fusion |
| Rab6 | No | Intra-Golgi | Intra-Golgi transport |
| Rab7 | Yes | Late endosome | Control late endocytic trafficking |
| Rab8 | Yes | Median Golgi and TGN | Basolateral protein transport from median Golgi and TGN |
| Rab9 | Yes | Late endosome | Transport from late endosomes to trans-Golgi |
| Rab10 | Yes | Median Golgi and TGN | Basolateral protein transport from median Golgi and TGN |
| Rab11 | No | Recycling endosomes | Transport from the Golgi, and apical and basolateral endocytic recycling |
| Rab12 | Not tested | Peripheral region of cell to perinuclear centrosomes | Transport from the peripheral region of cell to perinuclear centrosomes |
| Rab14 | Yes | Early endosomes and Golgi | Transport between early endosomes and Golgi |
| Rab15 | Not tested | Early and recycling endosomes | Inhibitor of endocytic internalization |
| Rab17 | Not tested | Epithelial specific; apical recycling endosome | Transport through apical recycling endosomes |
| Rab18 | Yes | ER and liposome | Liposome formation |
| Rab20 | No | Recycling endosome | In apical endocytosis/recycling |
| Rab21 | No | Cell membrane | Focal adhesion site transport |
| Rab22 | Yes | Early endosomes and TGN | Transport between TGN and early endosomes and vice versa |
| Rab23 | Yes | Plasma membranes and early endocytic vesicles | Transport between plasma membranes and early endocytic vesicles |
| Rab24 | Yes | ER/cis-Golgi region and on late endosomal structures | Autophagy-related processes |
| Rab25 | No | Apical recycling endosome | Transport through apical recycling endosomes |
| Rab26 | No | TGN-apico-lateral membranes | Exocytosis of secretory granules and vesicles from TGN to apico-lateral membranes |
| Rab31 | Yes | Early endosomes and TGN | Transport between TGN and early endosomes and vice versa |
| Rab32 | Yes | Melanosomes | Biogenesis of melanosomes |
| Rab38 | Yes | Melanosomes | Biogenesis of melanosomes |
| Rab35 | Yes | Recycling endosomes | Apical endocytic recycling |
| Rab43 | No | ER–Golgi intermediate | ER–Golgi trafficking |
| Autophagy | |||
| LC3 | Yes | Autophagosomes | Biogenesis of autophagosomes |
| Sample name | NIPAM (g) | AAc (g) | BIS (g) | SDS (g) | PolyFluor 570 (mg) |
|---|---|---|---|---|---|
| PN1 | 1.550 | 0 | 0.043 | 0.115 | 1 |
| PN2 | 1.550 | 0 | 0.043 | 0.055 | 1 |
| PN3 | 1.500 | 0 | 0.108 | 0 | 1 |
| PNA | 1.234 | 0.202 | 0.043 | 0.158 | 0 |
Drug loading and drug encapsulation efficiency
PNA nanogels were negatively charged, while DOX and CQ were positively charged in PBS (pH 7.4) due to their pKa values. The drugs were conjugated via electrostatic binding to carboxylic moieties provided by acrylic acid. Therefore, 1.50 g of PNA nanogels was dissolved in 100 mL of 0.01 M PBS pH 7.4, to which 0.3 g of DOX was added. The mixture was stirred overnight at 4 °C. To remove the unencapsulated drug, the mixture was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. After the supernatant was decanted, the precipitation was lyophilized. CQ was loaded in a similar way except that the unencapsulated drug was removed by dialysis against PBS for 4 hours. To determine the amount of the unencapsulated drug, the drug concentration in the supernatant or the dialysis media was measured spectroscopically. The DOX- and CQ-loaded nanogels were named DOX-PNA and CQ-PNA, respectively. Drug loading capacity (LC) and encapsulation efficiency (EE) were calculated using the following equations:
000 rpm. After the supernatant was decanted, the precipitation was lyophilized. CQ was loaded in a similar way except that the unencapsulated drug was removed by dialysis against PBS for 4 hours. To determine the amount of the unencapsulated drug, the drug concentration in the supernatant or the dialysis media was measured spectroscopically. The DOX- and CQ-loaded nanogels were named DOX-PNA and CQ-PNA, respectively. Drug loading capacity (LC) and encapsulation efficiency (EE) were calculated using the following equations:| LC = (md − md, u)/mg × 100% |
| EE = (md − md, u)/md × 100% |
In vitro drug release
Drug release mechanisms from nanogels were diffusion-controlled. The PNA nanogels are thermally sensitive. The PNA nanogels were in the swollen state during drug loading at 4 °C, but they shrank as the temperature increased, and the shrinking of the nanogels at 37 °C expelled the drugs. A predetermined amount of DOX-PNA or CQ-PNA was dissolved in 1 mL of PBS. The solution was transferred into a dialysis tube and immersed in 19 mL of PBS as release media. At suitable time intervals, the media was replaced with the fresh ones. The drug concentration in the release media was determined spectroscopically with a TU-1810PC UV-vis spectrophotometer (Purkinje General, China).Characterization of the nanogel particles
The hydrodynamic diameters (Dh) of the nanogel particles were measured by dynamic light scattering with a Brookhaven 90 Plus laser particle size analyzer. All the measurements were carried out at a scattering angle of 90°. The sample temperature was controlled with a build-in Peltier temperature controller. Zeta potential was measured on a Malvern Mastersizer 2000. The confocal images of the nanogel particles were acquired on a Leica TCS SP8. TEM images were acquired on a Tecnai G2 F20 transmission electron microscope (Table 3).| Size at 37 °C (nm) | PDI | ζ (mV) | LC (%) | EE (%) | |
|---|---|---|---|---|---|
| a In 0.01 M PBS pH 7.4 at 37 °C. | |||||
| PN1 | 52.5 ± 0.8 | 0.05 | −12.4 ± 2.0 | — | — |
| PN2 | 101.3 ± 0.2 | 0.07 | −16.6 ± 2.9 | — | — |
| PN3 | 299.8 ± 0.8 | 0.105 | −17.4 ± 0.2 | — | — |
| PNA | 176.3 ± 2.1 | 0.05 | −12.7 ± 1.4 | — | — |
| DOX-PNA | 206.2 ± 4.4 | 0.14 | −5.8 ± 0.1 | 17.3 | 92.8 |
| CQ-PNA | 200.4 ± 2.2 | 0.11 | −12.1 ± 1.4 | 9.0 | 45.0 |
Cell culture
MCF-7 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS).Intracellular trafficking of Rho-loaded nanogels
Rhodamine B (Rho) was used as a model fluorescent molecule, which was formulated in nanogels. The non-transfected or EGFP-Rab1–43 or DsRed-Rab1–43 were incubated with 1 mg mL−1 Rho-labeled nanogels at 37 °C for 2 h. For lysosome detection, the cells were incubated with Lyso-Tracker Red for 1 h. Then the cells were washed with PBS three times, and then fixed by using 4% paraformaldehyde for 20 min. Cells were stained with DAPI for 15 min and washed three times with PBS. Confocal microscopy was performed on a FLUO-VIEW laser scanning confocal microscope (Olympus, FV1000, Olympus Optical, and Japan) in sequential scanning mode using a 60–100× objective.Autophagy assays
MCF-7 cells were transfected with EGFP-LC3 or DsRed-LC3 for 12 h and then the cells were treated with 100 μg mL−1 Rho-labeled nanogels, drug-free nanogels, 1 μM DOX or DOX-loaded nanogels (equal to 1 μM DOX) for 24 h. Then, the cells were fixed in 4% paraformaldehyde. The percentage of cells with fluorescent dots representing EGFP-LC3 translocation were counted by confocal microscopy as described previously.23 The LC3II protein level was detected using the anti-LC3B antibody.24Immunoblotting
Immunoblotting was carried out as previously described.23 For abbreviation, MCF-7 cells were treated with DOX, and DOX-loaded nanogels as indicated were harvested and the lysates were subjected on 12% SDS-PAGE. The LC3B protein levels were detected by using LC3B and β-actin antibodies. The HRP conjugated secondary antibody was used to detect the primary antibody followed by enhanced chemiluminescence (ECL) detection (Thermo Scientific).Immunofluorescence assay
For abbreviation, the MCF-7 cells transfected with or without the pEGFP-Rab plasmid were treated with or without nanogels and were fixed in 4% paraformaldehyde. And then the cells were blocked by using 3% BSA. Then, the cells were incubated with the following primary antibodies: EEA1, Clathrin, LC3, Arf-6, flotillin, Cdc42, RhoA, EEA1, Caveolin as indicated. The TRITC and FITC labeled secondary antibodies were used to detect the primary antibody. Confocal microscopy was performed on a FLUO-VIEW laser scanning confocal microscope (Olympus, FV1000, Olympus Optical, and Japan) in sequential scanning mode using a 60–100× objective.In vivo anti-tumor assay with xenograft SCID mice model
Animal experiments were approved by the Administrative Committee of Animal Research in the Tsinghua University Shenzhen Graduate School. Female severe combined immunodeficient (SCID) mice were purchased from the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Science. MCF-7 cells (2 × 106 cells per mouse) were inoculated subcutaneously to mice at the right axilla. Tumors were measured by using a vernier calipers and the volume (V) was calculated to be V = d2 × D/2, where d and D are the shortest and the longest diameter of the tumor in mm respectively. When the tumor volume reached around 30 mm3 (designated as the 0 day), treatments were performed. The mice were randomly divided into five groups (each group had 5 animals, n = 5). Saline was used for the control experiment. 10 mg kg−1 DOX, DOX-loaded nanogels), 50 mg kg−1 CQ and DOX-loaded nanogels combined with CQ-loaded nanogels (at a single dose of 50 mg kg−1 CQ) were injected via the peritoneal cavity in PBS on days 0, 3, 6, 9, 12, 15 and 18. The mice were sacrificed by cervical decapitation 20 days after treatment. The terminal tumor weight (mg) was measured and utilized to evaluate the antitumor activity.Statistical analysis
All results are reported as the mean ± S.D. of three independent experiments. Comparisons were performed using a two-tailed paired Student's t test (*P < 0.05, **P < 0.01, ***P < 0.001).Results and discussion
Preparation and characterization of nanogels
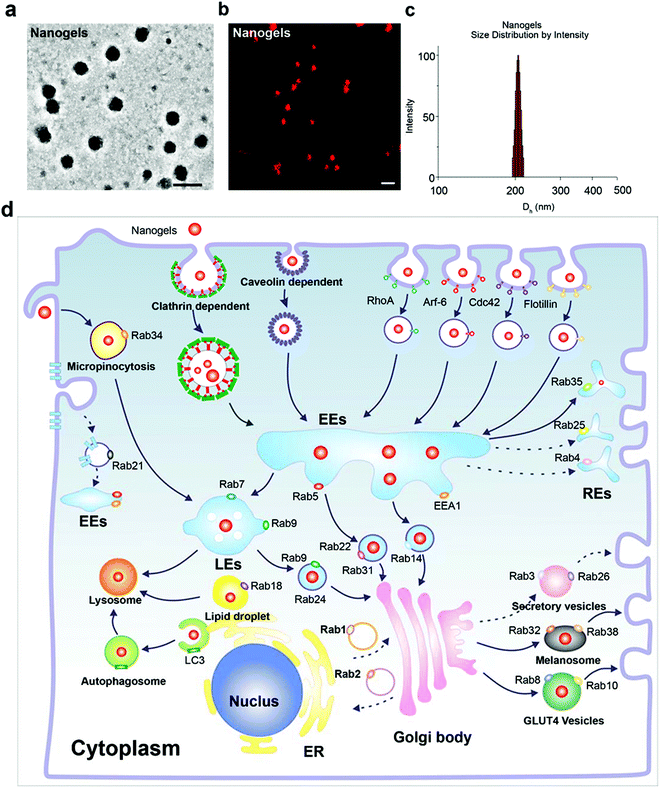
Poly(N-isopropylacrylamide) (PNIPAM) nanogels have found numerous biomedical applications, such as drug delivery. Upon external stimuli, such as changes in temperature, the nanogel particles swell or shrink, thus close or open the channel. We prepared three Rhodamine B (Rho)-labeled PNIPAM nanogels with different diameters (Fig. S1†). The size of these nanogels were around 70 (PN1), 150 (PN2), and 300 nm (PN3) at 37 °C, respectively (Fig. S1†). All the particles bear negative charges as indicated by their negative zeta potential (Table 2). In order to investigate the cellular trafficking of the nanogels, we treated the MCF-7 cells with these three types of nanogels. As expected, all the three nanogels with different diameters could be effectively taken up by the cells (data not shown). When the size of the nanoparticles is larger than 500 nm they are difficult to be taken up by the cells. Thus, we choose the PN2 which has a diameter of around 150 to 200 nm (size of the three PN nanogels in the centre) as a model nanogel to investigate the intracellular pathway. The morphology of the particles under a TEM is shown in Fig. 1a. The nanogels were also observed using a confocal microscope at room temperature. Both of the TEM and confocal images showed that the nanogels had a nearly spherical shape (Fig. 1a and b). Next, we detected the size distribution by dynamic light scattering. As expected, the diameter of the nanogels was around 200 nm as shown in Fig. 1c.Nanogels incepted by the cells through the endocytosis pathway
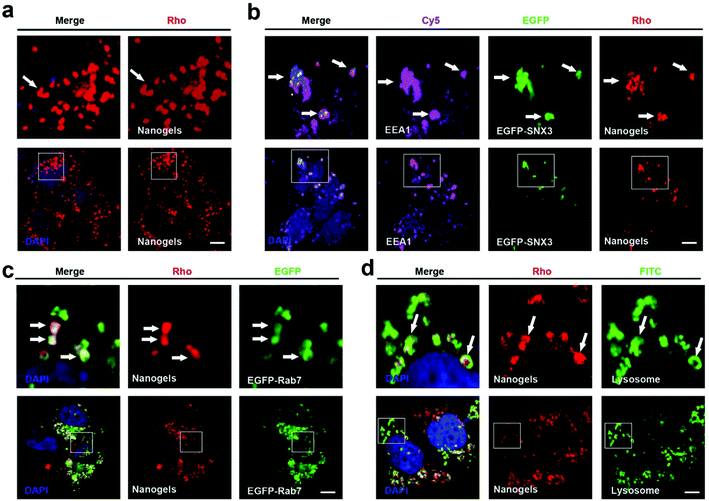
Endocytosis is a complex network in the cells which includes clathrin dependent and clathrin independent pathways. Besides those depending on clathrin proteins, there are many other kinds of clathrin independent endocytosis such as micropinocytosis, caveolin dependent and caveolin independent (Arf-6, flotillin, Cdc42 and RhoA-dependent) pathways (Fig. 1d). In order to detect the pathway employed by the cells to incept the nanogels, MCF-7 cells were treated with Rhodamine (Rho)-labeled PNIPAM nanogels for 20 hours. Then, we observed that there are a lot of Rho-positive vesicles containing nanogels within the cells (Fig. 2a). Next, we wanted to know whether there is co-localization of the Rho-positive vesicles with clathrin, caveolin, Arf-6, flotillin, Cdc42 and RhoA-positive endocytosis vesicles. The immunofluorescence assay was performed. We observed that Rho-positive vesicles co-localized with all of the six endocytosis marker positive vesicles (Fig. S2†). These data demonstrate that the cells incepted the nanogels through multiple endocytosis pathways to go across the cell membrane.Sorting nexins have functions in diverse processes, including endocytosis, endosomal sorting and endosomal signaling.25 SNX3 is localized on early endosomes and is responsible for the transportation from the cell membrane to early endosomes.25 EEA1 and Rab5 have been widely used as the marker of early endosomes, meanwhile Rab7 has been used as the marker of late endosomes.19 Therefore, we used EEA1 and EGFP-SNX3 to indicate EEs, and EGFP-Rab7 to indicate LEs. Next, we treated the vector and EGFP-Rab7 transfected MCF-7 cells with the Rho-labeled nanogels for 20 h. We found the Rho-labeled nanogel containing vesicles co-localized with EEA1 and EGFP-SNX3 labeled EEs (Fig. 2a and b). Rho-labeled nanogels containing vesicles also co-localized with EGFP-Rab7 positive LEs (Fig. 2c). To detect the lysosome, immunofluorescence assay with the LAMP1 antibody were carried out. We observed that the green lysosome contained the red Rho-labeled nanogels within the vesicles (Fig. 2d). These data show that the nanogels were incepted by the cells through endocytosis and then transported to LEs and EEs, and finally transported to lysosome for degradation.
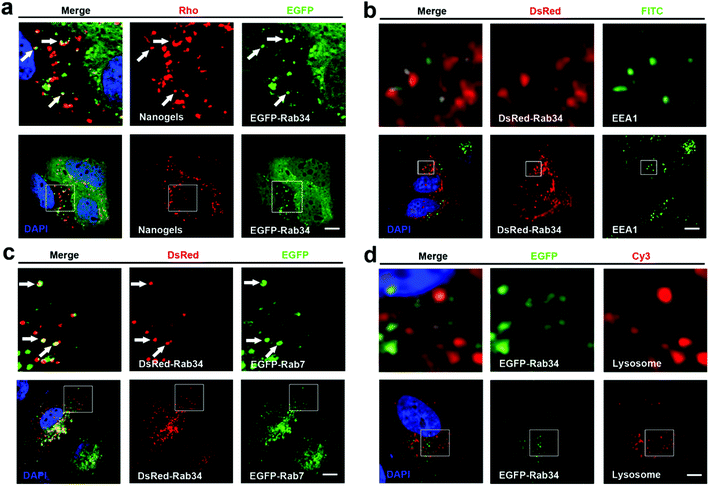
There are more than 30 types of Rab proteins that have been found within the cells which have their specific cellular function. The most important function of these Rab proteins is the regulation of the intracellular vesicle formation and trafficking.26 Rab5 and Rab7 proteins have been widely used to detect the intracellular tracking of many nanoparticles in the published literature. Rab18, 21, 23 and 34 also have been reported to regulate the endocytosis pathway.26,27 However, there is almost no report to consider these Rab proteins as the marker of endocytosis to investigate the trafficking pathways of the nanoparticles. Therefore, we incubated the EGFP-Rab18, 21, 23 and 34 transfected MCF-7 cells with Rho-labeled nanogels for 20 h. Then we observed that EGFP-Rab18, 23, 34 positive vesicles sequestered the red Rho labeled nanogels but not the Rab21 vesicles (Table 1). Rab34 controls the process of micropinocytosis and also is a specific marker of micropinocytosis.26,28 To detect whether micropinocytosis is involved in the uptake of the nanogels by the cells, we incubated the nanogels with MCF-7 cells. Interestingly, we observed that Rab34 positive vesicles also sequestered Rho-labeled nanogels (Fig. 3a). In order to detect the down-stream pathway of micropinocytosis, we determined the relationship between the DsRed-Rab34 positive vesicles and the classic endocytosis pathways (EEA1, EGFP-Rab7 and lysosome tracker). We found that Rab34 positive vesicles co-localize with EGFP-Rab7 positive LEs and lysosomes but not with EEA1 positive EEs (Fig. 3b and c). These data indicate that micropinocytosis is involved in the inception of the nanogels.
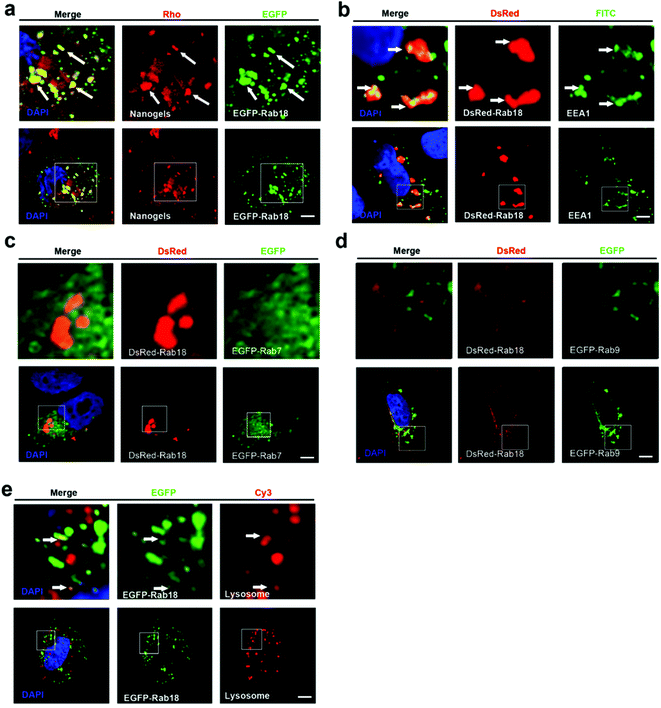
Lipid droplet formation is regulated by Rab18 in adipocytes.29 Rab18 regulates the release of lipids, which act as excellent markers for lipid droplet dynamics within the cells.28 Interestingly, we observed that Rab18 positive lipid droplets contain the Rho-labeled nanogels (Fig. 4a). To investigate the trafficking role of Rab18-positive vesicles in the intracellular pathways, we detected DsRed-Rab18 with the markers of the classic endocytosis pathway (EEA1, EGFP-Rab7 and lysosome). We observed that Rab18 has co-localized with EEA1 positive EEs and lysosomes. However, Rab18 did not merge with EGFP-Rab7 and EGFP-Rab9 positive LEs (Fig. 4b–e). Thus, we identified another novel endocytosis pathway involved in the transport of the nanogels: the EE-lipid droplet (Rab18 positive)-lysosome pathway.
Nanogels transported by the recycling endosome pathway
The protein receptors on the cell membranes are recycled by the recycling endosome pathway always.27 Rab4, 11, 15, 17, 20, 25 and 35 are the main regulators of the formation of the recycling endosomes.27,28 Rab11 and Rab35 regulate slow endocytic recycling derived from EEs.26,27 Rab4 mediates fast endocytic recycling from EEs.26,27 Rab15, 17, 20 and Rab25 control trafficking through the apical recycling endosomes to the apical plasma membrane.26,27,29 There is seldom report about the delivery of nanoparticles through the recycling endosome system. To detect whether nanogels could be transported through the recycling endosomes, the Rho-labeled nanogels were incubated with EGFP-Rab4, 11, 20, 25 and 35 transfected MCF-7 cells for 20 h. Rho-labeled nanogels localized with Rab35 positive slow recycling endosomes were observed (Fig. 4a and b). However, Rab4 positive fast recycling endosomes or Rab20 and Rab25 positive apical recycling endosomes does not contain the Rho-labeled nanogels (Fig. S4b–f†). These data suggested that nanogels may be delivered out of the cells through slow recycling endosomes. Therefore, we found that nanogels could be incepted by the MCF-7 cells through multiple proteins dependent endocytosis and Rab34 positive micropinocytosis (Fig. 1d). EE-LE-lysosome and EE-lipid droplet-lysosome pathways are two novel pathways responsive for the nanogel delivery within the cells (Fig. 1d). Moreover, nanogels appeared in the Rab positive slow and apical recycling endosomes indicated that the cells may use the recycling pathway to transport the particles out of the cells (Fig. S4†).Nanogels were delivered through the exocytosis pathway
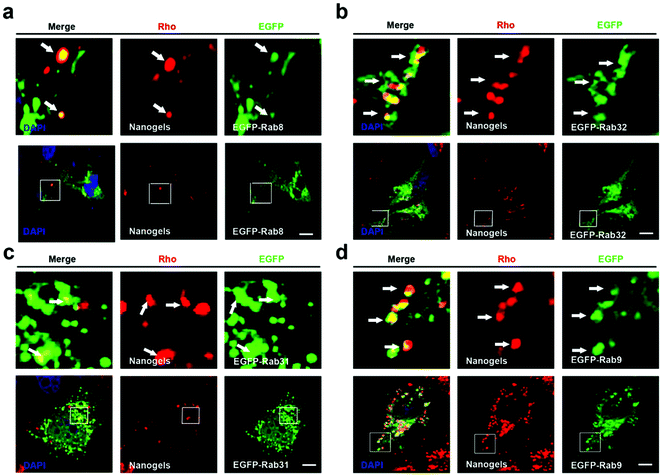
Cells employ exocytosis to deliver secretory vesicles into the extracellular space.30,31 Rab proteins such as Rab3, 8, 10, 14, 27, 32, 38 also regulate the formation, transport and fusion of these secretory vesicles.26,28 Among these Rab proteins, Rab3, Rab26, Rab27 and Rab37 mediate classic secretory vesicles.26,28 Rab8, Rab10 and Rab14 control the GLUT4 vesicle formation and delivery.26,28 Rab27, Rab32 and Rab38 regulate the melanosomes formation and delivery.26,27 To detect whether nanogels could be transported out of the cell through the exocytosis pathway, rho-labeled nanogels were incubated with EGFP-Rab3, Rab26, Rab37, Rab8, Rab10, Rab14, Rab27, Rab32, Rab38 transfected cells for 20 h (Table 1). We observed that DsRed-Rab8, 10 positive vesicles (GLUT4 vesicle) contain the Rho-labeled nanogels and Rab32, 38 positive melanosome homologous vesicles also contain the nanogels (Fig. 5a, b and S5a, b†). However, EGFP-Rab3 and 26 positive secretory vesicles did not sequester the nanogels (Fig. S5c and d†). These data indicated that nanogels may be secreted to the extracellular environment through GLUT4 vesicles and the melanosome homologue vesicles in MCF-7 cells (Fig. 1d).GLUT4 transport vesicles are derived from the TGN.26 The nanogels have to be transported to the Golgi from EEs and LEs.26 Rab14, Rab22 and Rab31 vesicles deliver cargoes from EEs to the TGN.26 Moreover, Rab9 positive vesicles transport proteins from LEs to the TGN.26 To investigate the pathways by which nanogels were transported to the TGN, Rho-labeled nanogels were incubated with EGFP-Rab14, 22, 31 transfected MCF-7 cells for 20 h. We observed that EGFP-Rab9, 14, 22, 31 positive vesicles contained the Rho-labeled nanogels (Fig. 5c and S6†). Moreover, to investigate whether the nanogels were also found in LEs, EGFP-Rab9 transfected MCF-7 cells were incubated with Rho-labeled nanogels for 20 h. Then, we observed that EGFP-Rab9 positive vesicles sequestered red nanogels (Fig. 5d). These data indicated that the nanogels were transported from EEs and LEs to the TGN, and finally delivered out of the cells through the exocytosis pathways.
Nanogels turnover by the autophagy pathway
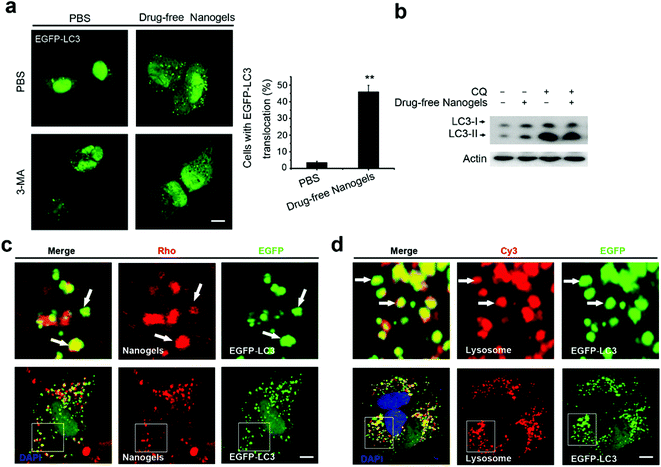
Autophagy is a cellular process for cells to under go stress condition. Intracellular cell membrane sequesters the aggregated proteins, damaged organelles and even the invaded pathogens such as viruses to autophagosomes and transports them to lysosome for degradation.20,32 Biodegradable nanoparticles, PLGA-based nanoparticles and micelles have been reported to be degraded through the autophagy pathway.33 The components and structure of nanogels are similar to the protein aggregates; therefore, autophagy may also sequester these nanogels.32 To observe autophagy, autophagosome (LC3-positive puncture) formation and the production of the LC3II protein was measured.21 LC3I proteins are cleaved on the C-terminal and LC3II proteins are produced which are finally transferred on the membrane of autophagosomes when autophagy is initiated.21 To detect whether the nanogels induce autophagy, EGFP-LC3 transfected MCF-7 cells were incubated with non-labeled drug-free nanogels for 20 h. After incubation, substantial autophagosomes were observed within the cells (Fig. 6a). The production of the LC3-II protein was also increased in the cells (Fig. 6b). To further investigate whether nanogels could be degraded through the autophagy pathway, we used the green fluorescence labeled LC3 protein (EGFP-LC3) to detect the autophagy of Rho-labeled nanogels. EGFP-LC3 transfected MCF-7 cells were incubated with the Rho-labeled nanogels for 20 h. After the incubation, the Rho-labeled nanogels were found to be sequestered within the EGFP-LC3 positive autophagosomes (Fig. 6c). Finally, the autophagosomes were delivered to lysosomes for the degradation of the nanogels and cargoes (Fig. 6d). These data indicate that autophagy pathways also could sequester the nanogels and degrade the cargoes by lysosome.Intracellular network of autophagy, endocytosis and exocytosis
Atg proteins are the main regulators of autophagy, while Rab proteins also show an important role in autophagy.34 Some proteins such as Rab5, Rab7, Rab8, Rab9, Rab11, Rab20, Rab32 and Rab33 have been shown to participate in the regulation of autophagy at different stages.34 Thus, the endocytosis and exocytosis vesicles may fuse with autophagosomes and have cross talk with autophagy.21 We observed that autophagosomes contained biodegradable nanoparticles such as PLGA-based NPs and micelles, previously.7,35 The autophagosomes may sequester the nanoparticles from the cytoplasm or there may be some other mechanism. The fusion of endocytosis vesicles with autophagosomes is another possible way for the autophagosomes to receive the nanoparticles besides direct sequestration from cell plasma. By screening the co-localization between the autophagosomes and the Rab positive endocytosis and exocytosis vesicles, we found that autophagosomes have co-localization with Rab7, Rab18, Rab23 and Rab34 positive vesicles (Fig. S7a–d†). These data suggested that autophagosomes may receive the nanogels by fusion with these endocytosis vesicles.We also detected the relationship between recycling endosomes and autophagosomes. MCF-7 cells transfected with EGFP-LC3 and DsRed-Rab4 (fast recycling endosomes), Rab11, Rab35 (slow recycling endosomes), Rab15, Rab17 and Rab25 (apical recycling endosomes) were observed by confocal microscopy. The Co-localization between DsRed-Rab11 and Rab35 with EGFP-LC3 positive autophagosomes was observed (Fig. S7e and f†). These data suggested that autophagosomes may also receive the nanogels through the recycling endosomes. To detect the relationship of autophagosomes with exocytosis vesicles, MCF-7 cells were transfected with EGFP-LC3 and DsRed-Rab3, Rab26, Rab37 (secretory vesicles), Rab8, Rab10, Rab14 (GLUT4 vesicles), Rab32, and Rab38 (melanosome). We observed that Rab8 and Rab10 positive GLUT4 vesicles merged with autophagosomes (Fig. S7g and h†). EGFP-LC3 positive autophagosomes did not co-localize with DsRed-Rab3, 26 positive secretory vesicles and Rab32, 38 positive melanosome homologue vesicles (Fig. S8a–c†). These data suggested that Rab11 positive slow recycling endosomes or Rab8 positive GLUT4 vesicles may deliver the nanogels to autophagosomes and degrade the nanogels through the autophagy pathways.
Autophagy inhibitor CQ and 3-MA inhibited DOX and DOX-loaded nanogels induced autophagy
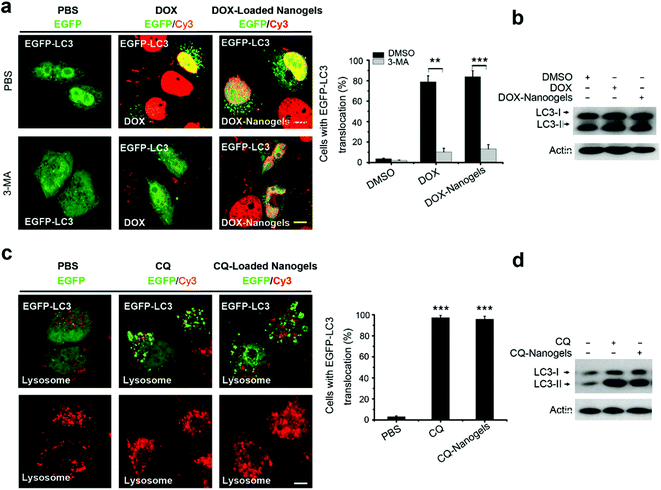
Most chemotherapeutic drugs could induce autophagy to arouse drug resistance.32,35–37 Autophagy is a double-edged sword. On one hand, autophagy can suppress tumorigenesis. On the other hand, however, autophagy can confer stress tolerance for survival of cancer cells once tumorigenesis occurs.20 We took doxorubicin (DOX) as an example drug to investigate whether autophagy is induced by both the drug and the drug loaded-nanogel NPs. We loaded DOX in nanogels and detected the drug release profile firstly. Compared to DOX, the release of CQ is faster. Over 50% of CQ was released in the beginning 2 hours, while only 15% DOX was released in the same time period (Fig. S9†). More than 95% was released in a 12-hour period, while only 50% of DOX were released in the same time period (Fig. S9†). Next, we investigated autophagy induction by DOX and DOX-loaded nanogels. As expected, after treatment of EGFP-LC3 transfected MCF-7 cells with DOX for 24 h, it was found that substantial autophagosomes were induced with the cells, and the protein level of LC3II also increased (Fig. 7a and b). Treatment of the cells with DOX-loaded nanogels also significantly induced autophagosome formation (Fig. 7a and b). Vps34 inhibitor 3-methyladenine (3-MA) could efficiently inhibit both DOX and DOX-loaded nanogel induction of autophagy. CQ is a lysosome inhibitor, which could disrupt lysosomes and block the autophagy flux.20 Thus, CQ could block the DOX induced autophagy, and inhibit the cells using autophagy to resist the apoptosis induced by DOX. Treatment of MCF-7 cells with CQ lead to the accumulation of autophagosomes by inducing lysosomal membrane permeabilization (LMP) (Fig. 7c). The protein levels of LC3II also increased (Fig. 7d). We loaded CQ in the nanogels alone or co-loaded CQ with DOX. Treatment of the MCF-7 cells with CQ-loaded nanogels could lead to the accumulation of autophagosomes by inducing LMP (Fig. 7c). Co-treatment of MCF-7 cells with CQ-loaded nanogels NPs and DOX-loaded nanogels significantly blocked the autophagy flux and lead to the accumulation of autophagosomes (Fig. 7d). These data demonstrated that co-treatment of these drugs with autophagy inhibitors may enhance the efficacy of nanomedicine.Autophagy inhibitor CQ enhances the tumor suppression effect of DOX by blocking the autophagy flux in vivo
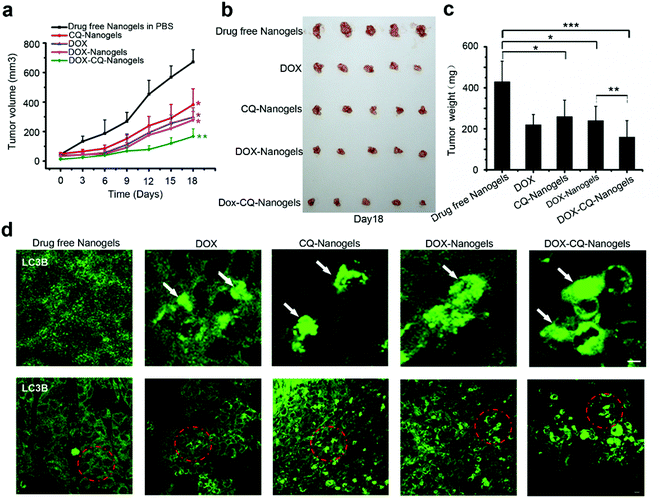
Autophagy inhibitors are used in combination with chemotherapeutic drugs to enhance intracellular drug delivery by nanoparticle formulation. In order to investigate the curative effect of chemotherapeutic drug-loaded nanogels in combination with autophagy inhibitors, the xenograft SCID mice model with the MCF-7 cell line implant was also used. The mice received intraperitoneal injections of drug-free nanogels, DOX, DOX-nanogels, CQ-nanogels and DOX-CQ-nanogels every 3 days for six consecutive cycles. The tumor size of the mice was recorded every 3 days until the 18th day. Control mice were injected with physiological saline. Tumor growth was surveyed for 18 days as shown in Fig. 8a. Injection of CQ-loaded nanogels had an inhibitory effect on the tumor growth (Fig. 8a–c). The DOX and DOX-loaded nanogels had almost the same effect in tumor growth suppression (Fig. 8a–c). Interestingly, the mice were treated with the CQ and DOX co-loaded nanogels, had significant suppression of the tumor growth (Fig. 8a–c). In order to confirm whether DOX could induce autophagy in tumor tissue and CQ could block the fusion of autophagosomes with the lysosome during treatment, immunofluorescence histochemistry assay was performed to detect the distribution of the endogenous LC3 protein. Interestingly, we found that substantial autophagosomes were induced in the tissue in the mice were treated with DOX and the DOX-loaded nanogels (Fig. 8d). However, when we treated the mice with the CQ and DOX co-loaded nanogels, the autophagosomes accumulated significantly (Fig. 8d). The xenograft tumor model demonstrated that inhibition of autophagy could significantly enhance the curative effect of chemotherapeutic drug-loaded nanogels. The co-loaded CQ with DOX significantly inhibited the cancer cell growth in vivo compared with that using the DOX-loaded nanogels alone. The tumor cells used autophagy to resist the DOX induced cell death. When we co-treated the mice with CQ, the fusion of autophagosomes with lysosomes was blocked, which lead to the accumulation of the autophagosomes within the tumor cells. Inhibition of autophagy could also cut down the tumor cells using autophagy to provide energy and nutrients for survival. Therefore, combining chemotherapeutic drugs with nanogels loaded with the autophagy inhibitor CQ holds great promise as a novel cancer treatment strategy.Conclusions
In this paper, we found that Rho-labeled nanogels were incepted by the cells mainly through micropinocytosis and multiple protein dependent endocytosis pathways. Besides the EE-LE-lysosome pathway, we found some other intracellular pathways: the micropinocytosis-LE-lysosome pathway and the EE-lipid droplet-lysosome pathway. Moreover, slow endocytic recycling, GLUT4 transport vesicles and melanosomes may transport the nanogels out of the cells. Besides endocytosis and exocytosis, nanogels were also found in autophagosomes (LC3 positive). The crosstalk between autophagosomes and endocytosis or exocytosis was identified. The fusion of these endocytosis and exocytosis vesicles with autophagosomes may provide another way for the autophagosomes to receive the nanogels (Fig. 9). The co-loaded CQ and DOX significantly inhibit cancer cell growth in vivo in the xenograft tumor model. The use of Rab proteins as vesicle markers to investigate the systematic intracellular trafficking of the nanogels will provide new targets to interfere with the cellular behaviour of the nanoparticles, and improve the therapeutic effect of nanomedicine.Author contributions
Z. Gu, L. Mei and Y. Zhang initiated and directed the study. X. Zhang, Z. Chen and X. Zeng designed the experiments. X. Zhang, X. Liang, J. Gu, D. Chang, J. Zhang, W. Tao and G. Liu performed the experiments. X. Zhang, Y. Ye, C. Wang interpreted the data. X. Zhang and L. Mei wrote the paper. The manuscript was written through contributions of all the authors. All the authors have given approval to the final version of the manuscript.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31270019, 51203085), the National Natural Science Foundation of China (Grants No. 21174070 and 21374048), the National Natural Science Foundation of China (No. 31270019), the Guangdong Natural Science Funds for Distinguished Young Scholar (No. 2014A030306036), the China Postdoctoral Science Foundation (No. 2015M580109), the Scientific and Technological Innovation Bureau of Nanshan District (No. KC2014JSCX0023A), and the Science, Technology & Innovation Commission of Shenzhen Municipality (No. JCYJ20150430163009479, JCYJ20150529164918738, CYZZ 20130320110255352) and Sloan Research Fellowship to Z. Gu.Notes and references
- F. Jia, X. Liu, L. Li, S. Mallapragada, B. Narasimhan and Q. Wang, J. Controlled Release, 2013, 172, 1020–1034 CrossRef CAS PubMed.
- J. Cao, R. Wang, N. Gao, M. Li, X. Tian, W. Yang, Y. Ruan, C. Zhou, G. Wang, X. Liu, S. Tang, Y. Yu, Y. Liu, G. Sun, H. Peng and Q. Wang, Biomater. Sci., 2015, 3, 1545–1554 RSC.
- Q. Wang, H. Cheng, H. Peng, H. Zhou, P. Y. Li and R. Langer, Adv. Drug Delivery Rev., 2015, 91, 125–140 CrossRef CAS PubMed.
- M. Hamidi, A. Azadi and P. Rafiei, Adv. Drug Delivery Rev., 2008, 60, 1638–1649 CrossRef CAS PubMed.
- S. V. Vinogradov, Curr. Pharm. Des., 2006, 12, 4703–4712 CrossRef CAS PubMed.
- S. Merino, C. Martin, K. Kostarelos, M. Prato and E. Vazquez, ACS Nano, 2015, 9, 4686–4697 CrossRef CAS PubMed.
- H. J. Kim, K. Zhang, L. Moore and D. Ho, ACS Nano, 2014, 8, 2998–3005 CrossRef CAS PubMed.
- R. Beyer, S. Iacopini, T. Palberg and H. J. Schope, J. Chem. Phys., 2012, 136, 234906 CrossRef PubMed.
- S. V. Vinogradov, T. K. Bronich and A. V. Kabanov, Adv. Drug Delivery Rev., 2002, 54, 135–147 CrossRef CAS PubMed.
- A. Albanese, A. K. Lam, E. A. Sykes, J. V. Rocheleau and W. C. Chan, Nat. Commun., 2013, 4, 2718 Search PubMed.
- B. H. Rihn and O. Joubert, ACS Nano, 2015, 9, 5634–5635 CrossRef CAS PubMed.
- E. A. Sykes, J. Chen, G. Zheng and W. C. Chan, ACS Nano, 2014, 8, 5696–5706 CrossRef CAS PubMed.
- S. Sindhwani, A. M. Syed, S. Wilhelm, D. R. Glancy, Y. Y. Chen, M. Dobosz and W. C. Chan, ACS Nano, 2016, 10, 5468–5478 CrossRef CAS PubMed.
- G. Sahay, D. Y. Alakhova and A. V. Kabanov, J. Controlled Release, 2010, 145, 182–195 CrossRef CAS PubMed.
- A. Akinc and G. Battaglia, Cold Spring Harbor Perspect. Biol., 2013, 5, a016980 CrossRef PubMed.
- G. Chang, C. Li, W. Lu and J. Ding, Macromol. Biosci., 2010, 10, 1248–1256 CrossRef CAS PubMed.
- D. M. Copolovici, K. Langel, E. Eriste and U. Langel, ACS Nano, 2014, 8, 1972–1994 CrossRef CAS PubMed.
- A. Chugh, F. Eudes and Y. S. Shim, IUBMB Life, 2010, 62, 183–193 CrossRef CAS PubMed.
- J. Huotari and A. Helenius, EMBO J., 2011, 30, 3481–3500 CrossRef CAS PubMed.
- N. Mizushima and M. Komatsu, Cell, 2011, 147, 728–741 CrossRef CAS PubMed.
- B. Levine and G. Kroemer, Cell, 2008, 132, 27–42 CrossRef CAS PubMed.
- A. H. Hutagalung and P. J. Novick, Physiol. Rev., 2011, 91, 119–149 CrossRef CAS PubMed.
- K. Matsunaga, T. Saitoh, K. Tabata, H. Omori, T. Satoh, N. Kurotori, I. Maejima, K. Shirahama-Noda, T. Ichimura, T. Isobe, S. Akira, T. Noda and T. Yoshimori, Nat. Cell Biol., 2009, 11, 385–396 CrossRef CAS PubMed.
- X. Zhang, H. Zhang, X. Liang, J. Zhang, W. Tao, X. Zhu, D. Chang, X. Zeng, G. Liu and L. Mei, Mol. Pharm., 2016, 13, 2578–2587 CrossRef CAS PubMed.
- P. J. Cullen, Nat. Rev. Mol. Cell Biol., 2008, 9, 574–582 CrossRef CAS PubMed.
- H. Stenmark, Nat. Rev. Mol. Cell Biol., 2009, 10, 513–525 CrossRef CAS PubMed.
- I. Jordens, M. Marsman, C. Kuijl and J. Neefjes, Traffic, 2005, 6, 1070–1077 CrossRef CAS PubMed.
- T. Bhuin and J. K. Roy, Exp. Cell Res., 2014, 328, 1–19 CrossRef CAS PubMed.
- S. Ozeki, J. Cheng, K. Tauchi-Sato, N. Hatano, H. Taniguchi and T. Fujimoto, J. Cell Sci., 2005, 118, 2601–2611 CrossRef CAS PubMed.
- T. C. Sudhof and J. Rizo, Cold Spring Harbor Perspect. Biol., 2011, 3(12), a005637 Search PubMed.
- S. Mochida, Neurosci. Res., 2011, 70, 16–23 CrossRef PubMed.
- Y. M. Kim, C. H. Jung, M. Seo, E. K. Kim, J. M. Park, S. S. Bae and D. H. Kim, Mol. Cell, 2015, 57, 207–218 CrossRef CAS PubMed.
- X. Zhang, X. Zeng, X. Liang, Y. Yang, X. Li, H. Chen, L. Huang, L. Mei and S. S. Feng, Biomaterials, 2014, 35, 9144–9154 CrossRef CAS PubMed.
- Z. Szatmari and M. Sass, Autophagy, 2014, 10, 1154–1166 CrossRef PubMed.
- X. Zhang, Y. Dong, X. Zeng, X. Liang, X. Li, W. Tao, H. Chen, Y. Jiang, L. Mei and S. S. Feng, Biomaterials, 2014, 35, 1932–1943 CrossRef CAS PubMed.
- M. Li, H. Yu, T. Wang, N. Chang, J. Zhang, D. Du, M. Liu, S. Sun, R. Wang, H. Tao, S. Geng, Z. Shen, Q. Wang and H. Peng, J. Mater. Chem. B, 2014, 2, 1619–1625 RSC.
- Y. Lu, A. Almetta, R. Langer and Z. Gu, Nat. Rev. Mater., 2016, 1, 16075 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6nr07866d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |