Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A set of robust fluorescent peptide probes for quantification of Cu(II) binding affinities in the micromolar to femtomolar range†
Tessa R.
Young
,
Chathuri J. K.
Wijekoon
,
Benjamin
Spyrou
,
Paul S.
Donnelly
,
Anthony G.
Wedd
and
Zhiguang
Xiao
*
School of Chemistry and The Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, Victoria 3010, Australia. E-mail: z.xiao@unimelb.edu.au; Fax: +61 3 9347 5180; Tel: +61 3 9035 6072
First published on 11th February 2015
Abstract
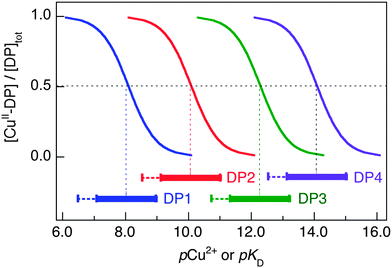
Reliable quantification of copper binding affinities and identification of the binding sites provide a molecular basis for an understanding of the nutritional roles and toxic effects of copper ions. Sets of chromophoric probes are now available that can quantify Cu(I) binding affinities from nanomolar to attomolar concentrations on a unified scale under in vitro conditions. Equivalent probes for Cu(II) are lacking. This work reports development of a set of four fluorescent dansyl peptide probes (DP1–4) that can quantify Cu(II) binding affinities from micromolar to femtomolar concentrations, also on a unified scale. The probes were constructed by conjugation of a dansyl group to four short peptides of specific design. Each was characterised by its dissociation constant KD, its pH dependence and the nature of its binding site. One equivalent of Cu(II) is bound by the individual probes that display different and well-separated affinities at pH 7.4 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −8.1, −10.1, −12.3 and −14.1, respectively). Intense fluorescence is emitted at λmax ∼ 550 nm upon excitation at ∼330 nm. Binding of Cu(II) quenches the fluorescence intensity linearly until one equivalent of Cu(II) is bound. Multiple approaches and multiple affinity standards were employed to ensure reliability. Selected examples of application to well-characterised Cu(II) binding peptides and proteins are presented. These include Aβ16 peptides, two naturally occurring Cu(II)-chelating motifs in human serum and cerebrospinal fluid with sequences GHK and DAHK and two copper binding proteins, CopC from Pseudomonas syringae and PcoC from Escherichia coli. Previously reported affinities are reproduced, demonstrating that peptides DP1–4 form a set of robust and reliable probes for Cu(II) binding to peptides and protein targets.
KD = −8.1, −10.1, −12.3 and −14.1, respectively). Intense fluorescence is emitted at λmax ∼ 550 nm upon excitation at ∼330 nm. Binding of Cu(II) quenches the fluorescence intensity linearly until one equivalent of Cu(II) is bound. Multiple approaches and multiple affinity standards were employed to ensure reliability. Selected examples of application to well-characterised Cu(II) binding peptides and proteins are presented. These include Aβ16 peptides, two naturally occurring Cu(II)-chelating motifs in human serum and cerebrospinal fluid with sequences GHK and DAHK and two copper binding proteins, CopC from Pseudomonas syringae and PcoC from Escherichia coli. Previously reported affinities are reproduced, demonstrating that peptides DP1–4 form a set of robust and reliable probes for Cu(II) binding to peptides and protein targets.
Introduction
Copper is a redox-active metal whose relevant oxidation states Cu(I) and Cu(II) bind with high affinities to many sites in biomolecules. These properties confer vital roles in cellular respiration, antioxidant defense, iron uptake, connective tissue formation, pigment synthesis and photosynthesis.1 However, when not under proper control, these same properties render copper toxic to cells. Then, undesirable redox chemistry may lead to catalytic generation of reactive oxygen species (ROS) while the dominant affinities result in displacement of other essential metals from their native sites. Consequently, a delicate balance must be maintained between deficiency and excess and specific homeostatic mechanisms have evolved to control copper metabolism.2–4 Errors in cellular copper handling appear to contribute to a range of inherited and acquired diseases. Genetic mutations invoke Menkes' and Wilson's diseases, conditions of, respectively, copper deficiency and overload. A range of neurodegenerative diseases have been linked to aberrant copper metabolism and include Alzheimer's, prion and Parkinson's diseases.5–9Reliable quantitative evaluation of copper–protein interactions for relevant oxidation states provides an essential key to understanding the molecular basis of nutritional roles and toxic effects. Such evaluation must be based on a combination of reliable affinity standards and sensitive detection probes.10 We have previously established a set of classic chromophoric ligands that are capable of quantifying Cu(I) binding in proteins with affinities within the range from nanomolar to attomolar concentrations in in vitro systems.11–15 Their establishment has unified the scattered literature data to a single affinity scale.10,13,15 Promising alternative probes for Cu(I) have been developed.16
In contrast, quantitative evaluation of binding affinities for Cu(II) sites in proteins and peptides currently rely largely on competition with a range of non-chromophoric ligands including those based on polydentate amine carboxylates such as ethylenediaminetetraacetate (Edta), ethylene glycol tetraacetate (Egta), N-(2-hydroxyethyl)ethylenediamine-N,N′,N′-triacetate (Hedta) and nitrilotriacetate (Nta) or amino acids such as glycine (Gly) and histidine (His).10 However, whilst the affinities of these ligands are well-documented, their Cu(II) complexes are ‘invisible’ and cannot act as direct detection probes. Their use depends upon observation of a change in an inherent property of the protein target itself upon the binding of Cu(II) (fluorescence, absorbance, circular dichroism).10,17 The availability of such properties depends on the target itself.
Isothermal titration calorimetry (ITC) has been employed extensively as a detection probe.18 The approach relies on the measurement of the heat generated during the course of metal-binding but suffers in complex systems from interfering heat release from accompanying processes such as dilution, precipitation or the presence of (unappreciated) competing ligands such as buffers. In addition, time constraints between the consecutive ITC titrations can be limiting as Cu(II) transfer tends to be sluggish. Such aspects seem to account for the scattered literature data for Cu(II) binding affinities, as highlighted by the cases of protein domains or peptides associated with neurodegenerative diseases.17 Consequently, there remains a need for reliable probes capable of direct quantification of Cu(II) binding with high sensitivity.
Recently, the Aβ16 peptide was modified by replacement of Tyr10 with the fluorescent Trp residue.19 This system proved to be a useful probe for differentiating the Cu(II) binding affinities of different forms of the Aβ peptide. However, it appears to be subject to interference from binding of a second equivalent of Cu(II) to a weaker binding site. The system was converted to a more sensitive and selective probe (denoted as Aβ16wwa) by replacement of Phe4 with Trp and His14 with Ala (conditional log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −9.8 at pH 7.4).20 However, the use of Aβ16wwa is also limited due to an interfering response from those protein targets that also contain Trp residues plus an ‘inner filter’ effect of Cu(II) species at ∼280 nm, its optimal excitation wavelength.
KD = −9.8 at pH 7.4).20 However, the use of Aβ16wwa is also limited due to an interfering response from those protein targets that also contain Trp residues plus an ‘inner filter’ effect of Cu(II) species at ∼280 nm, its optimal excitation wavelength.
Fluorescent probes for Cu(II) with red-shifted excitation have been reported (see recent reviews21,22) including several containing one or two sensitive dansyl groups (DNS) attached to primary amine group(s) in an amino acid or a short peptide,23–26 as well as several biosensors based on fluorescence-labelled carbonic anhydrases.27,28 However, these probes were either not demonstrated to be practical for quantification of Cu(II) affinities in biomolecules or not readily available for the practical applications.
This paper reports a set of four new sensitive and quantitative probes for Cu(II) that were constructed by conjugation of DNS to four different short peptides. They each bind one equiv. of Cu(II) cleanly with varying binding modes, binding affinities and pH dependency and are able to probe Cu(II) binding sites and define Cu(II) binding affinities from micromolar to femtomolar concentrations on a unified scale. Examples of their application to peptide and protein targets are provided.
Experimental section
Materials and general procedure
Ligands Gly, His, Egta, Edta and Hedta were purchased from Sigma-Aldrich and were used as received. Concentrations of Egta, Edta and Hedta were calibrated via Cu(II) titration into a mixture of ligand and probe DP2 (cf.Fig. 9 below). A Cu(II) standard was prepared by dissolving CuSO4 in Milli-Q water and its concentration calibrated via reaction with excess Cu(I) ligand bathocupröine disulfonate (Bcs) in buffer 3-(N-morpholino)propanesulfonate (MOPS) containing reductant NH2OH. Under such conditions, all copper ions were converted quantitatively to the well-defined chromophoric complex anion [CuI(Bcs)2]3− with a strong absorbance at 483 nm (ε = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).12 Proteins CopC from Pseudomonas syringae and PcoC from Escherichia coli were expressed and isolated as reported.29,30 Their concentrations were estimated from their solution absorbances at 280 nm using reported extinction coefficients of ε(280) = 8700 and 10
000 M−1 cm−1).12 Proteins CopC from Pseudomonas syringae and PcoC from Escherichia coli were expressed and isolated as reported.29,30 Their concentrations were estimated from their solution absorbances at 280 nm using reported extinction coefficients of ε(280) = 8700 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 M−1 cm−1, respectively.29,30 Peptides Aβ16 (amyloid-β peptide: DAEFRHDSGYEVHHQK), Ac-Aβ16 (N-terminus acetylated) and Aβ16wwa (sequence: DAEWRHDSGWEVHAQK) were synthesised on site by solid phase peptide techniques. Identity was verified by electrospray ionisation mass spectrometry (ESI-MS) while purity was confirmed by HPLC to be >98%. Peptide concentrations were estimated from absorbance maxima at ∼276 nm using εmax = 1410 M−1 cm−1 for those Aβ16 peptides containing a single tyrosine residue and ε = 11
100 M−1 cm−1, respectively.29,30 Peptides Aβ16 (amyloid-β peptide: DAEFRHDSGYEVHHQK), Ac-Aβ16 (N-terminus acetylated) and Aβ16wwa (sequence: DAEWRHDSGWEVHAQK) were synthesised on site by solid phase peptide techniques. Identity was verified by electrospray ionisation mass spectrometry (ESI-MS) while purity was confirmed by HPLC to be >98%. Peptide concentrations were estimated from absorbance maxima at ∼276 nm using εmax = 1410 M−1 cm−1 for those Aβ16 peptides containing a single tyrosine residue and ε = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 at 280 nm for the Aβ16wwa peptide containing two Trp residues. The concentrations obtained matched those estimated from fluorescence titrations with the copper standard assuming formation of a 1
000 M−1 cm−1 at 280 nm for the Aβ16wwa peptide containing two Trp residues. The concentrations obtained matched those estimated from fluorescence titrations with the copper standard assuming formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex in each case.
1 complex in each case.
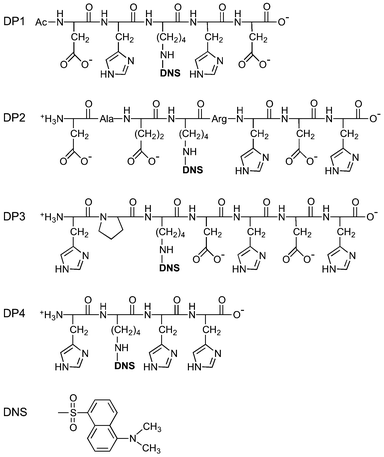
The four peptide probes listed in Table 1 were custom synthesised by GL Biochem (Shanghai), together with two shorter peptides with sequences GHK and DAHK. The dansyl fluorophore was introduced at the ε-amino group of the sole lysine residue in the sequence (denoted as KDNS; Fig. 1) using Fmoc-L-Lys(dansyl)-OH. The identity and purity (>98%) of each probe was confirmed by ESI-MS and HPLC (Table 1). The solution spectrum of each probe features an intense absorbance in the near UV region with λmax = 326 nm, characteristic of a single dansyl group (cf., Fig. 2, cyan trace for DP1–4 versus orange trace for DAHK). The maximal molar extinction coefficient at 326 nm was determined accurately via titration of each probe solution with a CuSO4 standard (Fig. S2, ESI†) to be 4500 M−1 cm−1 for each probe. This value is within the range reported previously for several dansyl derivatives31,32 and was used in this work to calibrate the probe concentration. The concentrations of GHK and DAHK were determined via titration with the CuSO4 standard using DP2 as a probe (vide infra; Fig. 9).
| Probe | Sequence | Net chargea | Molar mass (Da) | (F1/F0) at given pHb | |||
|---|---|---|---|---|---|---|---|
| Calcd | Found | 6.2 | 7.4 | 9.2 | |||
| a At pH 7.4. b F 1 and F0 are the fluorescence intensities upon binding 1.0 and 0.0 equiv. of Cu(II), respectively, under the same pH conditions and the F1 values were determined directly from the titration turning point unless otherwise indicated. c This F1 value was obtained from the best curve fitting of the experimental data to eqn (4) (see Fig. 4b). | |||||||
| DP1 | Ac-DH(KDNS)HD | −3 | 925.9 | 925.3 | — | 0.15c | 0.06 |
| DP2 | DAE(KDNS)RHDH | −2 | 1240.3 | 1240.5 | 0.22 | 0.17 | 0.17 |
| DP3 | HP(KDNS)DHDH | −2 | 1118.2 | 1118.4 | 0.13 | 0.13 | 0.13 |
| DP4 | H(KDNS)HH | 0 | 790.8 | 791.3 | 0.09 | 0.09 | 0.09 |
The experimental results were highly dependent on pH. To ensure accuracy and reliability, the solution pH was controlled within ±0.05 pH units and checked before and after each experiment.
Spectroscopic approaches
UV-visible spectra were recorded on a Varian Cary 300 spectrophotometer in dual beam mode with quartz cuvettes of 1.0 cm path length. All titrations with metal ions were performed in appropriate buffers and corrected for baseline and dilution. Fluorescence emission spectra were recorded on a Varian Cary Eclipse spectrophotometer with a band pass of 20 nm for both excitation and emission. The excitation wavelength for the dansyl probes was λ = 330 nm and the emission spectra were recorded between λ = 450–750 nm at a scale rate of λ = 600 nm min−1. The excitation wavelength for other peptides or proteins was 280 nm with a recorded spectral range between 295–500 nm. Visible CD spectra were recorded on a Chirascan-plus spectrometer (Applied Photophysics) using a 1.0 cm cell. Three scans were averaged and a baseline was subtracted from each recorded spectrum.Electron paramagnetic resonance (EPR) spectra were recorded on Bruker Elexsys E 500 EPR spectrometer. The samples were prepared by adding 0.9 equivalent Cu(II) into apo-peptides (each 0.5 mM) in an appropriate buffer (50 mM) containing ∼10% glycerol. The samples were snap-frozen in liquid nitrogen and the spectra recorded at 77 K in a liquid-nitrogen finger Dewar. The EPR parameters (g values and A values) were extracted directly from the recorded spectra.
Quantification of Cu(II) binding via direct metal ion titration
Each DP probe emits intense fluorescence at λmax ∼ 550 nm upon excitation at ∼330 nm. Cu(II) binding quenches the fluorescence intensity linearly until the binding site(s) are saturated with Cu(II). The binding stoichiometry was determined in each case by direct titration of the probe solution (2.0 mL) in an appropriate buffer with a CuSO4 standard at a concentration 40 times that of the probe. For those probes with weaker Cu(II) affinity, the titration was conducted with high probe concentration and/or in a buffer with elevated pH to promote quantitative binding of the added Cuaq2+ ions to achieve better definition of the titration turning point.When the titration was conducted with lower probe concentration, the turning point may become less well-defined, suggesting the existence of a binding equilibrium (eqn (1)) under the constraint of KD′ ∼ [P] and thus [CuIIP]/[Cu(II)]tot < 1 in eqn (2):20
| Cuaq2+ + 2P + ‘CuIIB’ ⇌ 2CuIIP + B | (1) |
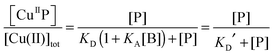 | (2) |
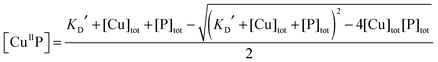 | (3) |
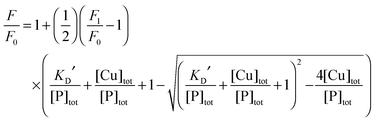 | (4) |
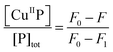 | (5) |
Quantification of Cu(II) binding affinities via ligand competition
For those probes with a sufficiently high affinity for Cu(II), the condition KD′ ∼ [P] cannot be met even at the experimentally accessible minimal probe concentrations (cf.eqn (1) and (2)). Then, a competition for Cu(II) with a suitable ligand of well-defined Cu(II) binding affinity is required for reliable quantification. In this work, the classic Cu(II) ligands L = Gly, His, Egta and Hedta (Fig. S1, ESI†) were employed. The two amino acid ligands react with Cu(II) to yield both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexes while the others form a 1
2 complexes while the others form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex only with known formation constants (Table S1, ESI†). They may compete for Cu(II) with probes P according to eqn (6a) or (6b) under conditions where the contributions of free Cuaq2+ and putative buffer complex ‘CuII–B’ to the total Cu(II) speciation are negligible. Then the free Cuaq2+ concentration in the competing solutions may be estimated viaeqn (7a) or (7b) and the pH-dependent conditional KD for CuII–P derived via curve-fitting of the experimental data to eqn (8):10
1 complex only with known formation constants (Table S1, ESI†). They may compete for Cu(II) with probes P according to eqn (6a) or (6b) under conditions where the contributions of free Cuaq2+ and putative buffer complex ‘CuII–B’ to the total Cu(II) speciation are negligible. Then the free Cuaq2+ concentration in the competing solutions may be estimated viaeqn (7a) or (7b) and the pH-dependent conditional KD for CuII–P derived via curve-fitting of the experimental data to eqn (8):10| CuIIP + 3L ⇌ P + CuIIL + CuIIL2 (L = Gly or His) | (6a) |
| CuIIP + L ⇌ P + CuIIL (L = Egta or Hedta) | (6b) |
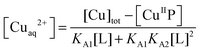 | (7a) |
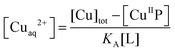 | (7b) |
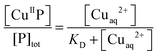 | (8) |
The experiments were conducted in buffers 2-(N-morpholino)ethanesulfonate (MES, pH 6.2), MOPS (pH 7.4) and N-cyclohexyl-2-aminoethanesulfonate (CHES, pH 9.2), respectively, to derive the corresponding KD values of each probe. The rates of reactions (6a) and (6b) vary considerably depending on the exact nature of P and L (Fig. S3, ESI†). Consequently, two experimental procedures were employed in this work:
Selected examples of applications
The four probes DP1–4 developed in this work bind Cu(II) with varying binding affinities and may be applied to quantify Cu(II)-binding properties (stoichiometry and affinity) of peptide/protein targets T (eqn (9)–(11)):| CuIIP + T ⇌ CuIIT + P | (9) |
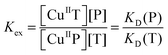 | (10) |
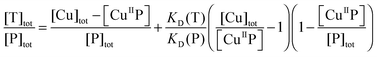 | (11) |
For a target whose Cu(II) affinity is comparable or stronger than that of the probe, titration of Cuaq2+ into a solution containing equimolar P and T quenched the probe fluorescence intensity until reaching a turning point at which the Cu(II) sites of both P and T were fully occupied. A control titration of the probe solution containing no protein target allowed determination of the stoichiometry of binding to the protein target since each probe is known to bind one equiv. of Cu(II) at the titration turning point (vide infra).
On the other hand, titration of a protein target into a solution containing the probe complex CuIIP may induce an effective competition satisfying eqn (9)–(11). The term [CuIIP] may be determined directly viaeqn (5) and then the protein KD(T) can be estimated via curve-fitting of the experimental data to eqn (11) using the known KD(P). The Cu(II)-exchange reaction of eqn (9) involving some targets is slow and takes ∼1 h to reach equilibrium (Fig. S3, ESI†). In such cases, procedure II was adopted. Briefly, a series of solutions were prepared which contained a fixed total concentration of the probe complex Cux–P (x ≤ 1.0) but varying total concentrations of target T in an appropriate buffer. Each solution was incubated until a stable fluorescence spectrum was reached (>1 h) and recorded. The data were compared and analysed to derive the affinity of the protein target as detailed above. The protein and peptide targets selected cover a wide range of affinities from micromolar to femtomolar and include Aβ-peptides Ac-Aβ16, Aβ16 and Aβ16wwa,20 two naturally occurring Cu(II)-chelating motifs in human serum and cerebrospinal fluid with sequences GHK and DAHK,33 and two copper binding proteins CopC from Pseudomonas syringae and PcoC from Escherichia coli.29,30
Results and discussion
Characterisation of the probes
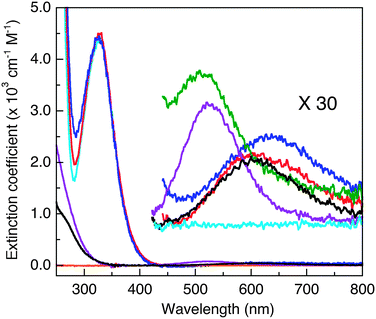
Titration of each probe with Cuaq2+ induced minor increases in absorbance at ∼280 nm (Fig. 2, red, blue traces), presumably arising from ligand-to-Cu(II) charge-transfer transitions. Absorbance changes were observed more clearly for the two dansyl-free Cu(II)–peptide complexes CuII–GHK and CuII–DAHK (Fig. 2, black and purple traces versus orange trace for apo-DAHK). The major absorbance at 326 nm assigned to the dansyl group remained essentially unchanged upon titration of Cuaq2+ into the four DP probe solutions (Fig. 2), consistent with the binding sites accessed by Cu(II) in each DP probe involving ligands derived from the peptide, not from the dansyl group itself.
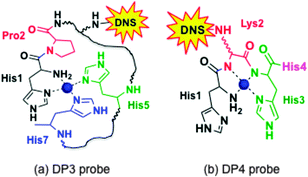
Weak absorbance peaks are seen in the visible region for the Cu(II) complexes, attributable to the Cu(II) d–d transitions. In particular, peaks at ∼525 nm (ε ∼ 100–20 M−1 cm−1)35 were observed for both CuII–DP4 and CuII–DAHK (Fig. 2, green and purple traces), suggestive of the presence of a so-called amino terminal Cu and Ni binding motif (i.e., ATCUN motif; see Fig. 3b).36 DAHK is a naturally-occurring Cu(II)-binding tetra-peptide found in human serum and its ability to bind Cu(II) in the ATCUN binding mode has been documented convincingly.37 Multiple additional pieces of evidence also support an ATCUN Cu(II) centre in Cu(II)–DP4 within the pH range 6.2–9.2 (Fig. 3b; vide infra and ESI†). Both peptides DP4 and DAHK possess the structural elements of the ATCUN motif: (i) a free N-terminus; (ii) a His residue in the third position; (iii) two intervening secondary amine peptide nitrogens.36
The other three probe complexes CuII–DP1–3 also exhibit similar peaks (ε < 100 M−1 cm−1) but at longer wavelengths (600–650 nm; Fig. 2). The behaviour is consistent with the presence of a type II Cu(II) centre in each complex. Similar d–d transitions were observed for CuII–GHK (Fig. 2) and CuII–PcoC.30
When excited at ∼330 nm, the dansyl fluorophore in each DP probe in solution emits intense fluorescence at λmax ∼ 550 nm (Fig. 4a). Titration with Cuaq2+ quenches the fluorescence intensity linearly until one equiv. of Cu(II) is added, suggesting that each probe binds one equiv. of Cu(II) preferentially (Fig. 4a and b; Fig. S2, ESI†). However, at sub-micromolar probe concentrations, the turning point of the titration curve for DP1 is lost (Fig. 4b), consistent with relatively weak Cu(II) binding. Titration of excess of a strong Cu(II) chelator such as Edta into solutions of each CuII–DP complex recovered the fluorescence intensity to that of the original metal-free form, consistent with the reversibility of Cu(II) binding and negligible inner-filter effects from other solution components. The absorbance at 326 nm can be attributed solely to the dansyl group (compare spectra of CuII–DP complexes versus CuII–DAHK in Fig. 2). The evidence suggests that the fluorescence quenching induced by Cu(II) binding arises primarily through paramagnetic interactions between the Cu(II) centre and the attached dansyl fluorophore, consistent with the previous observation of little change in absorbance at 326 nm upon Cu(II) binding (Fig. 2). This validates application of eqn (5) to the Cu(II) speciation analysis for the competing equilibriums of eqn (6) and (9) as there is no need to consider the possible inner-filter effects of the non-chromophoric ligands and metal ions. The system provides an excellent opportunity to develop a series of probes covering a range of affinities for Cu(II). Four such probes were developed in this work and are presented below, together with a brief comparison with our previous probe Aβ16wwa.
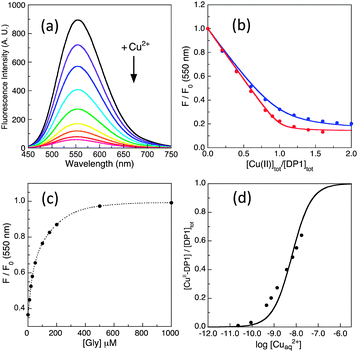 | ||
Fig. 4 Determination of apparent KD′ for CuII–DP1 by direct metal ion titration (a, b) and conditional KD by ligand competition (c, d) in MOPS buffer (5.0 mM unless otherwise indicated, pH 7.4): (a) quenching of fluorescence emission intensity (λmax = 550 nm) of apo-DP1 (2.0 μM) upon titration with Cuaq2+ solution (80 μM); (b) change in F/F0 for DP1 as a function of [Cu(II)]tot/[DP1]tot concentration: 2.0 μM (red) and 0.2 μM (blue in 0.5 mM buffer). The solid traces are fitting curves of the experimental data to eqn (4), providing apparent log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD′ = −8.0 ± 0.1; (c) recovery of F/F0 for CuII0.8–DP1 (2.0 μM) with increasing concentration of competing ligand Gly; (d) curve fitting of [CuII–DP1]/[DP1]totversus log[Cuaq2+] to eqn (8), providing conditional log KD′ = −8.0 ± 0.1; (c) recovery of F/F0 for CuII0.8–DP1 (2.0 μM) with increasing concentration of competing ligand Gly; (d) curve fitting of [CuII–DP1]/[DP1]totversus log[Cuaq2+] to eqn (8), providing conditional log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −8.1 ± 0.2 for CuII–DP1. KD = −8.1 ± 0.2 for CuII–DP1. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DP1 ∼ 1
DP1 ∼ 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 4b; eqn (1)) at pH 7.4 (MOPS buffer ≤ 5.0 mM) and provided an estimate of the apparent log
1 (Fig. 4b; eqn (1)) at pH 7.4 (MOPS buffer ≤ 5.0 mM) and provided an estimate of the apparent log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD′ = −8.0 ± 0.1 and specific F1/F0 values (Tables 1 and 2). It was also possible to determine a conditional log
KD′ = −8.0 ± 0.1 and specific F1/F0 values (Tables 1 and 2). It was also possible to determine a conditional log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −8.1 ± 0.2 at pH 7.4 for CuII–DP1 via ligand competition using Gly as a competing affinity standard (Fig. 4c and d), based on reported data given in Table S1 (ESI†). The consistency of these two independent approaches confirms negligible buffer effects under the experimental conditions and the reliability of the KD value determined for Cu(II)–DP1 here. This claim is consolidated by the fact that both probes DP1 and DP2 compete effectively for Cu(II) with the Ac-Aβ16 peptide and provide consistent outcomes (vide infra). Further details are given in the ESI.†
KD = −8.1 ± 0.2 at pH 7.4 for CuII–DP1 via ligand competition using Gly as a competing affinity standard (Fig. 4c and d), based on reported data given in Table S1 (ESI†). The consistency of these two independent approaches confirms negligible buffer effects under the experimental conditions and the reliability of the KD value determined for Cu(II)–DP1 here. This claim is consolidated by the fact that both probes DP1 and DP2 compete effectively for Cu(II) with the Ac-Aβ16 peptide and provide consistent outcomes (vide infra). Further details are given in the ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD) of CuII-probe complexes
KD) of CuII-probe complexes
| Complex | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD KD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a a |
Affinity std | Ref. | ||
|---|---|---|---|---|---|
| pH 6.2 | pH 7.4 | pH 9.2 | |||
a Unless stated otherwise, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD data were determined via ligand competition for Cu(II) between each probe and the specified affinity standard in respective buffer (50 mM) of MES (pH 6.2), MOPS (pH 7.4) and CHES (pH 9.2). The values in brackets refer to estimated deviations based on the quality of curve-fittings of the experimental data (see Fig. 4 and 5; Fig. S4–S6, ESI).
b Apparent log KD data were determined via ligand competition for Cu(II) between each probe and the specified affinity standard in respective buffer (50 mM) of MES (pH 6.2), MOPS (pH 7.4) and CHES (pH 9.2). The values in brackets refer to estimated deviations based on the quality of curve-fittings of the experimental data (see Fig. 4 and 5; Fig. S4–S6, ESI).
b Apparent log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD determined via direct metal ion titration.
c Determined in diluted buffers (5.0 mM MOPS for 2.0 μM DP1 and 0.5 mM MOPS for 0.20 μM DP1) to minimize possible buffer competition for Cu(II) with the weak binding site.
d Determined previously with fluorescence emission of Trp in Aβ16wwa as detection probe. KD determined via direct metal ion titration.
c Determined in diluted buffers (5.0 mM MOPS for 2.0 μM DP1 and 0.5 mM MOPS for 0.20 μM DP1) to minimize possible buffer competition for Cu(II) with the weak binding site.
d Determined previously with fluorescence emission of Trp in Aβ16wwa as detection probe.
|
|||||
| CuII–DP1 | — | −8.0(2)b,c | — | ‘H2O’ | Fig. 4a and b |
| — | −8.1(2)c | −11.6(1) | Gly | Fig. 4c and d | |
| CuII–DP2 | −8.0(1)c | −10.1(1) | −12.5(1) | Gly | Fig. 5b and c |
| −8.0(1)c | −10.0(1) | −12.4(1) | His | Fig. 5b and d | |
| CuII–DP3 | −10.2(1) | −12.4(1) | −13.4(1) | Gly | Fig. S4a and b (ESI) |
| −9.9(1) | −12.2(1) | −13.4(1) | His | Fig. S4a and c (ESI) | |
| CuII–DP4 | −10.7(2) | −14.1(1) | −18.0(2) | His | Fig. S5a and b (ESI) |
| −11.4(3) | −14.1(1) | −18.0(1) | Egta | Fig. S5a and c (ESI) | |
| CuII–Aβ16wwa | −7.7(1) | −9.9(1) | −12.5(1) | DP2 | Fig. S6c and d (ESI) |
| −7.9(2)b,c | — | — | ‘H2O’ | Fig. S6a and b (ESI) | |
| — | −9.8(2)d | — | Gly | 20 | |
An equivalent analysis in CHES buffer (5.0 mM, pH 9.2) provided log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −11.6 ± 0.1 for CuII–DP1 (Table 2). In addition, the fluorescence titration experiments revealed different F1/F0 values in the two buffers (Table 1). It is apparent that the Cu(II) binding site at pH 9.2 is different from that at pH 7.4. It is likely that, at higher pH, one or two deprotonated amides of the peptide links of His2 and/or His4 are involved in Cu(II) binding (cf., the coordination mode of His3 in Fig. 3b). Further log
KD = −11.6 ± 0.1 for CuII–DP1 (Table 2). In addition, the fluorescence titration experiments revealed different F1/F0 values in the two buffers (Table 1). It is apparent that the Cu(II) binding site at pH 9.2 is different from that at pH 7.4. It is likely that, at higher pH, one or two deprotonated amides of the peptide links of His2 and/or His4 are involved in Cu(II) binding (cf., the coordination mode of His3 in Fig. 3b). Further log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD estimates at pH 8.0 and 8.5 are included in Table S2 (ESI†).
KD estimates at pH 8.0 and 8.5 are included in Table S2 (ESI†).
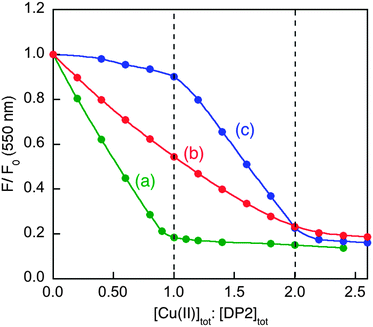
Titration of Cuaq2+ into solutions of DP2 (2.0 and 10 μM) in MOPS buffer (50 mM, pH 7.4) induced a linear quenching with a clear turning point at one equiv. of Cuaq2+ but further minor quenching was apparent at higher Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) probe ratios (Fig. 5a). There appears to be one higher affinity binding site with one or more sites of lower affinity, as observed previously for the Aβ16wwa probe.20
probe ratios (Fig. 5a). There appears to be one higher affinity binding site with one or more sites of lower affinity, as observed previously for the Aβ16wwa probe.20
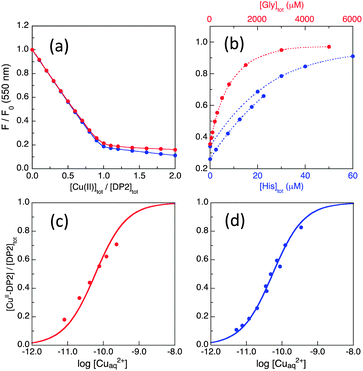 | ||
Fig. 5 Determination of conditional KD for CuII–DP2 by competition with ligand Gly or His in MOPS buffer (50 mM, pH 7.4): (a) comparison of the titration curves for DP2 at 2.0 μM (red) and 10.0 μM (blue); (b) recovery of F(550) of CuII0.8–DP2 or CuII–DP2 (10 μM) with increasing concentration of competing ligand Gly (in red on top scale) or His (in blue on bottom scale); (c, d) curve fittings of [CuII–DP2]/[DP2]totversus log[Cuaq2+] to eqn (8) derived an consistent estimate of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −10.1 ± 0.1 for CuII–DP2 with either competing ligand Gly (c) or His (d). KD = −10.1 ± 0.1 for CuII–DP2 with either competing ligand Gly (c) or His (d). | ||
The affinity of the first site is too high (KD too small) to be determined via direct metal ion titration due to the detection limit of the probe concentration (∼0.1 μM). Consequently, the affinity was determined via ligand competition with both Gly and His as affinity standards (Fig. 5b and c), as described for DP1 (see ESI†). Both analyses provided log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −10.1 ± 0.1 at pH 7.4, an affinity higher by two orders of magnitude than that of DP1 (Table 2).
KD = −10.1 ± 0.1 at pH 7.4, an affinity higher by two orders of magnitude than that of DP1 (Table 2).
Equivalent experiments were conducted at lower and higher pH values in buffers MES (50 mM, pH 6.2) and CHES (50 mM, pH 9.2) (see ESI†). The derived affinities were 2 orders of magnitude lower at pH 6.2 and 2–3 orders of magnitude higher at pH 9.2 (Table 2). These observations are consistent with competition between Cu(II) and protons at pH 6.2 for the N-terminal nitrogen (pKa = 8.0–8.5) as well as the two His ligands (pKa ∼ 6.5) and the involvement of at least one peptide backbone amido ligand at pH 9.2. Further log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD estimates at other pH values are provided in Table S2 (ESI†).
KD estimates at other pH values are provided in Table S2 (ESI†).
Supporting evidence is provided by the characteristic F1/F0 values being different in the solutions at lower and higher pH (Table 1). The spectroscopic evidence supports a Cu(II) binding site transition similar to that which occurs for the Aβ16 peptide at pH ∼ 7.8.38 The Aβ16 peptide itself and its derivative Aβ16wwa possess comparable Cu(II) affinities and similar pH dependencies to that of DP2 (vide infra). Confirming characterisation of Aβ16wwa via DP2 is included in the ESI.†
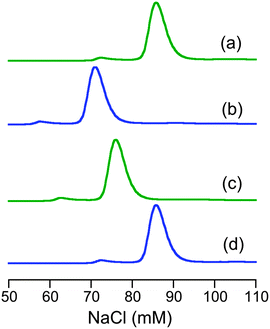
The characterisation data are consistent with the tetragonal site shown in Fig. 3a: (i) the complex CuII–DP3 eluted before apo-DP3 from an analytical anion exchange column equilibrated at pH 7.4 (Fig. 6), indicating that Cu(II) binding leads to a net gain of cationic charge, consistent with ionisation of the single N-terminal nitrogen only; (ii) DP3 is the only probe among the four whose log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD at various pH within the range 6.2–9.2 can be fitted satisfactorily to eqn (S2) (ESI†) (see footnote of Table S1, ESI†) assuming a Cu(II) site of {–NH2, 3NIm(His)} (Fig. 7). The derived parameters are the expected values: log(absKD) = −13.6 for the pH-independent absolute dissociation constant; pKa = 8.8 for the N-terminal amine; an average pKa = 6.5 for the three His side-chains. Apparently, the Cu(II) site in DP3 lacks peptide nitrogen anionic ligands with pKa > 9.2 within the pH range 6.2–9.2. Notably, DP3 binds Cu(II) with an unchanged quenching index F1/F0 = 0.13 (Table 1) and the Cu(II)–DP3 complex displays an indistinguishable EPR spectrum (Fig. S7 and Table S3, ESI†) within the pH range 6.2–9.2, suggesting a uniform Cu(II) site. The carboxylate side-chains of Asp4 and/or Asp6 may contribute as axial ligand(s) but will have little impact on the pH dependency of log
KD at various pH within the range 6.2–9.2 can be fitted satisfactorily to eqn (S2) (ESI†) (see footnote of Table S1, ESI†) assuming a Cu(II) site of {–NH2, 3NIm(His)} (Fig. 7). The derived parameters are the expected values: log(absKD) = −13.6 for the pH-independent absolute dissociation constant; pKa = 8.8 for the N-terminal amine; an average pKa = 6.5 for the three His side-chains. Apparently, the Cu(II) site in DP3 lacks peptide nitrogen anionic ligands with pKa > 9.2 within the pH range 6.2–9.2. Notably, DP3 binds Cu(II) with an unchanged quenching index F1/F0 = 0.13 (Table 1) and the Cu(II)–DP3 complex displays an indistinguishable EPR spectrum (Fig. S7 and Table S3, ESI†) within the pH range 6.2–9.2, suggesting a uniform Cu(II) site. The carboxylate side-chains of Asp4 and/or Asp6 may contribute as axial ligand(s) but will have little impact on the pH dependency of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD at pH > 6 and on the EPR spectrum. They are sensitive to equatorial ligands only.
KD at pH > 6 and on the EPR spectrum. They are sensitive to equatorial ligands only.
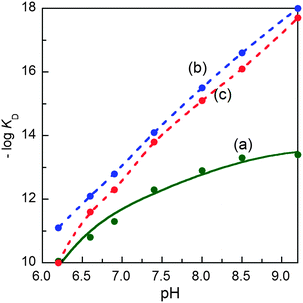 | ||
Fig. 7 Variation of conditional dissociation constant pKD (= −log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD) with solution pH for Cu(II) complexes of: (a) DP3; (b) DP4 and (c) DAHK. The solid trace in (a) showed the best fit of the experimental data to eqn (S2) (ESI†) based on a metal binding site of {–NH2, 3NIm(His)} in DP3 with the derived parameters log(absKD) = −13.6, pKa = 8.8 for –NH2 and an average pKa = 6.5 for the three NIm(His). The dashed traces in (b, c) are the simple interpolations of the experimental data points for DP4 and DAHK ligands since the data cannot be fitted to eqn (S2) (ESI†) within the pH window. KD) with solution pH for Cu(II) complexes of: (a) DP3; (b) DP4 and (c) DAHK. The solid trace in (a) showed the best fit of the experimental data to eqn (S2) (ESI†) based on a metal binding site of {–NH2, 3NIm(His)} in DP3 with the derived parameters log(absKD) = −13.6, pKa = 8.8 for –NH2 and an average pKa = 6.5 for the three NIm(His). The dashed traces in (b, c) are the simple interpolations of the experimental data points for DP4 and DAHK ligands since the data cannot be fitted to eqn (S2) (ESI†) within the pH window. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD for selected Cu(II) complexes determined with the set of four DP probes to those reported previously
KD for selected Cu(II) complexes determined with the set of four DP probes to those reported previously
| Complex | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KDa KDa |
Reported log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD at pH 7.4 unless otherwise indicated KD at pH 7.4 unless otherwise indicated |
Probe – affinity std | Ref. | ||
|---|---|---|---|---|---|---|
| pH 6.2 | pH 7.4 | pH 9.2 | ||||
a Unless stated otherwise, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD data reported in this work were determined via ligand competition for Cu(II) between each probe with affinity given in Table 2 and the specified Cu(II)-binding Target in respective buffer (50 mM) of MES (pH 6.2), MOPS (pH 7.4) and CHES (pH 9.2). The bracketed values refer to estimated deviations based on the quality of curve-fitting of the experimental data (cf.Fig. 10).
b Apparent affinity estimated via direct metal ion titration in KPi buffer (pH 7.0).
c Values determined in KPi buffer at pH 7.0.
d The affinity difference between DP4 and Hedta at pH 6.2 is too large to allow reliable estimation. KD data reported in this work were determined via ligand competition for Cu(II) between each probe with affinity given in Table 2 and the specified Cu(II)-binding Target in respective buffer (50 mM) of MES (pH 6.2), MOPS (pH 7.4) and CHES (pH 9.2). The bracketed values refer to estimated deviations based on the quality of curve-fitting of the experimental data (cf.Fig. 10).
b Apparent affinity estimated via direct metal ion titration in KPi buffer (pH 7.0).
c Values determined in KPi buffer at pH 7.0.
d The affinity difference between DP4 and Hedta at pH 6.2 is too large to allow reliable estimation.
|
||||||
| CuII–PcoC–H1F | −8.2(2) | DP1 – Gly | This work | |||
| ≥−6b | PcoC – ‘H2O’ | 30 | ||||
| CuII–AcAβ16 | −8.1(1) | DP1 – Gly | This work | |||
| −8.4(1) | DP2 – Gly/His | This work | ||||
| −8.3 | Aβ16wwa–Gly | 20 | ||||
| CuII–Aβ16 | −8.2(1) | −10.2(1) | −12.5(1) | DP2 – Gly/His | This work | |
| −8.1(1) | −10.1(1) | −12.3(2) | DP3 – Gly/His | This work | ||
| −10.0 | Aβ16wwa–Gly | 20 | ||||
| CuII–CopC | −12.3(2) | −13.7(1) | DP4 – His/Egta | This work | ||
| −13.1c | CopC – Egta | 29 | ||||
| CuII–PcoC | −11.3(3) | −13.5(3) | −14.0(3) | DP3 – Gly/His | This work | |
| −11.5(3) | −13.5(1) | DP4 – His/Egta | This work | |||
| −13.2c | PcoC – Egta | 30 | ||||
| GHK | −10.4(1) | −13.4(2) | DP3 – Gly/His | This work | ||
| −10.4(1) | −13.3(2) | −16.3(2) | DP4 – His/Egta | This work | ||
| −13.2 | ITC – Gly | 33 | ||||
| −13.0 | Potentiometry | 41 | ||||
| DAHK | −10.0(1) | −13.8(2) | −17.7(2) | DP4 – His/Egta | This work | |
| −13.6 | ITC – Gly | 33 | ||||
| −13.8 | Potentiometry | 42 | ||||
| CuII–Hedta | < −13d | −15.2(3) | −16.8(1) | DP4 – His/Egta | This work | |
| –13.7 | −16.7 | −14.9 | 44 | |||
The observed high pH dependency of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD beyond pH 9.2 is not compatible with a DP3-type binding mode for Cu(II). Further experimental evidence, with controls consisting of the ATCUN ligand DAHK37 and the non-ATCUN ligand DP3 (vide supra), confirm that DP4 binds Cu(II) in the classic ATCUN binding mode, as shown in Fig. 3b. A full documentation is provided in the ESI.†
KD beyond pH 9.2 is not compatible with a DP3-type binding mode for Cu(II). Further experimental evidence, with controls consisting of the ATCUN ligand DAHK37 and the non-ATCUN ligand DP3 (vide supra), confirm that DP4 binds Cu(II) in the classic ATCUN binding mode, as shown in Fig. 3b. A full documentation is provided in the ESI.†
Application of the probes
The four DP probes developed in this work are able to quantify Cu(II) binding properties of target proteins and peptides in the extended range from micromolar to femtomolar (Fig. 8). Their individual binding sites differ and so do the dependencies of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD on solution pH (Table 2 and Table S2, ESI†).
KD on solution pH (Table 2 and Table S2, ESI†).
Their potential of application was tested on a range of well-characterised targets. Results are summarised in Fig. 9 and 10 and Table 3. For example, the Cu(II) binding stoichiometries of peptides Aβ16, GHK and DAHK were determined by Cuaq2+ titration of a mixture containing equal molar concentrations of each target peptide and DP2, as detailed for Aβ16 and DAHK in Fig. 9. It is apparent that the latter peptide exhibits a single site of highest affinity for Cu(II).
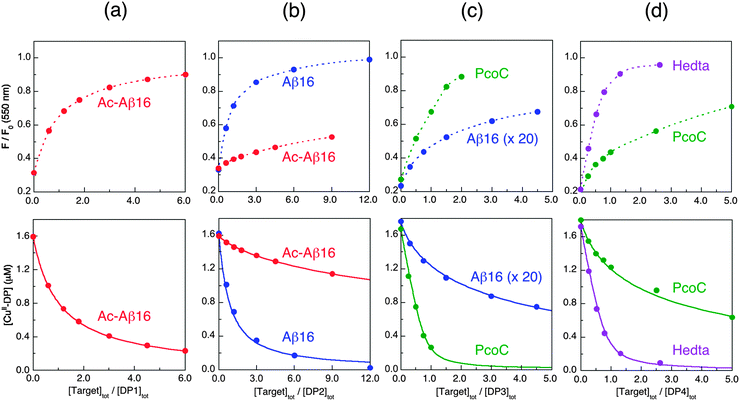 | ||
| Fig. 10 Determination of conditional KD for various protein/peptide targets in MOPS buffer (50 mM, pH 7.4): (a) Ac-Aβ16 with DP1 probe; (b) Ac-Aβ16 and Aβ16 with DP2 probe; (c) Aβ16 (note: 20× conc.) and PcoC with DP3 probe; (d) PcoC and Hedta with DP4 probe. Top panel showed recovering of the DP fluorescence intensity upon titration of each CuIIx–DP complex (x = 0.8–0.9) with related competing Target ligands where the dot traces are the simple interpolations of the experimental points, and bottom panel gave correlation of [CuII–DP] versus [target]tot/[DP]tot where the solid traces are the fitting curves of the experimental data to eqn (11) that allowed determination of conditional KD for each peptide/protein target as given in Table 3. | ||
To quantify affinities, a protein or peptide target was titrated into a solution containing the appropriate probe complex CuII–DP to initiate an effective competition for Cu(II) according to eqn (9). The solution was monitored for recovery of probe fluorescence intensity according to eqn (5) (see Fig. 10, top panel). This allowed Cu(II) speciation analysis to determine a correlation between [CuII–DP] and [target]tot/[DP]tot that was fitted to eqn (11) to allow estimation of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD for the target from the known log
KD for the target from the known log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD for the probe complex (Fig. 10, bottom panel; see Experimental section for details).
KD for the probe complex (Fig. 10, bottom panel; see Experimental section for details).
As the sensitive ranges of the probes overlap (Fig. 8), the affinities of many targets listed in Table 3 were consolidated with two different DP probes, highlighting the internal consistency of the approach. For example, both DP2 and DP3 provided estimates of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD for Aβ16 at pH 7.4 that were the same within experimental error to our previous value determined with the Aβ16wwa probe (Table 3). Acetylation of the N-terminus of Aβ16 leads to a decrease in affinity of ∼2 orders of magnitude, as confirmed with both DP1 and DP2. This confirms the key role of the N-terminal amine in Cu(II) binding. Variation of log
KD for Aβ16 at pH 7.4 that were the same within experimental error to our previous value determined with the Aβ16wwa probe (Table 3). Acetylation of the N-terminus of Aβ16 leads to a decrease in affinity of ∼2 orders of magnitude, as confirmed with both DP1 and DP2. This confirms the key role of the N-terminal amine in Cu(II) binding. Variation of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD with pH for Aβ16 is similar to that of DP2 within the whole pH range 6.2–9.2. It is comparable also to that of DP3 between pH 6.2–7.4, but very different between pH 7.4–9.2 (Table 3). This is consistent with a Cu(II) site transition in Aβ16 with a peptide amide nitrogen anionic ligand contributing to Cu(II) coordination at pH > 7.4, but not at pH < 7.4 (vide supra).38
KD with pH for Aβ16 is similar to that of DP2 within the whole pH range 6.2–9.2. It is comparable also to that of DP3 between pH 6.2–7.4, but very different between pH 7.4–9.2 (Table 3). This is consistent with a Cu(II) site transition in Aβ16 with a peptide amide nitrogen anionic ligand contributing to Cu(II) coordination at pH > 7.4, but not at pH < 7.4 (vide supra).38
CopC and PcoC are two copper binding proteins expressed to the periplasmic space of two different copper-resistant Gram-negative bacteria.39,40 They share highly conserved protein sequences, protein structures and metal-binding sites. They each feature two separated copper-binding sites specific, respectively, for Cu(I) and Cu(II).29,30 The affinities of their Cu(II) sites at pH 7.4 were determined, again indistinguishably, with the DP3 and DP4 probes. These values (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.7 and −13.5 at pH 7.4) are consistent with previous estimates (log
KD = −13.7 and −13.5 at pH 7.4) are consistent with previous estimates (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.1 and −13.2 at pH 7.0).30 Variation of log
KD = −13.1 and −13.2 at pH 7.0).30 Variation of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD with pH is somewhat comparable to that of DP3 (Table 4), consistent with the similarity of their Cu(II) binding sites (Fig. 3a). The Cu(II) site in CopC comprises a bidentate His1 ligand, the side-chain of His90 and a possible H2O molecule or a carboxylate side-chain as the fourth coordination ligand.29 Consequently, mutation of His1 to Phe1 decreases the Cu(II) affinity by more than five orders of magnitude, as determined in this work with probe DP1 (Table 3). A conserved proline residue in the second position of both protein sequences promotes the DP3 binding mode and prevents the DP4 ATCUN binding mode (Fig. 3).
KD with pH is somewhat comparable to that of DP3 (Table 4), consistent with the similarity of their Cu(II) binding sites (Fig. 3a). The Cu(II) site in CopC comprises a bidentate His1 ligand, the side-chain of His90 and a possible H2O molecule or a carboxylate side-chain as the fourth coordination ligand.29 Consequently, mutation of His1 to Phe1 decreases the Cu(II) affinity by more than five orders of magnitude, as determined in this work with probe DP1 (Table 3). A conserved proline residue in the second position of both protein sequences promotes the DP3 binding mode and prevents the DP4 ATCUN binding mode (Fig. 3).
| Complex | λ max (nm)/εmax (M−1 cm−1) for CuII(d–d)a | Δ(Q−) upon CuII binding at pH 7.4b |
Δ(log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD) with pH KD) with pH |
(N−)–CuII at pH | ||
|---|---|---|---|---|---|---|
| 6.2 → 7.4 | 7.4 → 9.2 | 7.4 | 9.2 | |||
| a Solution spectra recorded in MOPS buffer at pH 7.4 (see Fig. 2). b Change in net negative charge (Q−) upon formation of each Cu(II) complex was evaluated by comparative elution, from an analytical anion-exchange column, of metal-free and metal-bound forms of each peptide/protein molecule (see Fig. 6). c Quoted from ref. 30. d Increase in net positive charge upon Cu(II) binding as evidenced from comparative elution from a cation-exchange column. e Quoted from data in Table S1 (ESI). | ||||||
| CuII–DP1 | ∼650/∼30 | — | — | 3.5 | ?? | Yes |
| CuII–DP2 | 625/70 | — | 2.1 | 2.4 | No | Yes |
| CuII–DP3 | ∼600/∼45 | Decrease | 2.2 | 1.1 | No | No |
| CuII–DP4 | 525/120 | Increase | 3.0 | 3.9 | Yes | Yes |
| CuII–Aβ16 | 625/80 | — | 2.0 | 2.3 | No | Yes |
| CuII–PcoC | 610/110c | Decreasec,d | 2.1 | 0.5 | No | No |
| CuII–GHK | 625/50 | — | 3.0 | 3.0 | Yes | Yes |
| CuII–DAHK | 525/100 | — | 3.8 | 3.9 | Yes | Yes |
| CuII–(Egta) | 2.4e | 3.0e | ||||
| CuII–(Hedta) | 1.3e | 1.4e | ||||
Peptides GHK and DAHK are naturally-occurring Cu(II)-binding peptides present in human serum and cerebrospinal fluid.36 Their Cu(II) binding affinities were determined previously by classic potentiometric titration at log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.0 and −13.8, respectively.41,42 Both values were confirmed more recently by independent ITC approach with [CuII(Gly)2] stabilized by excess Gly at high concentrations from 2.8 mM to 50 mM (log
KD = −13.0 and −13.8, respectively.41,42 Both values were confirmed more recently by independent ITC approach with [CuII(Gly)2] stabilized by excess Gly at high concentrations from 2.8 mM to 50 mM (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.2 and −13.6).33 This work supported these values (log
KD = −13.2 and −13.6).33 This work supported these values (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.3 and −13.8) but with the simpler approach of ligand competition with the probe DP4 (Table 3). Furthermore, we confirm readily that the Cu(II) affinities of both peptides are highly pH-sensitive within the range pH 6.2–9.2 (Tables 3 and 4 and Fig. 7), consistent with peptide amide nitrogen(s) acting as key ligand(s).37 Interestingly, at pH 7.4, the Cu(II) affinities of DAHK (log
KD = −13.3 and −13.8) but with the simpler approach of ligand competition with the probe DP4 (Table 3). Furthermore, we confirm readily that the Cu(II) affinities of both peptides are highly pH-sensitive within the range pH 6.2–9.2 (Tables 3 and 4 and Fig. 7), consistent with peptide amide nitrogen(s) acting as key ligand(s).37 Interestingly, at pH 7.4, the Cu(II) affinities of DAHK (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.8)42 and the C-terminal amidated DAH-NH2 (log
KD = −13.8)42 and the C-terminal amidated DAH-NH2 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −13.7)43 are only marginally weaker than the Cu(II) affinity of our DP4 probe (log
KD = −13.7)43 are only marginally weaker than the Cu(II) affinity of our DP4 probe (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KD = −14.2; sequence H(KDNS)HH), supporting the proposed Cu(II) site in DP4 of Fig. 3b with a minimal contribution from the side-chains of His1 and His4 as possible axial ligand(s) only.
KD = −14.2; sequence H(KDNS)HH), supporting the proposed Cu(II) site in DP4 of Fig. 3b with a minimal contribution from the side-chains of His1 and His4 as possible axial ligand(s) only.
Last but not least, the Cu(II) affinity of a well-documented classic Cu(II) ligand Hedta was checked with the DP4 probe (Fig. 10d). Although the affinity of Hedta at pH 6.2 is too high to be accessed by the DP4 probe, the data acquired at pH 7.4 and 9.2 match the literature values (Table 3). This is a further endorsement of the reliability and robustness of the four DP probes developed in this work.
Concluding remarks
A set of four highly sensitive fluorescent probes DP1–4 were developed and established in this work. They were demonstrated to be a set of robust and complementary probes capable of exploring the Cu(II) chemistry of proteins and peptides. Each of the probes binds one equivalent of Cu(II) cleanly with minimal formation of ternary complexes with competing ligands, a pre-condition for determination of metal affinity by ligand competition (Fig. S2, ESI†). The probes were designed to display affinities in the micromolar to femtomolar concentration ranges at pH 7.4 (Table 2). The affinities were calibrated carefully in each case with at least two well-documented and independent affinity standards (Gly, His, Egta, Hedta) and were cross-checked directly or indirectly with other independent approaches such as direct metal ion titration, ITC and potentiometry (Tables 2 and 3).Similar to most other Cu(II) binding molecules, the affinities of the four DP probes are pH-dependent, as summarised in Table 4 and so great care must be excised to control the correct solution pH when these probes are used. On the other hand, a detailed comparison between the differences in such pH dependency among the four DP probes, plus other evidence, provides valuable information of the nature of the different Cu(II) sites in DP1–4 (cf., Fig. 3). Overall, this system provides a valuable platform to explore Cu(II) binding targets, as exemplified by Table 4.
Acknowledgements
This work was supported by fund from the Australian Research Council under grant DP130100728. We thank Mrs Sioe See Volaric (University of Melbourne) for providing technical support for recording EPR spectra.Notes and references
- R. A. Festa and D. J. Thiele, Copper: An essential metal in biology, Curr. Biol., 2011, 21, R877–R883 CrossRef CAS PubMed.
- S. Lutsenko, Human copper homeostasis: a network of interconnected pathways, Curr. Opin. Chem. Biol., 2010, 14, 211–217 CrossRef CAS PubMed.
- N. J. Robinson and D. R. Winge, Copper metallochaperones, Annu. Rev. Biochem., 2010, 79, 537–562 CrossRef CAS PubMed.
- S. Tottey, C. J. Patterson, L. Banci, I. Bertini, I. C. Felli, A. Pavelkova, S. J. Dainty, R. Pernil, K. J. Waldron, A. W. Foster and N. J. Robinson, Cyanobacterial metallochaperone inhibits deleterious side reactions of copper, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 95–100 CrossRef CAS PubMed.
- J. F. Mercer, The molecular basis of copper-transport diseases, Trends Mol. Med., 2001, 7, 64–69 CrossRef CAS.
- K. J. Barnham, C. L. Masters and A. I. Bush, Neurodegenerative diseases and oxidative stress, Nat. Rev. Drug Discovery, 2004, 3, 205–214 CrossRef CAS PubMed.
- P. S. Donnelly, Z. Xiao and A. G. Wedd, Copper and Alzheimer's disease, Curr. Opin. Chem. Biol., 2007, 11, 128–133 CrossRef CAS PubMed.
- E. Gaggelli, H. Kozlowski, D. Valensin and G. Valensin, Copper homeostasis and neurodegenerative disorders (Alzheimer's, prion, and Parkinson's diseases and amyotrophic lateral sclerosis), Chem. Rev., 2006, 106, 1995–2044 CrossRef CAS PubMed.
- D. J. Waggoner, T. B. Bartnikas and J. D. Gitlin, The role of copper in neurodegenerative disease, Neurobiol. Dis., 1999, 6, 221–230 CrossRef CAS PubMed.
- Z. Xiao and A. G. Wedd, The challenges of determining metal-protein affinities, Nat. Prod. Rep., 2010, 27, 768–789 RSC.
- Z. Xiao, F. Loughlin, G. N. George, G. J. Howlett and A. G. Wedd, C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking, J. Am. Chem. Soc., 2004, 126, 3081–3090 CrossRef CAS PubMed.
- Z. Xiao, P. S. Donnelly, M. Zimmermann and A. G. Wedd, Transfer of Copper between Bis(thiosemicarbazone) Ligands and Intracellular Copper-Binding Proteins. Insights into Mechanisms of Copper Uptake and Hypoxia Selectivity, Inorg. Chem., 2008, 47, 4338–4347 CrossRef CAS PubMed.
- Z. Xiao, J. Brose, S. Schimo, S. M. Ackland, S. La Fontaine and A. G. Wedd, Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1 and related proteins: detection probes and affinity standards, J. Biol. Chem., 2011, 286, 11047–11055 CrossRef CAS PubMed.
- P. Bagchi, M. T. Morgan, J. Bacsa and C. J. Fahrni, Robust Affinity Standards for Cu(I) Biochemistry, J. Am. Chem. Soc., 2013, 135, 18549–18559 CrossRef CAS PubMed.
- Z. Xiao, L. Gottschlich, R. van der Meulen, S. R. Udagedara and A. G. Wedd, Evaluation of quantitative probes for weaker Cu(I) binding sites completes a set of four capable of detecting Cu(I) affinities from nanomolar to attomolar, Metallomics, 2013, 5, 501–513 RSC.
- C. J. Fahrni, Synthetic fluorescent probes for monovalent copper, Curr. Opin. Chem. Biol., 2013, 17, 656–662 CrossRef CAS PubMed.
- I. Zawisza, M. Rózga and W. Bal, Affinity of copper and zinc ions to proteins and peptides related to neurodegenerative conditions (Aβ, APP, α-synuclein, PrP), Coord. Chem. Rev., 2012, 256, 2297–2307 CrossRef CAS PubMed.
- N. E. Grossoehme, A. M. Spuches and D. E. Wilcox, Application of isothermal titration calorimetry in bioinorganic chemistry, JBIC, J. Biol. Inorg. Chem., 2010, 15, 1183–1191 CrossRef CAS PubMed.
- D. Jiang, L. Zhang, G. P. G. Grant, C. G. Dudzik, S. Chen, S. Patel, Y. Hao, G. L. Millhauser and F. Zhou, The Elevated Copper Binding Strength of Amyloid-β Aggregates Allows the Sequestration of Copper from Albumin: A Pathway to Accumulation of Copper in Senile Plaques, Biochemistry, 2012, 52, 547–556 CrossRef PubMed.
- T. R. Young, A. Kirchner, A. G. Wedd and Z. Xiao, An Integrated Study of the Affinities of the Aβ16 Peptide for Cu(I) and Cu(II): Implications for the Catalytic Production of Reactive Oxygen Species, Metallomics, 2014, 6, 505–517 RSC.
- Y. Jeong and J. Yoon, Recent progress on fluorescent chemosensors for metal ions, Inorg. Chim. Acta, 2012, 381, 2–14 CrossRef CAS PubMed.
- M. Formica, V. Fusi, L. Giorgi and M. Micheloni, New fluorescent chemosensors for metal ions in solution, Coord. Chem. Rev., 2012, 256, 170–192 CrossRef CAS PubMed.
- Y. Zheng, K. M. Gattas-Asfura, V. Konka and R. M. Leblanc, A dansylated peptide for the selective detection of copper ions, Chem. Commun., 2002, 2350–2351 RSC.
- C. R. Lohani, J. M. Kim and K.-H. Lee, Two dansyl fluorophores bearing amino acid for monitoring Hg2+ in aqueous solution and live cells, Tetrahedron, 2011, 67, 4130–4136 CrossRef CAS PubMed.
- L. N. Neupane, P. Thirupathi, S. Jang, M. J. Jang, J. H. Kim and K.-H. Lee, Highly selectively monitoring heavy and transition metal ions by a fluorescent sensor based on dipeptide, Talanta, 2011, 85, 1566–1574 CrossRef CAS PubMed.
- B. Wang, H.-W. Li, Y. Gao, H. Zhang and Y. Wu, A Multifunctional Fluorescence Probe for the Detection of Cations in Aqueous Solution: the Versatility of Probes Based on Peptides, J. Fluoresc., 2011, 21, 1921–1931 CrossRef CAS PubMed.
- H.-H. Zeng, R. B. Thompson, B. P. Maliwal, G. R. Fones, J. W. Moffett and C. A. Fierke, Real-Time Determination of Picomolar Free Cu(II) in Seawater Using a Fluorescence-Based Fiber Optic Biosensor, Anal. Chem., 2003, 75, 6807–6812 CrossRef CAS PubMed.
- B. J. McCranor, H. Szmacinski, H. H. Zeng, A. K. Stoddard, T. Hurst, C. A. Fierke, J. R. Lakowicz and R. B. Thompson, Fluorescence lifetime imaging of physiological free Cu(II) levels in live cells with a Cu(II)-selective carbonic anhydrase-based biosensor, Metallomics, 2014, 6, 1034–1042 RSC.
- L. Zhang, M. Koay, M. J. Maher, Z. Xiao and A. G. Wedd, Intermolecular transfer of copper ions from the CopC protein of Pseudomonas syringae. Crystal structures of fully loaded Cu(I)Cu(II) forms, J. Am. Chem. Soc., 2006, 128, 5834–5850 CrossRef CAS PubMed.
- K. Y. Djoko, Z. Xiao, D. L. Huffman and A. G. Wedd, Conserved Mechanism of Copper Binding and Transfer. A Comparison of the Copper-Resistance Proteins PcoC from Escherichia coli and CopC from Pseudomonas syringae, Inorg. Chem., 2007, 46, 4560–4568 CrossRef CAS PubMed.
- G. Weber, Polarization of the fluorescence of macromolecules. II. Fluorescent conjugates of ovalbumin and bovine serum albumin, Biochem. J., 1952, 51, 155–167 CAS.
- R. F. Chen, Dansyl labeled proteins: Determination of extinction coefficient and number of bound residues with radioactive dansyl chloride, Anal. Biochem., 1968, 25, 412–416 CrossRef CAS.
- A. Trapaidze, C. Hureau, W. Bal, M. Winterhalter and P. Faller, Thermodynamic study of Cu2+ binding to the DAHK and GHK peptides by isothermal titration calorimetry (ITC) with the weaker competitor glycine, JBIC, J. Biol. Inorg. Chem., 2012, 17, 37–47 CrossRef CAS PubMed.
- There is a minor fluorescence contribution at 550 nm from high concentration of histidine (>1 mM). To avoid such contribution, recovery of the fluorescence intensity was monitored at 600 nm when a high concentration (>1 mM) of histidine was needed as a competing ligand.
- The higher intensity of the d–d transition at 525 nm for CuII–DP4 relative to that for CuII–DAHK is due to a background contribution of the dansyl absorbance in the former complex.
- C. Harford and B. Sarkar, Amino Terminal Cu(II)- and Ni(II)-Binding Motif of Proteins and Peptides: Metal Binding, DNA Cleavage, and Other Properties, Acc. Chem. Res., 1997, 30, 123–130 CrossRef CAS.
- C. Hureau, H. Eury, R. Guillot, C. Bijani, S. Sayen, P.-L. Solari, E. Guillon, P. Faller and P. Dorlet, X-ray and Solution Structures of CuII–GHK and CuII–DAHK Complexes: Influence on Their Redox Properties, Chem. – Eur. J., 2011, 17, 10151–10160 CrossRef CAS PubMed.
- C. Hureau, Coordination of redox active metal ions to the amyloid precursor protein and to amyloid-β peptides involved in Alzheimer disease. Part 1: An overview, Coord. Chem. Rev., 2012, 256, 2164–2174 CrossRef CAS PubMed.
- D. A. Cooksey, Molecular mechanisms of copper resistance and accumulation in bacteria, FEMS Microbiol. Rev., 1994, 14, 381–386 CrossRef CAS PubMed.
- D. L. Huffman, J. Huyett, F. W. Outten, P. E. Doan, L. A. Finney, B. M. Hoffman and T. V. O'Halloran, Spectroscopy of Cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon, Biochemistry, 2002, 41, 10046–10055 CrossRef CAS PubMed.
- M. J. A. Rainer and B. M. Rode, The complex formation of copper(II) with GHL and HSA, Inorg. Chim. Acta, 1984, 92, 1–7 CrossRef CAS.
- M. Sokolowska, A. Krezel, M. Dyba, Z. Szewczuk and W. Bal, Short peptides are not reliable models of thermodynamic and kinetic properties of the N-terminal metal binding site in serum albumin, Eur. J. Biochem., 2002, 269, 1323–1331 CrossRef CAS.
- K. S. Iyer, S. J. Lau, S. H. Laurie and B. Sarkar, Synthesis of the native copper(II)-transport site of human serum albumin and its copper(II)-binding properties, Biochem. J., 1978, 169, 61–69 CAS.
- A. E. Martell and R. M. Smith, NIST Critically Selected Stability Constants of Metal Complexes Database 46, Version 8.0, U.S. Dept. of Commerce, NIST Standard Reference Data Program, Gaithersburg, MD, 2004 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Characterisation of probes DP1-4; Table S1; eqn (S1) and (S2); Fig. S1–S6. See DOI: 10.1039/c4mt00301b |
| This journal is © The Royal Society of Chemistry 2015 |