Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In vitro gene expression within membrane-free coacervate protocells†
T.-Y.
Dora. Tang
a,
Dirk
van Swaay
b,
Andrew
deMello
b,
J. L.
Ross Anderson
c and
Stephen
Mann
*a
aCentre for Protolife Research, School of Chemistry, University of Bristol, Bristol, BS8 1TS, UK. E-mail: s.mann@bristol.ac.uk
bInstitute for Chemical and Bioengineering, ETH Zurich, Wolfgang-Pauli-Str. 10, 8093 Zurich, Switzerland
cSchool of Biochemistry, University of Bristol, Bristol, BS8 1TD, UK
First published on 18th June 2015
Abstract
Cell-free gene expression of a fluorescent protein (mCherry) is demonstrated within the molecularly crowded matrix of a polysaccharide/polypeptide coacervate.
The design, engineering and expression of novel gene architectures and circuitry provide a key overarching technology in synthetic biology. While top-down approaches based on the intelligent re-configuration of living organisms represent the main focus of current activity,1 there is growing interest in bottom-up strategies that seek to generate minimal representations of synthetic cellularity using biologically informed components and processes.2,3 As a consequence, advances in protocell engineering are contributing to a resurgence in the use and development of in vitro gene expression systems (IVGES),4,5 which provide an attractive approach to off-line biological information storage and processing in synthetic cell-free environments. Integration of in vitro transcription/translation modules in synthetic protocells has mainly focused on the synthesis of polynucleotides and proteins within phospholipid vesicles.6–8 This approach has provided important steps towards the construction of protocells with archetypal properties such as RNA replication,9 sequential gene expression,10 membrane-pore (α-hemolysin) assembly,7 gene switching behaviour,11 and extra-vesicular signalling to bacterial cells.12,13 Cell-free gene expression systems have also been encapsulated within water-in-oil emulsion droplets,14 as well as in non-lipid membrane-bound protocells produced by the spontaneous assembly of protein–polymer nanoconjugates (proteinosomes)15 or amphiphilic silica nanoparticles (colloidosomes),16 and in molecularly crowded environments such as aqueous dextran/polyethylene glycol (PEG) two-phase water-in-oil emulsion droplets17 and condensed droplets of a cell lysate.18 The latter systems are of particular interest as crowding agents can influence gene expression by volume exclusion effects that reduce diffusional rates, increase and decrease association and dissociation binding rates respectively, and provide mechanisms for fine tuning gene circuit dynamics and enhancing system robustness.19
We recently showed that coacervate micro-droplets prepared by complexation of cationic polyelectrolytes and anionic mono- or polynucleotides can be used as membrane-free, molecularly crowded protocells with a range of biomimetic characteristics.20–25 As cell-free gene expression has not been demonstrated in coacervate-based protocells, we embarked on developing appropriate methodologies to establish this platform technology in chemically enriched, molecularly crowded micro-droplets. Herein, we report the use of a carboxymethyl-dextran/polylysine (CM-dextran/PLys) coacervate for the sequestration and retention of a plasmid-containing IVGES, and demonstrate cell-free gene expression and folding of the red fluorescent protein mCherry at pH 8.
In vitro gene expression was undertaken within a polysaccharide/polypeptide coacervate prepared at pH 8 by mixing equimolar aqueous solutions of CM-dextran (Mw = 10–15 kDa) and PLys (Mw = 5–15 kDa) (final [CM-dextran monomer]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [PLys monomer] molar ratio ≈ 1
[PLys monomer] molar ratio ≈ 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) (ESI,† Methods). The coacervate suspension was centrifuged, the polyelectrolyte-depleted supernatant removed, and a small aliquot (typically 7 μL) of an ice-cold IVGES containing the pEXP5-NT/mCherry plasmid (IVGES
0.5) (ESI,† Methods). The coacervate suspension was centrifuged, the polyelectrolyte-depleted supernatant removed, and a small aliquot (typically 7 μL) of an ice-cold IVGES containing the pEXP5-NT/mCherry plasmid (IVGES![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) coacervate volume ratio = 0.75
coacervate volume ratio = 0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
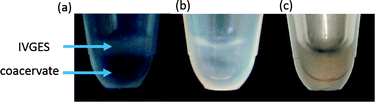
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) added (Fig. 1a). The mixture was kept on ice, and then raised to a temperature of 18 °C to initiate gene expression under continuous shaking. The resulting IVGES/pEXP5-NT/mCherry-containing coacervate was homogenous and transparent, and showed no evidence of phase separation over a period of at least 24 h. Significantly, the reaction mixture showed a progressive increase in red fluorescence with time consistent with mCherry synthesis and folding within the coacervate medium (Fig. 1b and c).
1) added (Fig. 1a). The mixture was kept on ice, and then raised to a temperature of 18 °C to initiate gene expression under continuous shaking. The resulting IVGES/pEXP5-NT/mCherry-containing coacervate was homogenous and transparent, and showed no evidence of phase separation over a period of at least 24 h. Significantly, the reaction mixture showed a progressive increase in red fluorescence with time consistent with mCherry synthesis and folding within the coacervate medium (Fig. 1b and c).
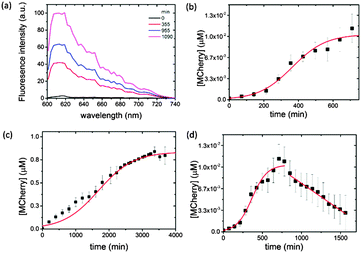
We used confocal microscopy to monitor the time-dependent increase in mCherry fluorescence in the coacervate phase at 18 °C (ESI,† Methods). To alleviate problems associated with background scattering, aliquots of the coacervate reaction mixture were removed at various time intervals and the extracted polysaccharide/polypeptide matrix disassembled by addition of a high ionic strength buffer containing the antibiotic Kanamycin to terminate protein expression. Each aliquot was centrifuged and the fluorescence spectrum of the supernatant recorded using an excitation wavelength of 594 nm. Emission spectra obtained as a function of time showed a peak maximum at 610 nm that was characteristic of folded mCherry, and which increased progressively in intensity over a period of approximately 12–18 h (Fig. 2a), indicating that transcription, translation and protein folding occurs within the coacervate matrix. The fluorescence intensities were converted to approximate protein concentrations using an appropriate calibration curve, and plots of mCherry concentration against time showed sigmoidal behaviour (Fig. 2b). The data were fitted to a logistic function given by [mCherry] = K/(1 + exp(−B(t − t1/2))) where K is the maximum protein concentration, B the maximum rate in μM min−1, and t1/2 the time in minutes at the maximum growth rate; i.e. the time where half the maximum protein concentration is reached.
Typically, a maximum protein concentration of 0.0103 ± 6.03 × 10−4 μM was observed at around t = 720 min (12.5 h), with a maximum rate of 0.0103 ± 0.0007 μM min−1 and t1/2 = 366 min. A sigmoidal-shaped profile was also observed for the in vitro gene expression of mCherry at 18 °C in buffered aqueous solution (Fig. 2c). Interestingly, expression in the control solution was accompanied by an approximately five-fold reduction in the maximum rate (B = 0.00191 ± 0.0001 μM min−1, t1/2 = 1687 ± 48 min), which is attributed to an increase in the local concentration of the biological components within the coacervate phase, combined with volume exclusion effects known to be associated with molecularly crowded environments.19
Although the pEXP5-NT/mCherry plasmid was expressed at a faster maximum rate in the coacervate, this behaviour was not sustained beyond a time frame of approximately 720 min (12.5 h). Fluorescence spectra recorded at later stages (up to 1500 min (25 h)) showed a progressive linear reduction in the intensity at 610 nm due to protein aggregation in the CM-dextran/PLys coacervate (Fig. 2d). As a consequence, the yield was much lower (K = 0.0103 × 10−4 μM) compared with the maximum protein concentration obtained in free solution (K = 0.9 μM after 4000 min (ca. 66 h)) (Fig. 2c). Indeed, the low yield of expressed mCherry required the use of a combination of antibody trap beads and Western blot analysis to extract, isolate and purify the expressed protein (ESI,† Methods and Fig. S1). Increasing the temperature of the reaction mixture from 18 to 37 °C increased the amount of protein aggregation, although sigmoidal behaviour was still observed over an initial period of 360 min (6 h) (data not shown). As we observed no apparent aggregation of mCherry in the presence of comparable concentrations of aqueous CM-dextran or PLys, we attribute the onset of protein aggregation in the molecularly crowded coacervate phase to a reduction in free volume as the polypeptide chain is continually translated. Occupation of the free space by the folded or unfolded mCherry increases the intermolecular interactions to produce aggregates of the expressed protein. Similar observations have been reported for the in vitro and in vivo over-expression of proteins within E. coli cells26 or artificial crowded environments,27 which lead to the formation of inclusion bodies.
We also undertook control experiments involving the cell-free gene expression of mCherry in aqueous solutions of 100 mM PLys or CM-dextran to assess the influence of the individual polyelectrolytes on protein expression and folding (ESI,† Methods). Fluorescence emission spectra indicated that there was a significant reduction in the yield of mCherry when compared with analogous reactions undertaken in buffered solution in the absence of the cationic polyelectrolyte (Fig. S2a and b, ESI†), suggesting that adverse interactions specifically between components of the IVGES and PLys were also responsible for the low yields, possibly by derailing the transcription/translation apparatus. In contrast, mCherry was successfully expressed and folded in a 100 mM aqueous solution of CM-dextran (Fig. S2b, ESI†).
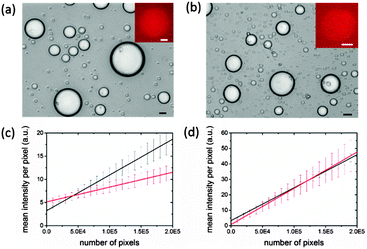
To study gene expression in individual membrane-free coacervate-based protocells, we used a PDMS microfluidic flow-focusing channel structure to disperse a continuous stream of the pEXP5-NT/mCherry plasmid-containing IVGES/CM-dextran/PLys coacervate into a laminar flow of aqueous buffer to produce a population of discrete membrane-free polysaccharide/polypeptide micro-droplets (ESI,† Methods).28 This procedure produced well-defined droplets that were between 5 and 60 μm in diameter and capable of incarcerating the gene expression system for up to 40 h (Fig. 3a and b). Significantly, fluorescence microscopy of individual droplets showed red fluorescence above the background level after 1 h (Fig. 3a and b). The fluorescence intensity was increased in the droplets compared to the surrounding solution, indicating that gene expression of mCherry and folding of the functional protein occurred within the molecularly crowded environment of the CM-dextran/PLys matrix. Fluorescence images were analysed to determine the mean fluorescence intensity of single droplets as a function of their projected surface area. We determined the mean fluorescence intensity per pixel (I) associated with the number of pixels per droplet (P), and plotted these values for droplets of different sizes imaged 1 or 16 h after collection in the microfluidic chamber. Linear fits to the data showed an increase in gradient (dI/dP) from values of 3.20 × 10−5 ± 0.6 × 10−5 to 7.71 × 10−5 ± 0.12 × 10−5 for mCherry-containing droplets imaged at t = 1 or 16 h, respectively (Fig. 3c), confirming gene expression and folding of the fluorescent protein within the protocells. In contrast, a similar analysis of IVGES/CM-dextran/PLys micro-droplets produced under identical conditions but containing a plasmid for expression of the non-fluorescent protein calmodulin (pEXP5-NT/CALML3) showed no change in the values for dI/dP after 1 and 16 h (2.13 × 10−5 ± 1.22 × 10−5 and 2.34 × 10−5 ± 5.0 × 10−5, respectively) (Fig. 3d), indicating a constant value for the background fluorescence in the droplets during the extended period of gene expression.
In conclusion, our results show that the myriad components of an in vitro gene expression ensemble can be isolated and contained within the crowded interior of a CM-dextran/PLys coacervate phase or membrane-free micro-droplets produced by microfluidic processing. The coacervate matrix facilitates sequestration and retention of the complex biological machinery and molecular pathways required for successful gene transcription and translation, and provides a supporting medium for polypeptide chain folding and post-translation construction of the chromophoric site in the functionally active protein. The maximum rate of expression of the pEXP5-NT/mCherry plasmid is enhanced compared to analogous conditions in free buffer solution, and we attribute this to increases in the local concentration of the biomolecular components, in combination with possible changes in RNA polymerase binding rates due to volume exclusion effects. Whilst our work represents a first step towards cell-free synthetic biology based on membrane-free genetically activated protocells, there are several immediate challenges that need to be addressed; notably, the relatively low yields due to the formation of protein aggregates after 12 h, coalescence of the micro-droplets over long time periods, sensitivity of the IVGES to polycationic components of the coacervate phase, and instability of the compartmentalized IVGES at relatively high ionic strength. Using coacervate-based hybrid protocells that are stabilized by organic24 or inorganic25 membranes could circumvent some of these problems, and provide a way forward towards the development of chemically enriched synthetic micro-systems capable of gene-mediated regulation, signalling and self-processing.
We thank the Engineering and Physical Sciences Research Council (EPSRC, UK), European Research Council (Advanced Grant) and the Royal Society (University Research Fellowship to JLRA) for financial support. We acknowledge the Wolfson Bioimaging Facility (Mr A. Leard) for assistance with confocal microscopy and thank Dr D. Jones (University of Cardiff) for the gift of pBAD/MCherry plasmid, Mr D. Watkins for cloning the mCherry gene into pEXT5/NT plasmid and Dr K. Charalambous (Kings College London) for discussions and assistance with Western Blot protocols.
Notes and references
- D. J. Mandell, M. J. Lajoie, M. T. Mee, R. Takeuchi, G. Kuznetsov, J. E. Norville, C. J. Gregg, B. L. Stoddard and G. M. Church, Nature, 2015, 518, 55–60 CrossRef CAS PubMed.
- J. C. Blain and J. W. Szostak, Annu. Rev. Biochem., 2014, 83, 615–640 CrossRef CAS PubMed.
- M. Li, X. Huang, T. D. Tang and S. Mann, Curr. Opin. Chem. Biol., 2014, 22, 1–11 CrossRef CAS PubMed.
- V. Noireaux, R. Bar-Ziv and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 12672–12677 CrossRef CAS PubMed.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS PubMed.
- T. P. de Souza, A. Fahr, P. L. Luisi and P. Stano, J. Mol. Evol., 2014, 79, 179–192 CrossRef PubMed.
- V. Noireaux and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17669–17674 CrossRef CAS PubMed.
- S. Matosevic and B. M. Paegel, J. Am. Chem. Soc., 2011, 133, 2798–2800 CrossRef CAS PubMed.
- H. Kita, T. Matsuura, T. Sunami, K. Hosoda, N. Ichihashi, K. Tsukada, I. Urabe and T. Yomo, ChemBioChem, 2008, 9, 2403–2410 CrossRef CAS PubMed.
- T. Sawasaki, T. Ogasawara, R. Morishita and Y. Endo, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14652–14657 CrossRef CAS PubMed.
- R. Lentini, M. Forlin, L. Martini, C. Del Bianco, A. C. Spencer, D. Torino and S. S. Mansy, ACS Synth. Biol., 2013, 2, 482–489 CrossRef CAS PubMed.
- R. Lentini, et al. , Nat. Commun., 2014, 5, 4012, DOI:10.1038/ncomms5012.
- P. M. Gardner, K. Winzer and B. G. Davis, Nat. Chem., 2009, 1, 377–383 CrossRef CAS PubMed.
- D. S. Tawfik and A. D. Griffiths, Nat. Biotechnol., 1998, 16, 652–656 CrossRef CAS PubMed.
- X. Huang, M. Li, D. C. Green, D. S. Williams, A. J. Patil and S. Mann, Nat. Commun., 2013, 4, 2239, DOI:10.1038/ncomms3239.
- M. Li, D. C. Green, J. L. R. Anderson, B. P. Binks and S. Mann, Chem. Sci., 2011, 2, 1739–1745 RSC.
- P. Torre, C. D. Keating and S. S. Mansy, Langmuir, 2014, 30, 5695–5699 CrossRef CAS PubMed.
- E. Sokolova, E. Spruijt, M. M. K. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. S. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- C. Tan, S. Saurabh, M. P. Bruchez, R. Schwartz and P. LeDuc, Nat. Nanotechnol., 2013, 8, 602–608 CrossRef CAS PubMed.
- S. Koga, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2011, 3, 720–724 CrossRef CAS PubMed.
- D. S. Williams, S. Koga, C. R. C. Hak, A. Majrekar, A. J. Patil, A. W. Perriman and S. Mann, Soft Matter, 2012, 8, 6004–6014 RSC.
- T. Y. D. Tang, M. Antognozzi, J. A. Vicary, A. W. Perriman and S. Mann, Soft Matter, 2013, 9, 7647–7656 RSC.
- J. Crosby, T. Treadwell, M. Hammerton, K. Vasilakis, M. P. Crump, D. S. Williams and S. Mann, Chem. Commun., 2012, 48, 11832–11834 RSC.
- T. D. Tang, C. R. C. Hak, A. J. Thompson, M. K. Kuimova, D. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2014, 6, 527–533 CrossRef PubMed.
- D. S. Williams, A. J. Patil and S. Mann, Small, 2014, 10, 1830–1840 CrossRef CAS PubMed.
- D. Hall and A. P. Minton, Biochim. Biophys. Acta, Proteins Proteomics, 2003, 1649, 127–139 CrossRef CAS.
- B. van den Berg, R. J. Ellis and C. M. Dobson, EMBO J., 1999, 18, 6927–6933 CrossRef CAS PubMed.
- D. Van Swaay, T.-Y. D. Tang, S. Mann and A. De Mello, Angew. Chem. DOI:10.1002/anie.201502886.
Footnote |
| † Electronic supplementary information (ESI): Experimental methods, protein extraction data, and fluorescence emission spectra. See DOI: 10.1039/c5cc04220h |
| This journal is © The Royal Society of Chemistry 2015 |